Abstract
Hedgehog (Hh) signaling is thought to occur in primary cilia, but the molecular basis of Gli2 and Gli3 activation by Hh signaling in cilia is unknown. Similarly, how ciliary gene mutations result in reduced Gli3 processing that generates a repressor is also not clear. Here we show that Hh signaling inhibits Gli2 and Gli3 phosphorylation by protein kinase A (PKA) in cilia. The cilia related gene Talpid3 (Ta3) mutation results in the reduced processing and phosphorylation of Gli2 and Gli3. Interestingly, Ta3 interacts and colocalizes with PKA regulatory subunit PKARIIβ at centrioles in the cell. The centriolar localization and PKA binding regions are located in the N- and C-terminal regions of Ta3, respectively. PKARIIβ fails to localize at centrioles in some Ta3 mutant cells. Therefore, our study provides the direct evidence that Gli2 and Gli3 are dephosphorylated and activated in cilia and that impaired Gli2 and Gli3 processing in Ta3 mutant is at least in part due to a decrease in Gli2 and Gli3 phosphorylation.
Keywords: Hedgehog, Gli2, Gli3, Talpid3, PKA, Cilia
1. Introduction
The Hedgehog (Hh) family of secreted signaling proteins plays fundamental roles in cell fate specification, proliferation, and differentiation. Loss of Hh signaling results in a wide range of birth defects, whereas aberrant activation of the Hh pathway causes several types of human cancer (Jiang and Hui, 2008). Hh signaling is initiated by the binding of Hh to its receptor Patched (Ptch) (Fuse et al., 1999; Stone et al., 1996), a twelve-pass membrane protein, followed by releasing Ptch inhibition to Smoothened (Smo), a G-protein coupled receptor (GPCR) (Riobo et al., 2006). In vertebrates, Smo then transduces signals downstream and ultimately actives the Gli2 and Gli3 zinc-finger-containing transcription factors, which in turn upregulate the expression of Hh targets, including Ptch1 and Gli1, another member of the Gli family (Goodrich et al., 1996; Marigo et al., 1996). Gli2 acts primarily as a transcriptional activator, whereas Gli3 largely serves as a repressor, though it also functions as a weak activator (Bai et al., 2004). Consistent with their functions, our previous studies showed that the majority of full-length Gli3 (Gli3FL) undergoes C-terminal processing to generate a repressor form (Gli3Rep), whereas only a small fraction of full-length Gli2 (Gli2FL) is processed (Gli2Rep) (Pan et al., 2006; Wang et al., 2000). Gli2/Gli3 processing is induced by multi-site phosphorylation at the C-termini, first by protein kinase A (PKA) and subsequently by glycogen synthase kinase 3 (GSK3) and casein kinase 1 (CK1). Phosphorylated Gli2/Gli3 proteins are recognized and ubiquitinated by the SCFβTrCP (Skp1-Cul1-F-box protein) E3 ubiquitin ligase, and finally processed by the proteasome (Pan and Wang, 2007; Wang and Li, 2006). Gli2FL and Gli3FL shuttle between the cytoplasm and nucleus, whereas Gli2Rep and Gli3Rep reside exclusively in the nucleus. Hh signaling suppresses Gli2 and Gli3 phosphorylation and processing, thus converting Gli2FL and Gli3FL into active forms (Kim et al., 2009; Niewiadomski et al., 2014; Pan et al., 2006; Wang et al., 2000).
The primary cilium is a non-motile microtubule-based protrusion from the cell surface and is found on most vertebrate cells. Primary cilia originate in the basal body, the specialized mother centriole of the centrosome, and are elongated and maintained by intraflagellar transport (IFT), which moves proteins and vesicles into and out of the cilia (Rosenbaum and Witman, 2002). The primary cilium serves not only as a key mechanosensory organelle but also as a site for signal transduction for several signaling pathways, including the Hh pathway (Goetz and Anderson, 2010). Thus, it is not surprising that defects in cilia structure and function result in a wide spectrum of structural birth abnormalities that include eye defects, kidney cysts, craniofacial and brain malformations, heart defects, and abnormal left-right patterning diseases, which are collectively known as ciliopathies (Novarino et al., 2011). Interestingly, many of these defects are due to the disruption of Hh signaling.
In support of Hh signaling in primary cilia, Ptch1 is localized to cilia in the absence of Hh. Hh stimulation leads to the exit of Ptch1 from cilia and subsequently the accumulation of Smo, Gli2, and Gli3 in cilia (Chen et al., 2009; Corbit et al., 2005; Haycraft et al., 2005; Rohatgi et al., 2007; Wen et al., 2010). Interestingly, Hh signaling also directs Gpr161, a ciliary localized GPCR, to be internalized from cilia. Gpr161 negatively regulates Hh signaling by increasing cAMP level, which in turn activates PKA. Consistent with this, Gli3 processing is reduced, and the Hh pathway is activated in the Gpr161 mutant embryos (Mukhopadhyay et al., 2013). Therefore, it is generally believed that Hh signaling inhibits Gli2 and Gli3 processing and activates Gli2FL and Gli3FL proteins by suppressing their PKA phosphorylation in cilia. However, to date there is no direct evidence that this is indeed the case.
All known cilia gene mutations impair signal transduction from Smo to Gli2/Gli3. The vast majority of cilia gene mutations results in reduced Gli3 processing and thus alters Gli3FL and Gli3Rep balance, which consequently leads to polydactyly phenotype. One example is Talpid3 (Ta3) mutation, which was initially identified in chicken, because the mutation results in polydactyly and central nervous system defect (Davey et al., 2006). Further studies showed that Ta3 is localized at centrioles and required for ciliogenesis (Yin et al., 2009). However, the molecular mechanisms underlying this reduced Gli3 processing in Ta3 and any other known ciliary gene mutants are unknown. In the present study, we show that Hh signaling inhibits PKA-mediated Gli2 and Gli3 phosphorylation in cilia. The Gli2 and Gli3 phosphorylation is significantly reduced in Ta3 mutant cells. Interestingly, Ta3 colocalizes with PKA in centrioles and interact with each other. PKA is mislocalized in some of the Ta3 mutant cells. Therefore, our study provides evidence that Hh signaling inhibits Gli2 and Gli3 processing and activates Gli2FL and Gli3FL proteins by suppressing Gli2 and Gli3 phosphorylation in cilia. The reduced Gli2 and Gli3 processing in Ta3 mutants is through a decrease in Gli2 and Gli3 phosphorylation, which is probably in part the result of the mislocalization of PKA in Ta3 mutant cells.
