Abstract.
Standard surgical resection of glioblastoma, mainly guided by the enhancement on postcontrast T1-weighted magnetic resonance imaging (MRI), disregards infiltrating tumor within the peritumoral edema region (ED). Subsequent radiotherapy typically delivers uniform radiation to peritumoral FLAIR-hyperintense regions, without attempting to target areas likely to be infiltrated more heavily. Noninvasive in vivo delineation of the areas of tumor infiltration and prediction of early recurrence in peritumoral ED could assist in targeted intensification of local therapies, thereby potentially delaying recurrence and prolonging survival. This paper presents a method for estimating peritumoral edema infiltration using radiomic signatures determined via machine learning methods, and tests it on 90 patients with de novo glioblastoma. The generalizability of the proposed predictive model was evaluated via cross-validation in a discovery cohort () and was subsequently evaluated in a replication cohort (). Spatial maps representing the likelihood of tumor infiltration and future early recurrence were compared with regions of recurrence on postresection follow-up studies with pathology confirmation. The cross-validated accuracy of our predictive infiltration model on the discovery and replication cohorts was 87.51% (odds ratio = 10.22, sensitivity = 80.65, and specificity = 87.63) and 89.54% (odds ratio = 13.66, sensitivity = 97.06, and specificity = 76.73), respectively. The radiomic signature of the recurrent tumor region revealed higher vascularity and cellularity when compared with the nonrecurrent region. The proposed model shows evidence that multiparametric pattern analysis from clinical MRI sequences can assist in in vivo estimation of the spatial extent and pattern of tumor recurrence in peritumoral edema, which may guide supratotal resection and/or intensification of postoperative radiation therapy.
Keywords: glioblastoma, tumor infiltration, radiomics, tumor recurrence, machine learning
1. Introduction
Glioblastoma is the most common and aggressive primary brain tumor with a median survival of 12 to 15 months.1 Surgical resection followed by chemoradiotherapy is the keystone in the management of these poor-prognosis tumors.2–4 The extent of resection is mainly guided by the boundary of the enhancing tumor (ET) on the postcontrast T1-weighted (T1CE) magnetic resonance imaging (MRI). The radiotherapy planning is also performed through ET delineation. Conformal radiotherapy, intensity-modulated radiation therapy, and proton therapy allow for an increased radiation dose to the tumor bed and surrounding margin while preserving critical brain structures.5,6 However, the malignant parenchyma extends beyond the enhancing borders of the tumor on T1CE,7,8 which is the primary target of the treatment. Stereotactic biopsies have demonstrated the presence of infiltrating glioma cells in peritumoral edema region (ED).9,10 The peritumoral edema is also a target for the planning of the postoperative radiotherapy,11,12 and aggressive extent of resection and radiation dose escalation in this region has improved survival.3,13
Peritumoral ED, a hyperintense T2-weighted signal around the ET, is a combination of infiltrating tumor cells14 and vasogenic edema,11 where fluid penetrates into the parenchymal extracellular space. It is not possible to distinguish infiltrating neoplasm from vasogenic edema with conventional imaging sequences. A noninvasive method that detects highly infiltrated tissues would be useful for understanding the behavior of the tumor and guiding treatment planning.
Multiparametric MRI noninvasively provides comprehensive information to characterize the tumor and surrounding abnormal tissues.15–18 Each imaging sequence reflects different characteristics of tissue, therefore, it would be beneficial to combine all this information in a single map to define areas of tumor infiltration in peritumoral ED. T1CE contains information about regional angiogenesis and integrity of the blood–brain barrier in the tumor. T2-weighted (T2) and T2-fluid attenuated inversion recovery (T2-FLAIR) sequences are helpful for assessing extracellular fluid in brain parenchyma.19 Measures derived from diffusion tensor imaging (DTI) provide information regarding the water diffusion process in the brain, affected in part by tumor cells architecture and density.20 Dynamic susceptibility contrast-enhanced (DSC)-MRI techniques reflect various aspects of perfusion in the brain,15 which provide quantitative measures of regional microvasculature and hemodynamics.21,22 These patterns are variably affected by infiltrating tumor cells and, independently, are not sufficiently specific to clearly define areas of tumor infiltration.7
The goal of this study is to leverage machine learning methods in order to elucidate multiparametric imaging signatures of heterogeneously infiltrated peritumoral brain tissue. Our work builds upon, and extends, previous attempts to identify infiltrated regions from imaging, which were lacking use of advanced radiomic features from multiparametric MRI sequences, and were mostly using image intensities.7,16,17 Moreover, we attempt to relate this imaging signature to possible underlying tumor biological processes. We hypothesize that the proposed method would capture and quantify subtle but important characteristics and otherwise visually imperceptible imaging variations of peritumoral tissue in patients with glioblastoma, which may relate to tumor infiltration and probability of relatively earlier tumor recurrence.
2. Materials and Methods
2.1. Participants
Institutional review board approval was obtained for this study. We first analyzed a discovery cohort of 31 patients with de novo glioblastoma, who were diagnosed at the Hospital of the University of Pennsylvania (UPenn) between 2006 and 2013. Inclusion of subjects was based on the following criteria: (i) age over 18 years, (ii) histopathological tissue diagnosis of glioblastoma (World Health Organization grade IV), (iii) clinical diagnosis of tumor recurrence proven with histopathologic analysis after repeat resection, (iv) no previous history of tumor and resection before the first surgery at University of Pennsylvania, (v) availability of 3 T scanner data, (vi) available preoperative MRI consisting of precontrast axial T1-weighted (T1), T1CE, T2, T2-FLAIR, DTI, and DSC-MRI, and (vii) available postoperative and recurrence time-point MRIs comprising at-least T1, T1CE, T2, and T2-FLAIR. More detail on image acquisition and data demographics can be found in Appendix A. Subjects who had residual tumor after surgical resection were excluded. Residual tumors were defined as any contrast-enhancing areas identified by neuroradiologist (M.B., 16 years of experience) on the immediate postoperative MRI scans captured no later than 48 h after the surgery. The replication cohort comprised 59 patients with de novo glioblastoma who met the aforementioned criteria. The same treatment, i.e., gross total resection of ET followed by temozolomide and radiotherapy was given to all the patients involved in the study.