2. Results
2.1. Hh signaling inhibits Gli2 and Gli3 phosphorylation by PKA in cilia
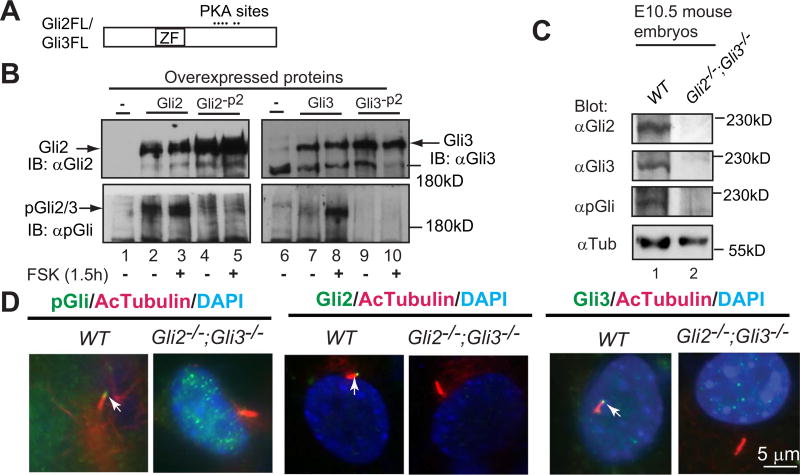
Gli2 and Gli3 each contain six PKA phosphorylation sites in their carboxyl (C) termini (Fig. 1A). The phosphorylation of the first four sites (P1–4) is required for Gli2 and Gli3 processing into the repressors, while phosphorylation of the fifth and sixth sites (P5–6) inhibits Gli2 and Gli3 transcriptional activity. Hh signaling inhibits Gli2 and Gli3 processing and activates Gli2FL and Gli3FL proteins by suppressing their phosphorylation (Niewiadomski et al., 2014; Pan et al., 2006, 2009; Wang et al., 2000). Although these Hh-mediated events are thought to occur in cilia, the direct evidence to support this hypothesis remains missing. To determine whether Hh signaling inhibits Gli2 and Gli3 phosphorylation in cilia, we generated an antibody (pGli) against a Gli2 peptide containing phosphorylation at the second PKA site (P2). Given that the Gli2 peptide is almost identical with Gli3 in the same region (see Materials and Methods for details), it was not surprising that this antibody weakly detected both Gli2 and Gli3 overexpressed in HEK293 cells by immunoblotting. Treatment of the cells with forskolin (Fsk), a chemical that increases cAMP levels to activate PKA, dramatically increased signals detected by the antibody, though it did not affect the levels of Gli2 and Gli3 expression shown by immunoblotting with Gli2 and Gli3 antibodies, respectively (Fig. 1B, compare lanes 2 to 3, 7to 8). In addition, the pGli antibody failed to detect Gli2 or Gli3 mutants (Gli2-P2 and Gli3-P2) with a mutation at the P2 site (Fig. 1B, lanes 4–5, 9–10), indicating that Gli2 and Gli3 detected by pGli antibody in the cells without Fsk treatment are also the phosphorylated form of the proteins. Similarly, this antibody also specifically detected the phosphorylated endogenous Gli2 and Gli3 in the wild type mouse embryos, as no signal was detected in Gli2;Gli3 double mutant embryos (Fig. 1C, compare lane 1 to lane 2). These results indicate that this antibody specifically recognizes the phosphorylated Gli2 and Gli3 at the P2 site.
Fig. 1.
A phosphopeptide antibody (pGli) specifically recognizes the phosphorylated Gli2 and Gli3 proteins at the second PKA site. (A) A diagram showing Gli2 and Gli3 proteins with the zinc-finger domain (ZF) and six PKA sites. (B) Immunoblots of overexpressed Gli2, Gli3, and their mutants at the second PKA site (Gli2-P2, Gli3-P2) with Gli2, Gli3, or pGli antibodies. Forskolin (FSK) induces Gli2/Gli3 phosphorylation. (C) Immunoblots of endogenous Gli2, Gli3, and phosphorylated Gli2/Gli3 using protein lysates prepared from wild type (WT) and Gli2;Gli3 double mutant (control) mouse embryos. (D) Gli2, Gli3, and PKA-phosphorylated Gli2 and Gli3 localize to primary cilia. Immunostaining of WT and Gli2;Gli3 double mutant MEFs for the indicated proteins. Note that pGli, Gli2, and Gli3 antibodies are specific, as no signals were detected in the protein lysates and cilia of the mutant cells. AcTubulin, acetylated tubulin, a cilia marker; DAPI, staining for nuclei. These experiments were performed at least two times.
Immunostaining also showed that the pGli antibody detected specific signals in cilia, as the immunofluorescence was only seen in wild type but not Gli2;Gli3 mutant MEFs. Similarly, both Gli2 and Gli3 antibodies, which specifically recognize their C-termini (Materials and Methods), detected Gli2FL and Gli3FL proteins in cilia, respectively (Fig. 1D). Given that immunoblotting results indicate that the pGli antibody is specific only for the phosphorylated form of Gli2 and Gli3 (Fig. 1B–C), taking together, we conclude that the signals detected by the pGli antibody in cilia are the phosphorylated form of endogenous Gli2 and Gli3 at the P2 site.
Mutagenesis studies showed that all six PKA sites in the Gli2 and Gli3 C-termini are phosphorylated by PKA in the cell without noticeable preference of one to another (Pan et al., 2006; Wang et al., 2000). Hh signaling activates Gli2 and Gli3 by inducing dephosphorylation of Gli2 and Gli3 at all six PKA sites, although the extent of dephosphorylation at P5–6 is slightly more than that of P1–4 (Niewiadomski et al., 2014). Therefore, although pGli antibody recognizes Gli2 and Gli3 phosphorylated only at the P2 site, the signals detected by the antibody serve as an indicator for the extent of Gli2 and Gli3 phosphorylation at P1–6 sites by PKA.
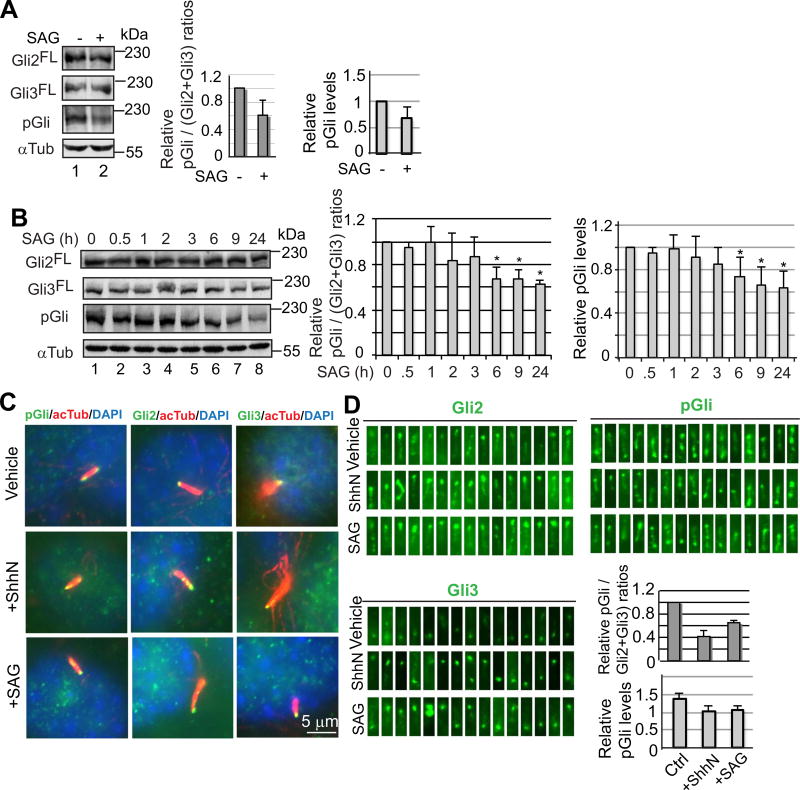
To determine whether Hh signaling inhibits the Gli2 and Gli3 phosphorylation, the Hh-responsive C3H10T1/2 cells were incubated with or without SAG, a synthetic Smo agonist that activates the Hh pathway (Chen et al., 2002), and subjected to immunoblotting with Gli2, Gli3, and pGli antibodies. Quantification of immunoblotting results revealed that after normalized against the levels of Gli2 and Gli3, the levels of the phosphorylated Gli2 and Gli3 after SAG treatment were about 0.6 of those without treatment. Quantification of phosphorylated Gli2/Gli3 levels alone also showed a similar level of reduction in average (Fig. 2A).
Fig. 2.
Hedgehog signaling inhibits PKA-mediated Gli2 and Gli3 phosphorylation in cilia. (A) Immunoblots showing the levels of Gli2FL, Gli3FL, and phosphorylated Gli2 and Gli3 (pGli) in C3H10T1/2 cells. Graphs show the relative pGli levels either with or without being normalized to those of Gli2FL and Gli3FL. Two-tailed Student t-test p-value = 0.034 < 0.5. (B) Immunoblots showing the time course of the levels of Gli2FL, Gli3FL, and pGli in response to SAG. Graphs to the right show the relative pGli/(Gli2+Gli3) or pGli values. P-values ≤ 0.0067 for bar graphs marked with *. (C) Representative images showing Gli2, Gli3, and pGli staining in cilia before and after treatment with ShhN or SAG. (D) Fifteen randomly chosen cilia that show positive staining for Gli2, Gli3, and pGli. The graphs show the arbitrary intensity of pGli staining per cilia by either with (upper graph) or without (lower graph) being normalized against Gli2FL and Gli3FL levels. Two-tailed Student t-test p-value ≤ 0.00057 (upper graph) or 0.036 (lower graph), respectively.
To determine the dynamics of Gli2 and Gli3 phosphorylation in response to SAG, the levels of phosphorylated Gli2 and Gli3 were examined in the cells that were treated with SAG for different periods of time. A significant decrease in the levels of Gli2 and Gli3 phosphorylation, which was measured by either being normalized against Gli2 and Gli3 levels or phosphorylated Gli2/Gli3 levels alone, was seen after treatment for 6 h. Treatment for 9 and 24 h did not significantly reduce the Gli2 and Gli3 phosphorylation levels further (Fig. 2B), suggesting that the Gli2 and Gli3 dephosphorylation levels reached almost the lowest by 6 h.
We next examined the levels of phosphorylated Gli2 and Gli3 proteins in cilia in response to ShhN, an active Sonic hedgehog N-terminal fragment, or SAG stimulation by immunostaining (Fig. 2C and D). Quantification of the immunofluorescent intensity in cilia showed that after normalized against the levels of Gli2 and Gli3, the levels of the phosphorylated Gli2 and Gli3 proteins decreased about a half. When the immunofluorescent intensity of pGli staining alone was measured, the reduction of phosphorylated Gli2/Gli3 levels in cilia upon ShhN or SAG stimulation was also significant, though it was smaller (Fig. 2D, graphs). Given that Gli2 and Gli3 dephosphorylation is correlated with their activation (Niewiadomski et al., 2014), our observations support the currently prevailing hypothesis that Gli2FL and Gli3FL are activated in cilia.
2.2. Ta3 mutation results in decreased processing and phosphorylation of Gli2 and Gli3
The vast majority of ciliary gene mutations affect Gli3 processing (Goetz and Anderson, 2010), but the mechanism is unknown. To address this question, we generated a Ta3 mutant allele in the mouse by targeted gene knockout approach (Fig. S1A–C), given that the Ta3 mutation in chick affects ciliogenesis and Hh signaling (Davey et al., 2006). Most of Ta3 mutant mouse embryos died around gestation day 10.5 (E10.5)(n > 30). The development of the Ta3 mutant embryos was often delayed (Fig. S1D). Hh signaling in Ta3 mutants was severely impaired as demonstrated by reduced Ptch1-lacZ expression (Goodrich et al., 1997) and loss of expression of ventral neural tube markers (Foxa2, Nkx2.2, Hb9, and Isl1) (Briscoe et al., 2000) (Fig. S1E–F). As expected, no cilia formed in Ta3 mutant MEFs (Fig. S1G). These results are similar to the phenotypes of chicken and another mouse Ta3 mutant that was previously reported (Bangs et al., 2011; Davey et al., 2006; Yin et al., 2009).
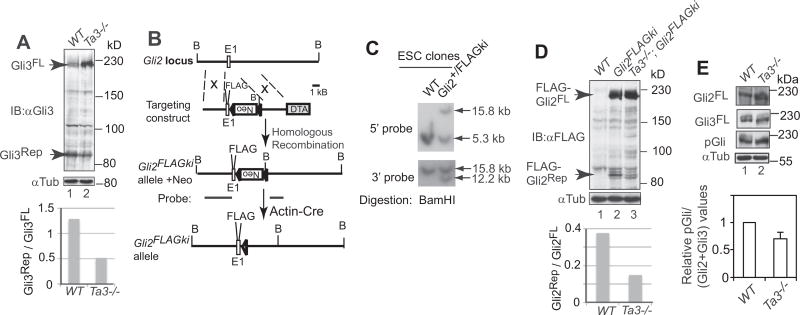
To determine whether Gli3 processing was reduced in the mouse Ta3 mutant, protein lysates of wild type and Ta3 mutant embryos were subjected to immunoblotting with a Gli3 N-terminal antibody, which recognizes both the Gli3FL and Gli3Rep (Wang et al., 2000). The results showed that Gli3 processing was reduced about a half in the mutant as compared to wild type (Fig. 3A).
Fig. 3.
The processing and PKA-mediated phosphorylation of Gli2 and Gli3 were diminished in Ta3 mutant cells. (A, D, E) Immunoblots for Gli3, FLAG-Gli2, Gli2, and pGli in wt and Ta3 mutant embryos. Tubulin immunoblots are loading controls. Graphs show the ratios of Gli3Rep to Gli3FL and FLAG-Gli2Rep to FLAG-Gli2FL and relative pGli to (Gli2+ Gli3) values. Two-tailed Student t-test p-value = 0.0136 for E. from three independent experiments. (B) The gene targeting strategy to create Gli2FLAGki allele. E1, exon 1; FLAG, 3×FLAG tag; Neo, pGkneo cassette in a reverse orientation; triangle, loxP site; DTA, diphtheria toxin A; B, BamHI site. (C) Southern blot showing wt and a representative Gli2FLAGki ES cell clone with probes shown in B.
To date, there is no evidence that Gli2 processing is also affected by ciliary gene mutations, largely because Gli2Rep levels are normally very low and hardly detectable (Pan et al., 2006). To overcome this obstacle, we inserted the FLAG tag in frame right after the initiation codon of Gli2 in the Gli2 locus using targeted homologous recombination approach (Fig. 3B–C). This allele was named Gli2FLAGki. To determine whether Gli2 processing is reduced in Ta3 mutant embryos, the Ta3 mutant allele was crossed into Gli2FLAGki mice. Immunoblotting with a FLAG antibody showed that the levels of the processed Gli2 protein, FLAG-Gli2Rep, in Ta3 mutant were markedly lower than those in wild type Ta3 embryos (Fig. 3D), indicating that Gli2 processing is also impaired in Ta3 mutant. This is the first evidence that Gli2 processing is reduced in a cilia related gene mutant.
Gli2 and Gli3 phosphorylation by PKA is essential for Gli2 and Gli3 processing (Pan et al., 2006, 2009; Wang et al., 2000). To understand the molecular basis underlying the reduced Gli2 and Gli3 processing, the levels of Gli2 and Gli3 phosphorylation were examined in the wild type and Ta3 mutant embryos by immunoblotting. The data showed that Gli2 and Gli3 phosphorylation levels in the mutant embryos were reduced to about 0.7 of those in wild type (Fig. 4D). Thus, the reduced Gli2 and Gli3 processing is at least in part due to a decrease in Gli2 and Gli3 phosphorylation levels.
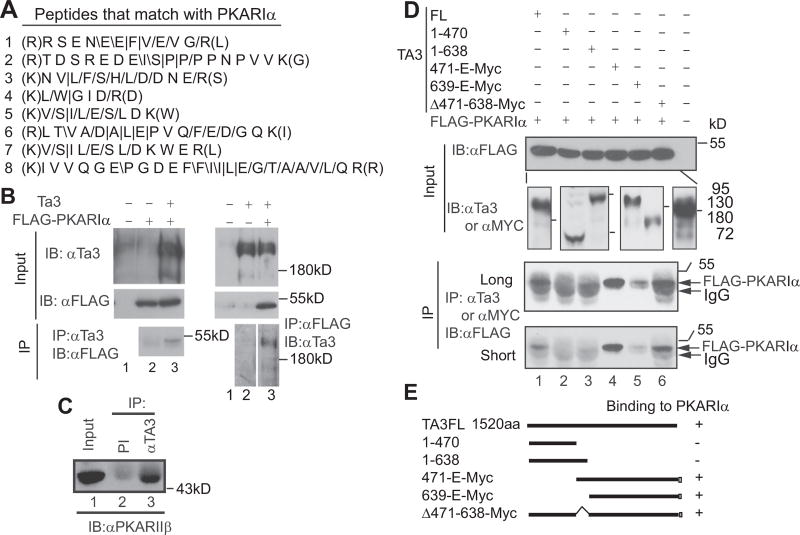
Fig. 4.