2.2. Preprocessing, Segmentation, and Calculation of Perfusion and Diffusion Derivatives
The fractional anisotropy (FA), radial diffusivity (RAD), axial diffusivity (AX), and apparent diffusion coefficient (ADC) were derived from DTI, and relative cerebral blood volume (rCBV) was derived from DSC-MRI. All preoperative MRIs of each patient were coregistered, smoothed, corrected for magnetic field in-homogeneities, and skull stripped.23–25 The computer-based glioma image segmentation and registration (GLISTR) algorithm26 was used to segment peritumoral ED and two distinct regions of tumor core (TC), i.e., ET and nonenhancing tumor core (NET) (Fig. 1).
Fig. 1.
Brain tumor segmentation via GLISTR by using input (a) T1CE, (b) T2-FALIR, (c) T1, and (d) T2 sequences. The segmented pertitumoral edema/tumor infiltration, enhancing tumor, and nonenhancing tumor core are seen overlaid on (e) axial, (f) sagittal, and (g) coronal view of T1CE sequence.
The GLISTR algorithm uses T1, T1CE, T2, and T2-FLAIR sequences. It simultaneously registers a probabilistic atlas of a healthy population to the MRIs with glioblastoma and segments the sequences into ET, NET, ED, and healthy tissue labels. This method uses the expectation maximization (EM) algorithm and incorporates a tumor growth model, a process which modifies the original atlas into a sequence with ET, NET, and ED adapted to best match a given set of patient sequences. The modified atlas is registered into the patient sequences, and then GLISTR utilizes this process for estimating the posterior probabilities of various tissue labels. EM iteratively refines the estimates of the posterior probabilities of tissue labels, the deformation field, and the tumor growth model parameters. The deformation field takes into account the mass effect produced by the tumor. Therefore, GLISTR has a framework for joint segmentation and nonlinear registration that is guided by the atlas. The inverted deformation field warps the patient sequences into the common atlas space. The mass effect gets relaxed in the atlas-normalized sequences; hence, the ventricles get decompressed compared to the original patients’ sequences. This method thus incorporates a tumor growth model to adapt the normal atlas into the anatomy of the patient brain.
2.3. Radiomic Features
Ideally, one would want to have ground truth of peritumoral tissue that is not infiltrated and tissue that is most-heavily infiltrated, in order to train a classifier. In the absence of such a ground truth, we took an alternative approach, which his discussed next. The heterogeneity of edematous tissue was assessed by defining two regions of interest (ROIs) within the peritumoral edema: near-tumor ROI (N-ROI), adjacent to TC, and far-from-tumor ROI (F-ROI), farthermost from the TC but still within the peritumoral bright-FLAIR ED (Fig. 2). The hypothesis behind using N-ROI and F-ROI was that they are expected to have relatively higher and lower infiltration and, therefore, can serve as references to model relatively more- and relatively less-infiltrated regions, respectively.7,27 There was not any fixed length of the ROIs; however, F-ROI was generally three times larger in size compared to N-ROI because more space was available for drawing farther from the tumor. The F-ROIs were subsampled at equal intervals to have a balanced training dataset with equal number of voxels of N-ROI and F-ROI.
Fig. 2.
Regions of interest: N-ROI, adjacent to TC; and F-ROI at the most distant edge of edema.
A set of radiomic features was extracted at each voxel of the N-ROI and F-ROI by using all the imaging sequences. The features were categorized into five different groups; (i) intensity features: signal intensity of preoperative MRIs; (ii) distance features: distance of a voxel from TC; (iii) statistical features: local mean and median of a voxel in preoperative MRIs; (iv) texture features: local contrast and entropy of a voxel in preoperative MRIs; (v) perfusion temporal dynamics: perfusion time series of each voxel was summarized by five principal components accounting for more than 95% of the signal’s variance. More detail on the features can be found in Appendix B. Prior to feature extraction, all the images were normalized in 0 to 255 range.
2.4. Discovery and Replication Evaluation
A multidimensional pattern classifier was trained on the above-mentioned features of the voxels of N-ROI and F-ROI using support vector machines (SVM)28 with a Gaussian kernel function. The trained model was then applied on test voxels to calculate the infiltration scores. The infiltration score of each voxel was converted to pseudoprobability of that voxel belonging to N-ROI or F-ROI class by using the Platt’s calibration method.29 In our modeling, this voxelwise map signifies spatial pseudoprobability of infiltration that was named the infiltration index. It may be noted that the infiltration index of a voxel has a value between 0 and 1 representing estimated least-infiltrated and most-infiltrated tissues, respectively.
In the discovery cohort, leave-one-out crossvalidation strategy was adopted wherein an SVM classifier was trained on ( for discovery cohort) subjects and was tested on the left out subject. This process was reiterated times, each time leaving a different subject out. In order to provide realistic estimates of how well the predictive models were likely to generalize to new populations, a model was prepared on all the subjects of the discovery cohort, i.e., and was tested in a totally independent replication cohort of de novo glioblastoma subjects. In particular, the principal component analysis transform and -score parameters (mean, standard deviation) were learned from the training set and then applied to test set (using crossvalidation on discovery cohort and using direct evaluation on replication cohort).