The Ta3 C-terminus interacts with PKA regulatory subunits. (A) The peptides obtained from mass spectrometry match with PKARIα. /, /, and | are referred to as y-ions, b-ions, and both, respectively. They were detected in the tandem mass spectrometry analysis of the peptides. (B) Coimmunoprecipitation showing that Ta3 interacts with FLAG-PKARIα in transfected HEK293 cells. (C) Coimmunoprecipitation showing that endogenous Ta3 interacts with PKARIIβ. (D) Coimmunoprecipitation showing that the C-terminal region of Ta3 interacts with PKARIα. Subpanels in the second panel from the top are from different immunoblots. The molecular weight markers correspond to the lines in each subpanel in the order. Two lower panels are the same with different exposure time. (E) Diagrams showing constructs and summary in D. IP, immunoprecipitation; IB, immunoblot; PI, preimmune serum.
2.3. Ta3 interacts and colocalizes with PKA
To understand how Ta3 mutation results in a decrease in Gli2 and Gli3 phosphorylation by PKA, we created NIH3T3 cells that stably expressed Ta3 tagged with FLAG and HA (FH) epitopes. Immunoaffinity purification was performed using these cells. The precipitated proteins were then subjected to mass spectrometric analysis, which showed that several tryptic peptides obtained from the precipitated proteins matched with PKA regulatory subunit type Iα, PKARIα (Fig. 4A). The interaction between Ta3 and PKARIα was confirmed by coimmunoprecipitation using Ta3 and FLAG tagged PKARIα overexpressed in HEK293 cells (Fig. 4B).
There are four PKA regulatory subunits in vertebrates, PKARIα/β and PKARIIα/β, which share a high level of sequence identity (Taylor et al., 2012). To determine whether the endogenous Ta3 interacts with one of the PKAR subunits, we chose PKARIIβ because there is a good commercial antibody available for this subunit. Ta3 antibody, but not pre-immune serum, could readily coimmunoprecipitate PKARIIβ (Fig. 4C), indicating that Ta3 also specifically interacts with PKARIIβ.
We next wanted to map the PKARIα binding region(s) in Ta3. To this end, several Ta3 mutant constructs were generated and coexpressed with FLAG-PKARIα in HEK293 cells. The protein lysates made from the cells were subjected to immunoprecipitation with either Ta3 or Myc (for Myc-tagged Ta3 mutants) antibodies, followed by immunoblotting with FLAG antibody to detect FLAG-PKARIα. The results showed that Ta3-1–470 and Ta3-1–638, the two N-terminal fragments, failed to co-precipitate FLAG-PKARIα, indicating that the N-terminal region is not sufficient to bind PKARIα. In contrast, Ta3FL (the full-length protein), Ta3-471-E-Myc (amino acid position 471 to C-terminal end), Ta3-639-E-Myc, and Ta3Δ471–638-Myc were coimmunoprecipitated with FLAG-PKARIα (Fig. 4D, two lower panels). It should be noted that less amount of FLAG-PKARIα coprecipitated with Ta3-639-E-myc is likely due to slightly lower levels of Ta3-639-E-myc expression (Fig. 4D, compare lanes 4–5 in the upper second panel). Therefore, these results indicate that the C-terminal region (639-E), but not the middle (471−638) or N-terminal regions, is required for PKARIα binding.
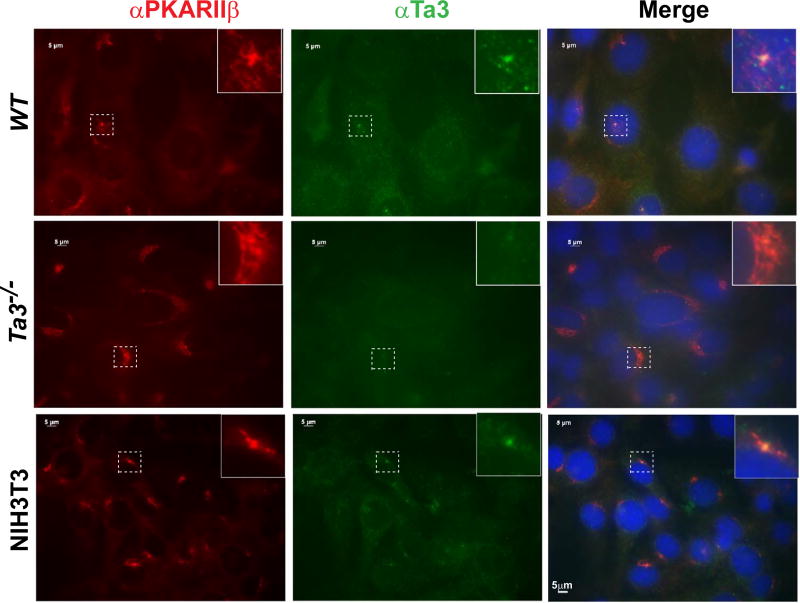
Given that Ta3 interacts with PKARIIβ, we next wanted to determine whether Ta3 is colocalized with PKARIIβ in the cell. Previous studies showed that both Ta3 and PKARIIβ were localized at centrioles (Kobayashi et al., 2014; Lignitto et al., 2011; Wu et al., 2014; Yin et al., 2009), but their colocalization has not been reported. Coimmunostaining wild type MEFs for Ta3 and PKARIIβ demonstrated that the two proteins indeed colocalized at centrioles. The staining for Ta3 was specific, as it was negative in Ta3 mutant cells. The Ta3 centriolar colocalization was also observed in NIH3T3 cells (Fig. 5). Together, the data support the finding that Ta3 interacts with PKARIIβ.
Fig. 5.
Ta3 and PKARIIβ colocalize at centrioles in the cell. Wild type (WT) and Ta3 mutant MEFs and NIH3T3 cells were stained for PKARIIβ, Ta3, and nuclei (DAPI, blue). Ta3 staining is specific, as no signals were detected in Ta3 mutant cells. Insets are the enlargement of the framed areas with dash lines.