2.5. Evaluation Method
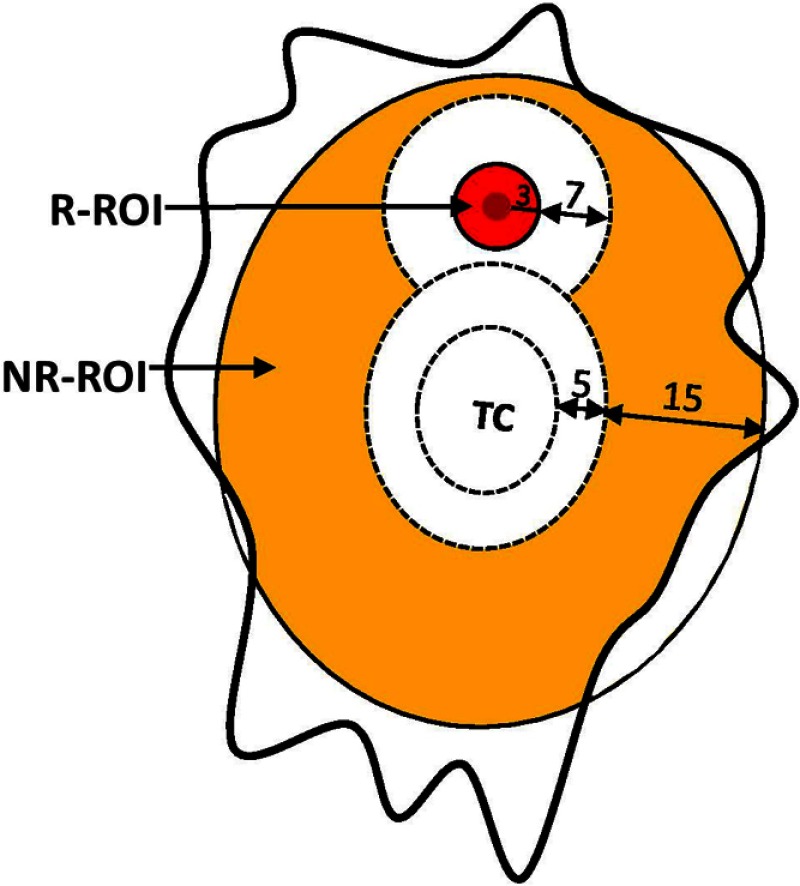
The evaluation method following the guidelines of Ref. 7 was used here. In brief, evaluation was performed by comparing the estimated infiltration map to two ROIs: recurring ROI (R-ROI) and nonrecurring ROI (NR-ROI). Figure 3 provides a cartoon representation of a tumor with the annotation of recurrence sites. It shows the regions (R-ROI and NR-ROI, both colored) used in the evaluation process.
Fig. 3.
Regions used in the evaluation process of the proposed method. R-ROI: region demarcated by the radiologists and additional 3 mm margin around it. White area in the figure shows regions excluded from the evaluation either due to close proximity to TC or to recurrence. NR-ROI: region other than TC, R-ROI, and the regions not included in evaluation due to close proximity (color figure online).
-
1.
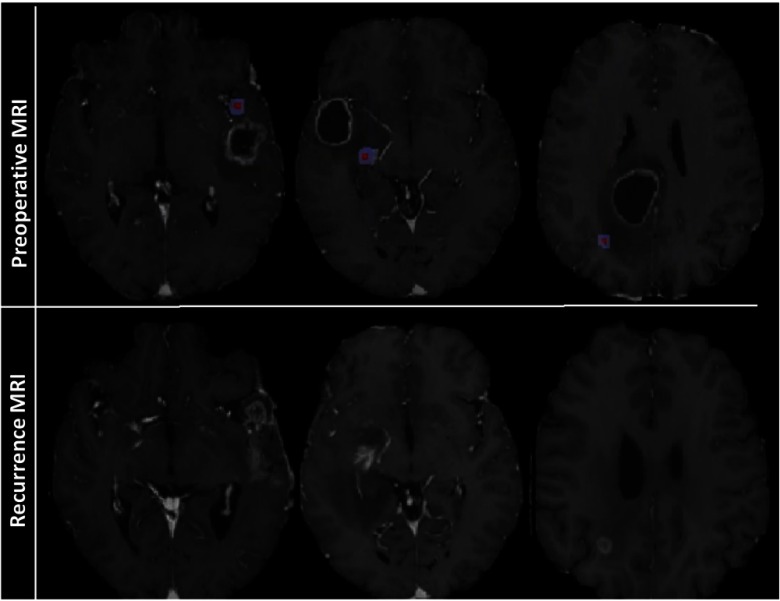
The R-ROIs were manually drawn by experts (H.A., M.B.) with the intention of selecting a small region on the preoperative MRI that best corresponded to the location of pathology-proven recurrence. This was a best approximation due to preoperative mass effect, resection, and inexact registration between preoperative and follow-up recurrence scans (Fig. 4). Hence, a 3 mm margin around the manually estimated origin-of-recurrent tumor region was included in the R-ROIs, assuming that they were likely to be heavily infiltrated but also accounting for registration uncertainties in placing the R-ROIs (Fig. 4).
-
2.
NR-ROIs were defined as all the remaining peritumoral EDs between 5 and 20 mm around the TC.
Fig. 4.
Top-row shows preoperative MRI sequences and the R-ROI (red) demarcated by the radiologists. The extended blue region shows the 3-mm margin that was also added to R-ROI to account for registration errors. The bottom-row shows recurrence MRI sequences, which were used as a reference to demarcate R-ROI (color figure online).
The following regions were excluded from the evaluation process due to several reasons.
-
1.
A margin between 3 and 10 mm (7 mm) around the R-ROI was excluded from any labeling as recurrence or nonrecurrence because of its immediate proximity to actual recurrence. Even if there were no registration uncertainty in the mapping from preoperative to follow-up scans, this tissue would naturally be deemed likely to be heavily infiltrated and to recur soon, due to its proximity to actual recurrence.
-
2.
The region beyond 20 mm distance from TC was excluded from evaluation to avoid artificially overestimating the predictive value of our analysis, because distant edema is naturally unlikely to recur early; therefore, a model predicting this correctly would have limited added value over common clinical knowledge. Similarly, the 5-mm margin around the TC was also excluded in the evaluation for several reasons. First, some peritumoral nonenhancing tissue is typically removed at our institution during surgery. Second, registration uncertainties in mapping recurrence to preoperative scans are maximal around the tumor, due to preoperative mass effect. Third, the natural history of glioblastoma shows that immediate peritumoral tissue is the most likely to be heavily infiltrated and to present the earliest recurrence; therefore, the practical value of a predictive model in that region is limited.
To assess the performance of the proposed infiltration prediction method, the R-ROI and NR-ROI served as positive and negative regions. The binary label maps were obtained by applying threshold of 0.5 on the continuous infiltration probability map (Fig. 5), and true positive (TP), false positive (FP), true negative (TN), and false negative (FN) were calculated by comparing the label map with positive and negative regions. Sensitivity, specificity, accuracy [Eqs. (1)–(3)], and area-under-the-curve (AUC) were calculated from the TP, FP, TN, and FN. In order to account for the different sizes of R-ROIs and NR-ROIs, we calculated balanced accuracy, which is the average value of sensitivity and specificity [Eq. (4)].
| (1) |
| (2) |
| (3) |
| (4) |
Fig. 5.
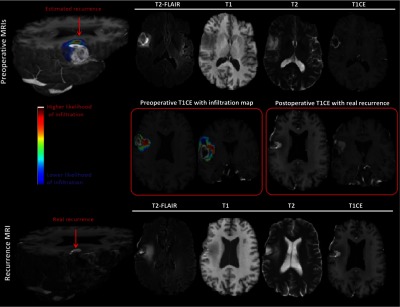
An estimated map for tumor infiltration from preoperative MRIs. Top row (preoperative MRIs): MRIs, comprising T1CE, T2-FLAIR, T1, and T2, on the right, and 3-D view of an estimated infiltration map overlaid on the T1CE image on the left. Red arrow points to the area of high risk. Bottom row (recurrence MRIs): MRIs, comprising T1CE, T2-FLAIR, T1, and T2, on the right, and 3-D view of real recurrence via T1CE image on the left. Middle row: Detailed view of preoperative T1CE MRI with the overlaid predicted infiltration map on the left, and detailed view of recurrence T1CE MRI on the right (color figure online).
In addition, odds ratio30 was calculated to quantify how strongly the estimated preoperative infiltration maps were associated with subsequent recurrence [Eq. (5)]. We considered the infiltration map as a risk factor (exposure) and calculated its association with the outcome, which is a recurrence in our study. The R-ROI and NR-ROI served as outcome of our model, whereas binary label map provided exposed (EX, ) and not exposed (NEX, ) regions (Table 1).
Table 1.
Outcome and exposure of the proposed infiltration prediction model.
| R-ROI | NR-ROI | |
|---|---|---|
| EX | ||
| NEX |
The exposure and outcome were then used to calculate odds ratio.
| (5) |
2.6. Software Availability
The proposed method is implemented in Insight Toolkit and is integrated in the Cancer Imaging Phenomics Toolkit (CaPTk), which is available in Ref. 31.
3. Results
3.1. Infiltration Scores
Table 2 shows the results separately for the subjects of the discovery and replication evaluations. Results for the discovery cohort were obtained using leave-one-out crossvalidation, whereas the results for replication cohort were based on single model trained on the discovery cohort. Classification results include a mean sensitivity of 80.65%, specificity of 87.63%, accuracy of 87.51%, and recurrence odds ratio estimates of 10.22 for tissue predicted to be relatively more infiltrated in the discovery cohort. Similar results were observed for the replication cohort. The proposed method showed a mean sensitivity of 97.06%, specificity of 76.73%, accuracy of 89.54%, and recurrence odds ratio estimates of 13.66 for the replication cohort.
Table 2.
Performance of the proposed infiltration prediction model for discovery and replication cohorts.
| Discovery cohort | Replication cohort | |
|---|---|---|
| Odds ratio | 10.22 | 13.66 |
| Accuracy | 87.51 | 89.54 |
| Sensitivity | 80.65 | 97.06 |
| Specificity | 87.63 | 76.73 |
| Balanced accuracy | 85.00 | 87.00 |
| AUC | 0.83 | 0.91 |
Figure 5 shows an infiltration map generated for an example subject. The left part of the middle row shows a preoperative T1CE sequence of the subject, and the predicted infiltration map is overlaid on it. The right part shows the T1CE sequence of the same subject with real recurrence. A side-by-side view of both the sequences shows that the predicted recurrence (red region in left) matches well with the real recurrence (bright region in right).
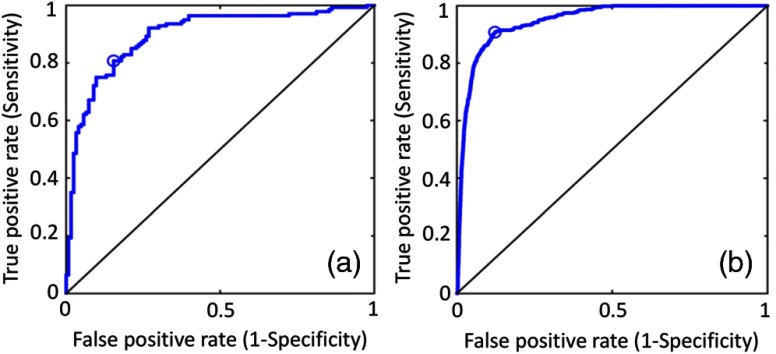
The predictive power of the developed infiltration model was also accessed via receiver operating characteristic (ROC) curve and corresponding AUC. The AUC was 0.83 and 0.91, for discovery and replication cohort, respectively (Fig. 6).
Fig. 6.
ROC curves in (a) discovery and (b) replication cohorts. The ROC curves are compared with chance (the black diagonal line).