2.4. Ta3 full-length protein, but not its mutants, rescues ciliogenesis in Ta3 mutant cells
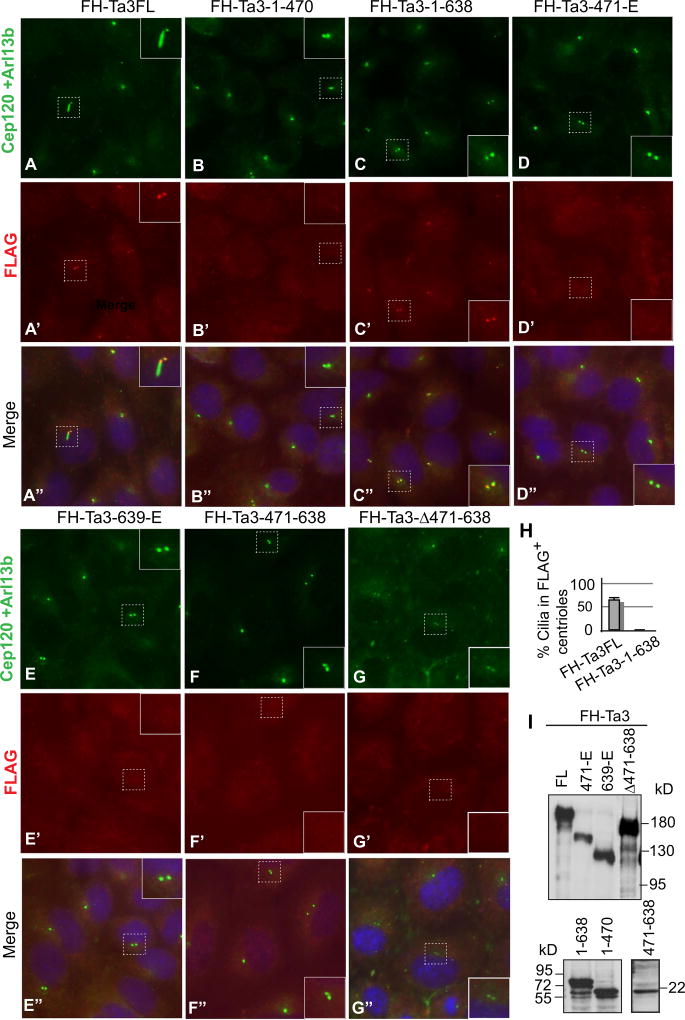
To better understand the relationship between Ta3 centriolar localization and Ta3 binding to PKARIIβ, the subcellular localization of exogenous Ta3 mutant proteins and their ability to rescue ciliogenesis were examined. For this purpose, we created the same Ta3 constructs in Fig. 4E in a murine retroviral vector that contains FH epitope tags. The retroviral vector was chosen because it exhibits higher transduction efficiency and lower level of expression as compared to a CMV-promoter based plasmid. After Ta3 mutant MEFs were separately infected with the virus carrying these constructs, the cells were co-immunostained for FH-Ta3 and its mutant proteins, Cep120 (labeling two centrioles) (Mahjoub et al., 2010), and Arl13b (a cilia marker) (Caspary et al., 2007). The results showed that only Ta3FL and Ta3-1–638, but not any other mutants, were localized at centrioles (Fig. 6A–G). The lack of centriolar localization of these mutants was not the results of the failed protein expression, since immunoblotting analysis showed that they were expressed (Fig. 6I). Interestingly, 60% of the cells in which FH-Ta3 localized at centrioles formed cilia, whereas none of the cells in which FH-Ta3-1–638 localized to centrioles generated any cilia (Fig. 6H). Therefore, the Ta3 full-length protein is required for the rescue of ciliogenesis, and Ta3-1–638 is responsible for Ta3 centriolar localization.
Fig. 6.
Expression of Ta3FL, but not Ta3 mutants, rescues ciliogenesis in Ta3 mutant cells. (A–G”) Ta3 mutant MEFs were transduced with the retrovirus carrying the constructs indicated above the panels. Following serum starvation, the cells were stained with FLAG, Cep120, and Arl13b antibodies and counterstained with DAPI for nuclei. FLAG labels Ta3 and mutants. Cep120 marks centrioles, and Arl13b is a cilia marker. Insets are enlargement of the framed areas with dash lines. Note that although both FH-Ta3FL and FH-Ta3-1–638 localize at centrioles, only FH-Ta3FL expression rescues cilia formation. (H) A graph showing that the percent cells that are FLAG-staining positive at centrioles form cilia. Two-tailed Student t-test p-value = 0.0001 (n=72). (I) Immunoblots showing the expression of Ta3 and mutant proteins.
2.5. PKARIIβ is not localized at centrosome in some of Ta3 mutant cells
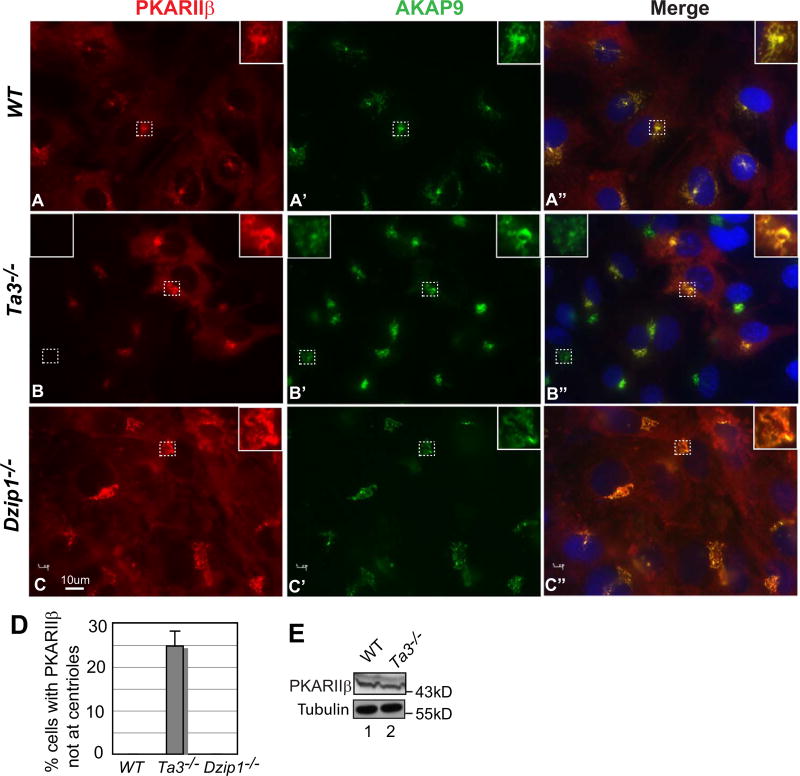
PKARIIβ is normally concentrated at Golgi-centrosome and perinuclear region (Lignitto et al., 2011). So is the A-kinase anchoring protein AKAP9 (Larocca et al., 2004; Schmidt et al., 1999). To understand the molecular and cellular mechanisms by which Gli2 and Gli3 phosphorylation is reduced in Ta3 mutant cells, PKARIIβ subcellular localization in both wild type and Ta3 mutant cells were investigated. As expected, PKARIIβ was colocalized with AKAP9 to centrosome-Golgi in all the wild type cells examined (Fig. 7As). However, PKARIIβ was not colocalized with AKAP9 in about a quarter of Ta3 mutant cells (Fig. 7Bs and D, no colocalization, left inset; colocalization, right inset). The absence of PKARIIβ at centrosome was not because of the difference in the protein levels between wild type and Ta3 mutant cells, as shown by immunoblot (Fig. 7E). This was also specific for Ta3 mutant cells, as PKARIIβ was normally localized at centrosome and Golgi in Dzip1 (a ciliary gene encoding a centrosomal protein) mutant cells that also lack cilia (Wang et al., 2013). Thus, Ta3 mutation disrupts PKARIIβ localization to centrosome-Golgi at least in some cells.
Fig. 7.
PKARIIβ is not colocalized with AKAP9 at centrosome in some Ta3 mutant cells. (A–C) WT, Ta3, and Dzip1 mutant MEFs were stained for PKARIIβ, AKAP9, and nuclei (DAPI, blue). Insets are the enlargement of the framed areas with dash lines. Insets in the right show colocalization, whereas an inset in the left shows a lack of colocalization. (D) The percentage of Ta3 mutant cells that lack PKARIIβ staining at centrioles. The graph was derived from three independent experiments. Note that PKARIIβ mislocalization is only detected in Ta3 but not Dzip1 mutant cells. P-value = 0.0002 (n ≥ 60). (E) Immunoblot showing the similar PKARIIβ levels in wt and Ta3 mutant cells.
3. Discussion
In the present study, we show that Hh signaling inhibits Gli2 and Gli3 phosphorylation by PKA in cilia. Ta3 mutation also results in decrease in the processing and phosphorylation of both Gli2 and Gli3. It should be noted that this is the first time to show that Gli2 processing is impaired in a ciliary gene mutant. Ta3 interacts and colocalizes with PKA regulatory subunits at the centrosome. Interestingly, PKARIIβ is not localized at the centrosome in some of the Ta3 mutant MEFs. Our findings provide the direct evidence that Hh-dependent Gli2 and Gli3 dephosphorylation occurs in cilia and support the hypothesis that the reduced Gli2 and Gli3 processing in Ta3 mutant is due to a decrease in Gli2 and Gli3 phosphorylation probably partially resulted from PKARIIβ mislocalization.