3.2. Imaging Features of Regions Involved in Training and Evaluation
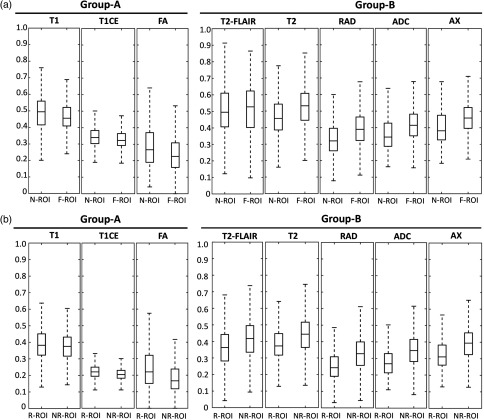
Considering the variation in the imaging profile of R-ROI and NR-ROI, we sought to better understand the primary imaging characteristics (features), which distinguish these regions (Fig. 7). The imaging features were significantly different between R-ROI and NR-ROI [Fig. 7(b)], and between N-ROI and F-ROI [Fig. 7(a)]. The imaging features were consistent between N-ROI (hypothesized to be more infiltrated) and R-ROI (recurrent region), and between F-ROI (hypothesized to be less infiltrated) and NR-ROI (nonrecurrent region). The major finding in this direction is that R-ROI, when compared with NR-ROI, revealed higher vascularity and cellularity. In addition, R-ROI has lower signal intensity on T2 and T2-FLAIR sequences and higher signal intensity on T1, thereby suggesting lower water concentration compared to NR-ROI. Each imaging feature contained significant discriminating information as can be seen by the separation between different regions in Figs. 7(a) and 7(b), which when integrated via a machine learning algorithm yielded the reported infiltration predictions. The discriminating power of various individual intensity features calculated by ROC curve analysis was: T1 (), T1CE (), T2 (), T2-FLAIR (), ADC (), FA (), AX (), and RAD ().
Fig. 7.
Imaging characteristics of the regions involved in training and evaluation of the proposed model. (a) N-ROI and F-ROI, (b) R-ROI, and NR-ROI. The -axis shows the regions and the -axis shows the values of the voxels (scaled in the range of 0 to 1) in each imaging sequence. The value of ANOVA test showed a significant difference between the two groups in all sequences ().
The proposed model was also evaluated using conventional imaging sequences such as T1, T1CE, T2-FLAIR, and T2 only, and intensity, texture, statistical, and distance features were used to build the model. The experiment was performed under the same conditions, i.e., leave-one-out crossvalidation on the discovery cohort () and then applying the model built on discovery cohort to an independent replication cohort (). The performance of the model for both the cohorts was lower (odds ratio: discovery = 6.59, replication = 8.65) compared to the performance obtained using all the imaging sequences (odds ratio: discovery = 10.22, replication = 13.66). This observation underscores the advantage of using a multiparametric MRI model that integrates synergistic imaging features extracted from various imaging sequences. However, this result also indicates that useful predictions can be obtained via conventional MRI, which is available in all clinics, as long as rich radiomic feature sets are extracted.
4. Discussion
Despite the therapeutic advancements over the past decade, the median survival for glioblastoma still remains around 14 months.32 The standard clinical practice for glioblastoma resection is removal of enhancing core of the tumor, mainly guided by the preoperative T1CE MRI, thereby leaving the majority of the infiltrating tumor mostly unresected. Similarly, the current radiation procedure involves radiation on the resection bed and a variable margin around the resection bed, which both normally receive the reduced and spatially uniform radiation dose. There is a need to quantify the heterogeneity within the peritumoral region and assess the spatial pattern and extent of tumor infiltration within the peritumoral region in order to guide these surgical processes and pave the way for targeted intensification of local therapy to the infiltrated peritumoral region. In this study, we aim to leverage multiparametric MRIs along with advanced machine learning methods to address this critical and unresolved need in the field of glioblastoma therapy and demarcate the regions at highest risk for tumor recurrence.
The multiparametric modeling has been previously leveraged to investigate the imaging surrogates of peritumoral infiltration. For instance, Akbari et al. used conventional and advanced imaging sequences to identify basic MR imaging features suggestive of glioblastoma infiltration, yielding a predictive model for recurrence.7 Our investigation builds on the described previous work for multiparametric analysis by implementing comprehensive quantitative analysis of distance, texture, statistical, and signal strength measures of the infiltrated and noninfiltrated regions from a large cohort of de novo glioblastoma patients using conventional and advanced imaging sequences. The better performance of our model compared to previous studies7,11,12 can be attributed to the comprehensive feature set and availability of a larger cohort.
4.1. Implications for Surgical and Radiation Planning
The ability to predict the site of tumor recurrence has numerous potential clinical ramifications. Neurosurgeons have proposed the concept of supratotal resection, in which they administer agents (e.g., Ref. 33) that identify areas of microscopic tumor infiltration at the time of surgical resection of a primary glioma, allowing for increased rates of gross total resection and improved progression-free survival.34 Adding a noninvasive tool to the armament of neurosurgeons may further increase these surrogates for overall survival. The proposed method would enable intensive, yet targeted, surgery and radiotherapy, thereby potentially delaying recurrence and prolonging survival.
Radiation dose escalation trials in the twentieth century uniformly failed to show a survival benefit.35,36 However, there is renewed interest in these types of trials in the modern era of concurrent chemoradiation with temozolomide. It is theorized that radiotherapy delivery techniques may improve the therapeutic ratio in dose escalation. Identification of the region of interest for radiation dose escalation remains challenging, as a balance is needed between normal tissue toxicity and therapeutic doses. Quantifying a region at highest risk of tumor recurrence provides a potential target for dose escalation based on the very spatial heterogeneity of disease in the peritumoral edema. The high risk areas, found in the infiltration maps, which often appear adjacent to ET or cavity but sometimes are a distance away, may serve as potential regions that could be included in a target volume for radiation dose escalation. We believe that using predictive models to better delineate the region of highest risk can influence the pattern of relapse in patient with glioblastoma, with the potential to improve clinical outcomes such as progression-free survival and overall survival, by directing high-dose radiation primarily toward regions likely to present earlier recurrence while offering relative preservation of lower-risk brain tissue. The proposed method will assist personalization of treatment regimens based on patient-specific features rather than a one-size-fits-all delineation of high-risk regions of interest.
4.2. Biological Interpretation of the Radiomic Signatures
The main findings of the derived radiomic signature (Fig. 7) indicate that R-ROI (when compared with NR-ROI) has lower signal intensity on native-T2 and T2-FLAIR signals, thereby suggesting lower water concentration. Further, the R-ROI shows relatively higher T1 signal intensity, which would also be consistent with lower water concentration. Finally, the R-ROI shows slightly higher T1CE, which suggests relatively more compromised blood–brain barrier in tissue, also consistent with the characteristics of infiltrating tumor. The diffusion measures provide information that relates to cell density of the peritumoral tissue. Regions with high cellularity tend to have lower ADC37 and higher FA.38 Consistent with the existing literature, the obtained radiomic signature suggests that the R-ROI (when compared with NR-ROI) has lower mean diffusivity and higher FA. The features calculated from DSC-MRI signal, which relate to aspects of tissue vascularization, perfusion, and permeability of blood vessels, were also different among R-ROI and NR-ROI.
Individual assessment of these radiomic signatures displayed differences between R-ROI and NR-ROI, as can be seen via visual inspection in Fig. 7. However, appropriate integration of these radiomic signatures via machine learning methods yielded much better sensitivity and specificity, both in crossvalidation and in independent cohort evaluation, underlining the value of multivariate pattern analysis approaches.
4.3. Limitations
A limitation of this study is that the data were acquired from a single institution, whereas multicenter data would be beneficial to further and externally validate our infiltration prediction model. However, the use of independent discovery and replication cohorts, along with the use of clinically available imaging sequences, provides confidence that this radiomic signature will generalize well to other institutions and patient populations.
5. Conclusion and Future Work
The postcontrast T1 imaging cannot delineate the surrounding infiltrating tumor for glioblastoma patients, therefore is not sufficient to guide the surgical and radiation procedures. The current study provides an in vivo noninvasive and reproducible method for preoperative assessment of the pattern of tumor infiltration within the peritumoral region, suggestive of subsequent tumor recurrence, which can offer significant advantages over the current clinical practice by guiding surgical and radiation treatment planning and paving the way for personalized targeted treatment.
While radiomic features used in this study provided promising indication of infiltration, future studies would benefit from using more sophisticated radiomic features to emphasize the biologic heterogeneity in glioblastoma. In fact, the use of machine-learning algorithms to implement both supervised and unsupervised feature detection may allow the model to account for potential complex and nonlinear radiographic-histologic relationships. Finally, the proposed model may generalize to a host of other tumor types; however, any model predictions other than glioblastoma would entail a replication evaluation similar to that performed in the current study.
Acknowledgments
This work was supported by the following two National Institutes of Health Grants: R01-NS042645 and U24-CA189523, and by a grant by Penn’s Abramson Cancer Center and the State of Pennsylvania.
Biographies
Saima Rathore received her BS degree in software engineering from Fatima Jinnah Women University, Rawalpindi, Pakistan, in 2006, and her MS degree in computer engineering from the University of Engineering and Technology, Taxila, Pakistan, in 2008. She completed her PhD in computer science from Pakistan Institute of Engineering and Applied Sciences, Islamabad, Pakistan, in 2015. Currently, she is working as a postdoctoral researcher in Perelman School of Medicine, University of Pennsylvania, USA. She is the coauthor of more than 40 research publications. Her research interests include medical image analysis, segmentation, classification, and evolutionary algorithms.
Biographies for the other authors are not available.
Appendix A: Additional Dataset Details
A.1. Image Acquisition Protocol
Preoperative MRIs were acquired using a 3-T scanner. Obtained for all patients prior to surgery were: T1-weighted: matrix ; resolution ; repetition time (TR): 1760 ms; echo time (TE): 3.1 ms; T1CE: matrix ; resolution ; TR: 1760 ms; TE: 3.1 ms. T2-weighted: matrix ; resolution ; TR: 4680 ms; TE: 85 ms. T2-FLAIR: matrix ; resolution ; TR: 9420 ms; TE: 141 ms. DTI: matrix ; resolution ; 30 gradient directions. DSC-MRI, gradient echo type echo planar imaging (GRE EPI) = field of view 22 cm ; resolution ; TR: 2000 ms; TE: 45 ms. An initial loading dose of one-quarter of the total contrast dose was administered first to help minimize errors due to potential contrast leakage out of intravascular space, and DSC-MRI data were acquired during a second bolus of the remaining contrast dose after a 5-min delay for a total of or 1.5 times single dose MultiHance (gadobenate dimeglumine). For postprocessing, blood volume maps were created on a Leonardo workstation (Siemens) using the neuro perfusion evaluation task card as per clinical routine.
A.2. Data Demographics
The discovery and replication cohort, respectively, comprised 31 and 59 patients. The demographics for both the cohorts are given in Table 3.
Table 3.
Demographics of the discovery () and replication () cohort of glioblastoma subjects.
| Characteristics | Category | Discovery | Replication |
|---|---|---|---|
| Age (years) | |||
| MEAN | 59.30 | 62.59 | |
| Std. deviation | 12.63 | 11.87 | |
| Gender | |||
| Male, (%) | 17 (54.83) | 30 (50.84) | |
| Female, (%) | 14 (45.16) | 29 (49.15) |
Appendix B: Radiomic Features
A set of radiomic features was extracted at each voxel by using all the imaging sequences (Table 4). The features were categorized into five different groups:
Intensity measures: The signal intensity of all the imaging sequences.
Texture measures: For the texture features, the imaging sequences were first normalized to 32 different gray levels, and then a bounding box of radius 2 voxels was used for all the voxels of each image. Subsequently, a gray level co-occurrence matrix was filled with the intensity values within a radius of 1 voxel to extract entropy and correlation.39
Statistical measures: The statistical features comprise the mean and median of the intensities from each imaging sequence within a radius of 1 for each voxel.
Distance measures: Shortest distance of a voxel from the tumor (ET+NET). The computer-based glioma image segmentation and registration algorithm26 was used to segment tumor.
Perfusion temporal dynamics: Perfusion time series of each voxel was summarized by five principal components accounting for more than 95% of the signal’s variance.