It is generally believed that Hh signaling occurs in cilia. In the absence of Hh signaling, both Ptch1, the Hh receptor that inhibits Smo activity, and Gpr161, a GPCR that antagonizes Hh signaling by activating PKA, are localized to cilia. Hh signaling results in the removal of Ptch1 and Gpr161 from cilia (Mukhopadhyay et al., 2013; Rohatgi et al., 2007) and concomitantly the accumulation of Smo, Gli2, and Gli3 in cilia (Chen et al., 2009; Corbit et al., 2005; Haycraft et al., 2005; Wen et al., 2010). Hh signaling ultimately inhibits Gli2 and Gli3 processing that generates repressors and converts Gli2FL and Gli3FL into activators. Given that Gli2 and Gli3 processing is dependent on the phosphorylation by PKA at P1–4 sites (Pan et al., 2006; Wang et al., 2000), whereas their activation is associated with dephosphorylation of all six PKA sites P1–6 or PKA gene mutations (Niewiadomski et al., 2014; Tuson et al., 2011), understanding where and how Gli2 and Gli3 are phosphorylated and dephosphorylated is greatly important. Here we show that Hh signaling inhibits Gli2 and Gli3 phosphorylation in cilia (Fig. 2C–D) and provide the first direct evidence that Gli2 and Gli3 are activated in cilia.
Most of the Ta3 mutant phenotypes, including polydactyly and the lack of the specification of ventral neural cell types in the neural tube, are the result of the failed activation and reduced processing of Gli2FL and Gli3FL proteins (Fig. S1E and F, 3A and D)(Bangs et al., 2011; Davey et al., 2006). It is not known how Gli2 and Gli3 processing is impaired and why Gli2FL and Gli3FL are inactive in Ta3 and, in fact, any other known ciliary gene mutants. We demonstrate here that Gli2 and Gli3 phosphorylation is reduced in Ta3 mutant (Fig. 3E). This explains why Gli2 and Gli3 processing is impaired in the mutant. However, this does not explain why Gli2FL and Gli3FL are inactive, since the decrease in phosphorylation has been shown to be associated with Gli2FL and Gli3FL activation and thus is expected to increase Gli2FL and Gli3FL activity (Niewiadomski et al., 2014). Therefore, Gli2FL and Gli3FL activation needs more than just dephosphorylation. For example, desumoylation and acetylation of Gli2 and Gli3 are likely required, as they have been shown to increase Gli2 and Gli3 activity (Han et al., 2012). In addition, the translocation of Gli2FL and Gli3FL into the nucleus is another mechanism essential for Gli2 and Gli3 activation by Hh signaling (Kim et al., 2009; Niewiadomski et al., 2014), which could be altered in Ta3 mutant cells.
The C-terminally truncated Ta3-1-638 is localized to centrioles, but fails to restore ciliogenesis. Ta3-471-E neither localizes at centrioles nor rescues ciliogenesis. Not only can Ta3FL rescue ciliogenesis but also localizes at centrioles (Fig. 6As, Cs, and Ds). These results indicate that the N-terminal region determines Ta3 centriolar localization, while the C-terminal region is required for ciliogenesis. The Ta3 centriolar localization is dependent on the C-terminal region (471-638aa) of the N-terminal fragment, as both Ta3Δ471–636 and Ta3-1-470 fail to localize at centrioles. However, Ta3-471-638 is not sufficient to direct Ta3 to the centrioles in MEFs (Fig. 6Ds, Fs, and Gs). The observation that Ta3-471-E fails to rescue ciliogenesis is inconsistent with a previous study showing that a similar Ta3 mutant was able to rescue ciliogenesis in chick embryos (Yin et al., 2009). The discrepancy between the two studies may be due to the experimental systems used—MEFs versus chick embryos, or the sequence difference in the mouse and chick Ta3 proteins. Additional studies are needed to distinguish between the two possibilities.
Ta3 is the first known ciliary protein that is shown to bind PKA. The PKA binding region is mapped to the Ta3 C-terminal region (Fig. 4D and E). In support of the interaction between Ta3 and PKA, the Ta3 and PKARIIβ colocalize at centrioles (Fig. 5). Thus, Ta3 serves as an AKAP to coordinate PKA-dependent Gli2 and Gli3 phosphorylation. Loss of Ta3 results in decrease in the Gli2 and Gli3 phosphorylation (Fig. 3E). The observation that PKARIIβ is absent at centrosome and Golgi in some of Ta3 mutant MEFs suggests that PKARIIβ mislocalization at least partially accounts for the reduced Gli2 and Gli3 phosphorylation in Ta3 mutant cells. This is specific for Ta3 mutant, since PKARIIβ centriolar localization is normal in Dzip1 and other ciliary gene mutant cells examined (Fig. 7, data not shown). We currently do not know why PKARIIβ mislocalization is found only in some of the Ta3 mutant cells. One possibility is that given that Ta3 mutant MEFs are heterogeneous, Ta3 mutation may affect PKARIIβ localization to different extent depending on cell types. It is possible that PKARIIβ localization has also been altered in the Ta3 mutant cells that seem to have normal PKARIIβ localization. Such alteration is just too subtle to be clearly detected by immunostaining. Further studies are necessary to determine why PKARIIβ fails to localize at centrioles in only some of Ta3 mutant cells.
4. Materials and methods
4.1. Mouse strains and the generation of a Ta3 mutant and Gli2-FLAGki alleles
BAC clones containing mouse Ta3 or Gli2 genomic DNA sequences were purchased from the BACPAC Resources Center (Oakland, CA, USA) and used to create Ta3 or Gli2FLAGki targeting constructs. The Ta3 construct was engineered by replacing the first three exons of the Ta3 gene with the pGKneo cassette flanked by loxP sites (Soriano, 1997) (Fig. S1A). The Gli2FLAGki construct was generated by inserting 3×FLAG and pGKneo cassette into the first ATG and first AvrII restriction site right after the first exon of the Gli2 locus, respectively (Fig. 3B). The linearized constructs were electroporated into W4 ES cells (Taconic Biosciences). The targeted Ta3 mutant ES cell clones were identified by digestion of ES cell genomic DNA with EcoRV (5′ homologous arm) or BglII (3′ homologous arm), followed by a Southern blot analysis using two probes as indicated (Fig. S1B). The targeted Gli2FLAGki mutant ES cell clones were determined by digestion of genomic DNA with BamHI, followed by a Southern blot analysis with two different probes, one for 5′- and the other for 3′-homologous recombination (Fig. 3B and C). Two for each of Ta3 and Gli2FLAGki targeted ES cell clones were injected into C57BL/6 blastocysts to generate chimeric founders, which were then bred with C57BL/6 to establish F1 heterozygotes. The Ta3 mutant heterozygotes and Gli2FLAGki homozygotes were maintained in 129/SVE, C57BL/6, and SW mix background. PCR (polymerization chain reaction) analysis was used for routine genotyping Ta3 with the following primers: forward primer BW725F, 5′-GTGATTATCTGTTCGTCAGTGC-3′ and reverse prime BW725R, 5′-CGCCTTTATGTTTGGACACAGT −3′ for the wild type (wt) Ta3 allele, which produced a 220 bp fragment; and forward primer BW725F and reverse primer BW128, 5′-TGCTAAAGCGCATGCTCCAG-3′ on pGKneo for the targeted Ta3 allele, which produced a 260 bp fragment. The primers used to genotyping Gli2FLAGki are BW573F, 5′-TGTCTGTGTCCTTTCCTCCAG-3′, and BW709R, 5′-GCAGAGGCACTGCCCTCCATA-3′, to produce a 300 bp fragment for wt and a 370 bp fragment for Gli2FLAGki. The animal work was approved by Institutional Animal Care and Use Committee at Weill Medical College.
4.2. Cell lines and cell culture
Wild type (wt), Ta3, and Gli2;Gli3 mutant primary mouse embryonic fibroblasts (pMEFs) were prepared from E11.5 or E12.5 mouse embryos. The pMEFs were cultured in DMEM supplemented with 10% FBS (fetal bovine serum)(Atlanta Biologicals), penicillin, and streptomycin (Corning). The immortalized wt and Ta3 mutant MEFs were generated by incubating the pMEFs in the same growth medium at the higher density for many passages until the cells passed crises and gained normal growth rate. HEK293 and C3H10T1/2 cells were originally obtained from ATCC and incubated in the same growth medium. NIH3T3 obtained from Xin-Yun Huang lab at Weill Medical College and its related cells were grown in DMEM supplemented with 10% calf serum (CS), penicillin, and streptomycin. Mycoplasma test was completed when the cells lines were received and was negative. To make ShhN conditioned medium, a pRK-ShhN expression construct was transfected into HEK293cells by calcium phosphate precipitation method (Wang et al., 2000). Two days post-transfection, the medium was collected. ShhN conditioned medium (1:10 dilution) and SAG (300 nM) (Cayman Chemical) were used to stimulate C3H10T1/2 for overnight or times as indicated. To induce Gli2 and Gli3 phosphorylation by PKA, the transfected HEK293cells were incubated with either vehicle (DMSO control) or Forskolin (Fsk, 40 µM)(Calbiochem) for 1.5 h before the cells were lysed.
4.3. cDNA constructs, cloning, and transfection
The mouse Ta3 full-length cDNA was created by combining three cDNA fragments that were amplified from a cDNA library by PCR. Murine retroviral pLNCX-FH-Ta3 or its mutant cDNA constructs (numbers are amino acid positions) were created by inserting the cDNA fragments into pLNCX-FH retroviral vector by PCR and general cloning techniques. pLNCX-FH was engineered by inserting FH, FLAG and HA double tags, into pLNCX (Clontech). pRK-Ta3 and its mutant cDNA constructs were generated by inserting the cDNA fragments into a CMV based pRK or pRK-myc vector using PCR and general molecular cloning techniques. PKARIα cDNA was amplified from a mouse cDNA library by PCR and inserted into the pCMV-FLAG (Sigma) vector to create pCMV-FLAG-PKARIα. The constructs were verified by DNA sequencing. Virus carrying FH-Ta3 or its mutant constructs was generated by cotransfecting Phoenix-Eco cells (ATCC) with each of the viral constructs and pEco packaging construct using the calcium phosphate precipitation method (Wang et al., 2013). pRK-Gli2, pRK-Gli3, pRK-Gli2-P2, pRK-Gli3-P2 were described previously (Pan et al., 2006; Wang et al., 2000).
4.4. Embryo section immunofluorescence and whole mount lacZ staining
For immunofluorescence of neural tube sections, mouse embryos at 10.5 days post coitus (E10.5) were dissected, fixed in 4% paraformaldehyde (PFA)/PBS for 1 h at 4 °C, equilibrated in 30% sucrose/PBS overnight at 4 °C, and embedded in OCT. The frozen embryos were transversely cryosectioned at the forelimb areas (10 µm/section). Tissue sections were immunostained with antibodies against Foxa2 (concentrated), Nks2.2, Hb9, Isl1, Pax6 (Developmental Study Hybridoma Bank (DSHB), Iowa) as described (Pan et al., 2009). Whole mount lacZ staining of mouse embryos was performed as described (Hogan et al., 1994).
4.5. Immunofluorescence and Microscopy
For cell ciliation studies, cells were plated on coverslips coated with 0.1% gelatin for at least overnight and serum starved with 0.1% FBS for 24 h to arrest the cells. For centrosome staining, cells were fixed in −20 °C cold methanol for 5 mins. For the cytoplasmic and cilia staining, cells were fixed in 4% PFA/PBS for 15 mins. After washed with PBS, the cells were incubated with blocking solution (PBS/0.2% Triton X-100/4% heat inactivated calf serum) for 20 mins. The cells were then incubated with primary antibodies in blocking solution for 1 h at room temperature. The cells were washed with PBS, incubated with secondary antibodies in blocking solution for 1 h at room temperature. After wash three times with PBS, the coverslips were mounted to glass slides with Vectashield mounting fluid with DAPI (Vector Labs). The staining was visualized using a Zeiss Axiovert fluorescent microscope. The fluorescence intensity in cilia was quantified using NIH imageJ. Two-tailed Student t-test was used to calculate p-values.
4.6. Antibodies
Arl13b, AKAP9, Gli2, Gli3, and pGli antibodies were generated by Covance or Biosynthesis. Rabbits were immunized with purified bacterially expressed His-tagged mouse Arl13b, His-tagged mouse AKAP9 fragment (2768–3116 aa), His-tagged mouse Gli2C-terminal fragment (605–1460 aa), GST-tagged Gli3 C-terminal fragment (1051–1179 aa), or the synthetic Gli2P2 phospho-peptide (NH2-CAYTVSRRS-(pS)-GISP-OH), in which residue C is not from Gli2 and was used for conjugation of the peptide to Keyhole Limpet Hemocyanin (KLH). It is worth noting that the sequence for Gli3 in the same region is AYLSSRRSSGISP with only the 3rd and 4th residues different from the Gli2. The phosphorylation site is conserved between Gli2 and Gli3. The antibodies were used in a 1:1000 dilution. Other antibodies include: Gli2 N-terminal, Gli3 N-terminal, Ta3, Cep120 (all 1:1000) (Pan et al., 2006; Wang et al., 2000; Wu et al., 2014), acetylated tubulin (1:4000), FLAG (T6793, F1804, Sigma), PKARIIβ (610625, BD Biosciences), and Myc (sc-788, Santa Cruz Biotechnology). Secondary antibodies Alexa Fluor 488-conjugated goat anti-rabbit IgG (111−545-144) and Cy3-conjugated goat anti-mouse IgG (115−165-146) were purchased from Jackson Immunoresearch, Inc.
4.7. Immunoblotting, coimmunoprecipitation, immunoaffinity purification, and mass spectrometry analysis
E10.5 mouse embryos used to detect FLAG-Gli2 or Gli3 were lysed in RIPA buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1%Triton X-100, 1% sodium deoxycholate, 0.1% SDS, protease inhibitors). Cultured cells used to detect Ta3, PKARIα, PKARIIβ were lysed in lysis buffer (50 mM HEPES (pH7.4), 150 mM NaCl, 10% glycerol, 1% NP40, protease inhibitors). Immunoblotting and coimmunoprecipitation were performed as described (Wang et al., 2000). The intensity of immunoblot bands was quantified using NIH imageJ. Two-tailed Student t-test was used to determine p-values.
For immunoaffinity purification, twenty 15 cm plates of NIH3T3 cells stably transduced with pLNCX-FH-Ta3 virus or NIH3T3control cells were lysed by Dounce homogenization in lysis buffer (50 mM HEPES (pH7.4), 150 mM NaCl, 10% glycerol, 0.5% NP40, freshly add DTT (1 mM), protease inhibitor cocktail). After cleared by centrifugation, the protein lysates were incubated with 200 µl FLAG antibody conjugated with Sepharose beads (A2220, Sigma) by rotation for 2 h at 4 °C. The beads were washed with the lysis buffer for at least 4 times. The immunoprecipitated proteins were denatured with SDS loading buffer and resolved by SDS-PAGE. The gel lanes were sliced into 8 pieces, which were subjected to digestion with trypsin. The resulting peptides were eluted and subjected to mass spectrometric analysis as described (Xu et al., 2010).
Supplementary Material
Acknowledgments
We thank Yong Pan for helping characterize the pGli antibody. Monoclonal antibodies, Foxa2, Isl1, Hb9, and Pax6 were purchased from the Developmental Studies Hybridoma Bank maintained by the University of Iowa, Department of Biological Sciences, Iowa City, Iowa 52242, under contract NO1-HD-7–3263 from the NICHD. This study was supported by National Institutes of Health (USA) (R01GM114429) to B.W., National Natural Science Foundation of China (31570721), Chinese Ministry for Foreign Experts (GDW20143100069), and Science and Technology Commission of Shanghai Municipality (14521100700 and 14520720200) to Q.M., National Natural Science Foundation of China (31470772) and Jiangsu Key Laboratory of Translational Research and Therapy for Neuro-Psycho-Diseases (BM2013003) to G.X. J.L. is a recipient of Scholarship from Chinese Scholarship Council.