Table 4.
Description of the features used for building infiltration model.
| Feature name | Feature description |
|---|---|
| INT_T2FLAIR | Intensity of T2-FLAIR image |
| INT_T1CE | Intensity of T1 post-contrast image |
| INT_T1 | Intensity of T1-weighted image |
| INT_T2 | Intensity of T2-weighted image |
| INT_AX | Intensity of AX image |
| INT_FA | Intensity of FA image |
| INT_RAD | Intensity of RAD image |
| INT_ADC | Intensity of ADC image |
| INT_RCBV | Intensity of RCBV image |
| Distance | Shortest distance of a voxel from tumor (ET+NET) |
| PC1_PERF | First principal component of perfusion signal |
| PC2_PERF | Second principal component of perfusion signal |
| PC3_PERF | Third principal component of perfusion signal |
| PC4_PERF | Fourth principal component of perfusion signal |
| PC5_PERF | Fifth principal component of perfusion signal |
| TXT_CO_T1CE | Texture measure of contrast in T1CE image |
| TXT_CO_T2FLAIR | Texture measure of contrast in T2-FLAIR image |
| TXT_EN_T1CE | Texture measure of entropy in T1CE image |
| TXT_EN_T2FLAIR | Texture measure of entropy in T2-FLAIR image |
| MN_T1CE | Mean value of a voxel in T1CE image |
| MN_T2FLAIR | Mean value of a voxel in T2FLAIR image |
| MD_T1CE | Median value of a voxel in T1CE image |
| MD_T2FLAIR | Median value of a voxel in T2FLAIR image |
Disclosures
The authors declare no conflict of interest.
References
- 1.Yang I., Aghi M. K., “New advances that enable identification of glioblastoma recurrence,” Nat. Rev. Clin. Oncol. 6(11), 648–657 (2009).https://doi.org/10.1038/nrclinonc.2009.150 [DOI] [PubMed] [Google Scholar]
- 2.Brandsma D., et al. , “Clinical features, mechanisms, and management of pseudoprogression in malignant gliomas,” Lancet Oncol. 9(5), 453–461 (2008).https://doi.org/10.1016/S1470-2045(08)70125-6 [DOI] [PubMed] [Google Scholar]
- 3.Sanai N., et al. , “An extent of resection threshold for newly diagnosed glioblastomas: clinical article,” J. Neurosurg. 115(1), 3–8 (2011).https://doi.org/10.3171/2011.2.JNS10998 [DOI] [PubMed] [Google Scholar]
- 4.Stupp R., et al. , “Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma,” N. Engl. J. Med. 352(10), 987–996 (2005).https://doi.org/10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- 5.Heesters M., et al. , “Brain tumor delineation based on CT and MR imaging. Implications for radiotherapy treatment planning,” Strahlenther. Onkol. 169(12), 729–733 (1993). [PubMed] [Google Scholar]
- 6.Lorentini S., et al. , “IMRT or 3D-CRT in glioblastoma? A dosimetric criterion for patient selection,” Technol. Cancer Res. Treat. 12(5), 411–420 (2013).https://doi.org/10.7785/tcrt.2012.500341 [DOI] [PubMed] [Google Scholar]
- 7.Akbari H., et al. , “Imaging surrogates of infiltration obtained via multiparametric imaging pattern analysis predict subsequent location of recurrence of glioblastoma,” Neurosurgery 78(4), 572–580 (2016).https://doi.org/10.1227/NEU.0000000000001202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konukoglu E., et al. , “Extrapolating glioma invasion margin in brain magnetic resonance images: suggesting new irradiation margins,” Med. Image Anal. 14(2), 111–125 (2010).https://doi.org/10.1016/j.media.2009.11.005 [DOI] [PubMed] [Google Scholar]
- 9.Yamahara T., et al. , “Morphological and flow cytometric analysis of cell infiltration in glioblastoma: a comparison of autopsy brain and neuroimaging,” Brain Tumor Pathol. 27(2), 81–87 (2010).https://doi.org/10.1007/s10014-010-0275-7 [DOI] [PubMed] [Google Scholar]
- 10.Guo J., et al. , “The relationship between Cho/NAA and glioma metabolism: implementation for margin delineation of cerebral gliomas,” Acta Neurochir. 154(8), 1361–1370 (2012).https://doi.org/10.1007/s00701-012-1418-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang E. L., et al. , “Evaluation of peritumoral edema in the delineation of radiotherapy clinical target volumes for glioblastoma,” Int. J. Radiat. Oncol. Biol. Phys. 68(1), 144–150 (2007).https://doi.org/10.1016/j.ijrobp.2006.12.009 [DOI] [PubMed] [Google Scholar]
- 12.Oppitz U., et al. , “3D-recurrence-patterns of gliobastomas after CT-planned postoperative irradiation,” Radiother. Oncol. 53(1), 53–57 (1999).https://doi.org/10.1016/S0167-8140(99)00117-6 [DOI] [PubMed] [Google Scholar]
- 13.Tanaka M., et al. , “High-dose conformal radiotherapy for supratentorial malignant glioma: a historical comparison,” Lancet Oncol. 6(12), 953–960 (2005).https://doi.org/10.1016/S1470-2045(05)70395-8 [DOI] [PubMed] [Google Scholar]
- 14.Barajas R. F., et al. , “Regional variation in histopathologic features of tumor specimens from treatment-naive glioblastoma correlates with anatomic and physiologic MR Imaging,” Neuro-Oncology 14(7), 942–954 (2012).https://doi.org/10.1093/neuonc/nos128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akbari H., et al. , “Pattern analysis of dynamic susceptibility contrast MRI reveals peritumoral tissue heterogeneity,” Radiology 273(2), 502–510 (2014).https://doi.org/10.1148/radiol.14132458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blystad I., et al. , “Quantitative MRI for analysis of peritumoral edema in malignant gliomas,” PLoS One 12(5), e0177135 (2017).https://doi.org/10.1371/journal.pone.0177135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang P. D., et al. , “A Multiparametric model for mapping cellularity in glioblastoma using radiographically localized biopsies,” Am. J. Neuroradiol. 38(5), 890–898 (2017).https://doi.org/10.3174/ajnr.A5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galban C. J., et al. , “Prospective analysis of parametric response map-derived MRI biomarkers: identification of early and distinct glioma response patterns not predicted by standard radiographic assessment,” Clin. Cancer Res. 17(14), 4751–4760 (2011).https://doi.org/10.1158/1078-0432.CCR-10-2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurki T., Lundbom N., Valtonen S., “Tissue characterisation of intracranial tumours: the value of magnetisation transfer and conventional MRI,” Neuroradiology 37(7), 515–521 (1995).https://doi.org/10.1007/BF00593707 [DOI] [PubMed] [Google Scholar]