Footnotes
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2017.06.012.
References
- Bai CB, Stephen D, Joyner AL. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev. Cell. 2004;6:103–115. doi: 10.1016/s1534-5807(03)00394-0. [DOI] [PubMed] [Google Scholar]
- Bangs F, Antonio N, Thongnuek P, Welten M, Davey MG, Briscoe J, Tickle C. Generation of mice with functional inactivation of talpid3, a gene first identified in chicken. Development. 2011;138:3261–3272. doi: 10.1242/dev.063602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev. Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc. Natl. Acad. Sci. Usa. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Wilson CW, Li YJ, Law KK, Lu CS, Gacayan R, Zhang X, Hui CC, Chuang PT. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009;23:1910–1928. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- Davey MG, Paton IR, Yin Y, Schmidt M, Bangs FK, Morrice DR, Smith TG, Buxton P, Stamataki D, Tanaka M, Munsterberg AE, Briscoe J, Tickle C, Burt DW. The chicken talpid3 gene encodes a novel protein essential for Hedgehog signaling. Genes Dev. 2006;20:1365–1377. doi: 10.1101/gad.369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse N, Maiti T, Wang B, Porter JA, Hall TM, Leahy DJ, Beachy PA. Sonic hedgehog protein signals not as a hydrolytic enzyme but as an apparent ligand for patched. Proc. Natl. Acad. Sci. USA. 1999;96:10992–10999. doi: 10.1073/pnas.96.20.10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat. Rev. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich LV, Johnson RL, Milenkovic L, McMahon JA, Scott MP. Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev. 1996;10:301–312. doi: 10.1101/gad.10.3.301. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- Han L, Pan Y, Wang B. Small ubiquitin-like Modifier (SUMO) modification inhibits GLI2 protein transcriptional activity in vitro and in vivo. J. Biol. Chem. 2012;287:20483–20489. doi: 10.1074/jbc.M112.359299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryos—A Laboratory Manual Second edition. Cold Spring Harbor Laboratory Press, USA; 1994. [Google Scholar]
- Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev. Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kato M, Beachy PA. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc. Natl. Acad. Sci. USA. 2009;106:21666–21671. doi: 10.1073/pnas.0912180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Kim S, Lin YC, Inoue T, Dynlacht BD. The CP110-interacting proteins Talpid3 and Cep290 play overlapping and distinct roles in cilia assembly. J. Cell Biol. 2014;204:215–229. doi: 10.1083/jcb.201304153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocca MC, Shanks RA, Tian L, Nelson DL, Stewart DM, Goldenring JR. AKAP350 interaction with cdc42 interacting protein 4 at the Golgi apparatus. Mol. Biol. Cell. 2004;15:2771–2781. doi: 10.1091/mbc.E03-10-0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lignitto L, Carlucci A, Sepe M, Stefan E, Cuomo O, Nistico R, Scorziello A, Savoia C, Garbi C, Annunziato L, Feliciello A. Control of PKA stability and signalling by the RING ligase praja2. Nat. Cell Biol. 2011;13:412–422. doi: 10.1038/ncb2209. [DOI] [PubMed] [Google Scholar]
- Mahjoub MR, Xie Z, Stearns T. Cep120 is asymmetrically localized to the daughter centriole and is essential for centriole assembly. J. Cell Biol. 2010;191:331–346. doi: 10.1083/jcb.201003009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marigo V, Johnson RL, Vortkamp A, Tabin CJ. Sonic hedgehog differentially regulates expression of GLI and GLI3 during limb development. Dev. Biol. 1996;180:273–283. doi: 10.1006/dbio.1996.0300. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Wen X, Ratti N, Loktev A, Rangell L, Scales SJ, Jackson PK. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell. 2013;152:210–223. doi: 10.1016/j.cell.2012.12.026. [DOI] [PubMed] [Google Scholar]
- Niewiadomski P, Kong JH, Ahrends R, Ma Y, Humke EW, Khan S, Teruel MN, Novitch BG, Rohatgi R. Gli protein activity is controlled by multisite phosphorylation in vertebrate Hedgehog signaling. Cell Rep. 2014;6:168–181. doi: 10.1016/j.celrep.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novarino G, Akizu N, Gleeson JG. Modeling human disease in humans: the ciliopathies. Cell. 2011;147:70–79. doi: 10.1016/j.cell.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol. Cell. Biol. 2006;26:3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Wang B. A novel protein-processing domain in Gli2 and Gli3 differentially blocks complete protein degradation by the proteasome. J. Biol. Chem. 2007;282:10846–10852. doi: 10.1074/jbc.M608599200. [DOI] [PubMed] [Google Scholar]
- Pan Y, Wang C, Wang B. Phosphorylation of Gli2 by protein kinase A is required for Gli2 processing and degradation and the Sonic Hedgehog-regulated mouse development. Dev. Biol. 2009;326:177–189. doi: 10.1016/j.ydbio.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riobo NA, Saucy B, Dilizio C, Manning DR. Activation of heterotrimeric G proteins by Smoothened. Proc. Natl. Acad. Sci. USA. 2006;103:12607–12612. doi: 10.1073/pnas.0600880103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Schmidt PH, Dransfield DT, Claudio JO, Hawley RG, Trotter KW, Milgram SL, Goldenring JR. AKAP350, a multiply spliced protein kinase A-anchoring protein associated with centrosomes. J. Biol. Chem. 1999;274:3055–3066. doi: 10.1074/jbc.274.5.3055. [DOI] [PubMed] [Google Scholar]
- Soriano P. The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development. 1997;124:2691–2700. doi: 10.1242/dev.124.14.2691. [DOI] [PubMed] [Google Scholar]
- Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, Scott MP, Pennica D, Goddard A, Phillips H, Noll M, Hooper JE, de Sauvage F, Rosenthal A. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Ilouz R, Zhang P, Kornev AP. Assembly of allosteric macromolecular switches: lessons from PKA. Nat. Rev. Mol. Cell Biol. 2012;13:646–658. doi: 10.1038/nrm3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuson M, He M, Anderson KV. Protein kinase A acts at the basal body of the primary cilium to prevent Gli2 activation and ventralization of the mouse neural tube. Development. 2011;138:4921–4930. doi: 10.1242/dev.070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- Wang B, Li Y. Evidence for the direct involvement of {beta}TrCP in Gli3 protein processing. Proc. Natl. Acad. Sci. USA. 2006;103:33–38. doi: 10.1073/pnas.0509927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Low WC, Liu A, Wang B. Centrosomal protein DZIP1 regulates Hedgehog signaling by promoting cytoplasmic retention of transcription factor GLI3 and affecting ciliogenesis. J. Biol. Chem. 2013;288:29518–29529. doi: 10.1074/jbc.M113.492066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Lai CK, Evangelista M, Hongo JA, de Sauvage FJ, Scales SJ. Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Mol. Cell. Biol. 2010;30:1910–1922. doi: 10.1128/MCB.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Yang M, Li J, Wang C, Cao T, Tao K, Wang B. Talpid3-binding centrosomal protein Cep120 is required for centriole duplication and proliferation of cerebellar granule neuron progenitors. PloS One. 2014;9:e107943. doi: 10.1371/journal.pone.0107943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 2010;28:868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Bangs F, Paton IR, Prescott A, James J, Davey MG, Whitley P, Genikhovich G, Technau U, Burt DW, Tickle C. The Talpid3 gene (KIAA0586) encodes a centrosomal protein that is essential for primary cilia formation. Development. 2009;136:655–664. doi: 10.1242/dev.028464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.