- 20.Lu S., et al. , “Peritumoral diffusion tensor imaging of high-grade gliomas and metastatic brain tumors,” Am. J. Neuroradiol. 24(5), 937–941 (2003). [PMC free article] [PubMed] [Google Scholar]
- 21.Wintermark M., et al. , “Comparative overview of brain perfusion imaging techniques,” J. Neuroradiol. 32(5), 294–314 (2005).https://doi.org/10.1016/S0150-9861(05)83159-1 [DOI] [PubMed] [Google Scholar]
- 22.Tykocinski E. S., et al. , “Use of magnetic perfusion-weighted imaging to determine epidermal growth factor receptor variant III expression in glioblastoma,” Neuro-Oncol. 14(5), 613–623 (2012).https://doi.org/10.1093/neuonc/nos073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenkinson M., et al. , “Fsl,” NeuroImage 62(2), 782–790 (2012).https://doi.org/10.1016/j.neuroimage.2011.09.015 [DOI] [PubMed] [Google Scholar]
- 24.Jenkinson M., Smith S., “A global optimisation method for robust affine registration of brain images,” Med. Image Anal. 5(2), 143–156 (2001).https://doi.org/10.1016/S1361-8415(01)00036-6 [DOI] [PubMed] [Google Scholar]
- 25.Smith S. M., “Fast robust automated brain extraction,” Human Brain Mapping 17(3), 143–155 (2002).https://doi.org/10.1002/(ISSN)1097-0193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon D., et al. , “Combining generative models for multifocal glioma segmentation and registration,” in Medical Image Computing and Computer-Assisted Intervention (MICCAI ’14), pp. 763–770 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamahara T., et al. , “Morphological and flow cytometric analysis of cell infiltration in glioblastoma: a comparison of autopsy brain and neuroimaging,” Brain Tumor Pathol. 27(2), 81–87 (2010).https://doi.org/10.1007/s10014-010-0275-7 [DOI] [PubMed] [Google Scholar]
- 28.Chang C.-C., Lin C.-J., “LIBSVM: a library for support vector machines,” ACM Trans. Intell. Syst. Technol. (TIST) 2(3), 27 (2001). [Google Scholar]
- 29.Platt J., “Probabilistic outputs for support vector machines and comparison to regularized likelihood methods,” in Advances in Large Margin Classifiers, Smola A. J., et al., Eds., MIT Press, Cambridge, Massachusetts: (2000). [Google Scholar]
- 30.Cornfield J., “A method for estimating comparative rates from clinical data. Applications to cancer of the lung, breast, and cervix,” J. Natl. Cancer Inst. 11, 1269–1275 (1951).https://doi.org/10.1093/jnci/11.6.1269 [PubMed] [Google Scholar]
- 31.Davatzikos C., et al. , “Cancer imaging phenomics toolkit: quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome,” J. Med. Imaging 5(1), 011018 (2018). https://doi.org/10.1117/1.JMI.5.1.011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Meir E., et al. , “Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma,” CA: Cancer J. Clin. 60(3), 166–193 (2010).https://doi.org/10.3322/caac.20069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stummer W., et al. , “Fluorescence-guided surgery with 5-aminolevulinic acid for resection of malignant glioma: a randomised controlled multicentre phase III trial,” Lancet Oncol. 7(5), 392–401 (2006).https://doi.org/10.1016/S1470-2045(06)70665-9 [DOI] [PubMed] [Google Scholar]
- 34.Esquenazi Y., et al. , “The survival advantage of ‘supratotal’ resection of glioblastoma using selective cortical mapping and the subpial technique,” Neurosurgery 81(2), 275–288 (2017).https://doi.org/10.1093/neuros/nyw174 [DOI] [PubMed] [Google Scholar]
- 35.Nelson D. F., et al. , “Combined modality approach to treatment of malignant gliomas–re-evaluation of RTOG 7401/ECOG 1374 with long-term follow-up: a joint study of the Radiation Therapy Oncology Group and the Eastern Cooperative Oncology Group,” NCI Monogr. (6), 279–284 (1988). [PubMed] [Google Scholar]
- 36.Chan J. L., et al. , “Survival and failure patterns of high-grade gliomas after three-dimensional conformal radiotherapy,” J. Clin. Oncol. 20(6), 1635–1642 (2002).https://doi.org/10.1200/JCO.2002.20.6.1635 [DOI] [PubMed] [Google Scholar]
- 37.Chen L., et al. , “The correlation between apparent diffusion coefficient and tumor cellularity in patients: a meta-analysis,” PLoS One 8(11), e79008 (2013).https://doi.org/10.1371/journal.pone.0079008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beppu T., et al. , “Fractional anisotropy value by diffusion tensor magnetic resonance imaging as a predictor of cell density and proliferation activity of glioblastomas,” Surg. Neurol. 63(1), 56–61 (2005).https://doi.org/10.1016/j.surneu.2004.02.034 [DOI] [PubMed] [Google Scholar]
- 39.Haralick R. M., Shanmugam K., Dinstein I. H., “Textural features for image classification,” IEEE Trans. Syst., Man, Cybern. SMC-3(6), 610–621 (1973).https://doi.org/10.1109/TSMC.1973.4309314 [Google Scholar]