Abstract
Taurine upregulated 1 gene (TUG1) is a long non-coding RNA associated with several types of cancer. Recently, differential expression of TUG1 was found in cancerous breast tissues and associated with breast cancer malignancy features. Although this is evidence of a potential role in breast cancer, TUG1 expression could not be associated with different subtypes, possibly due to the small number of samples analyzed. Breast cancer is a heterogeneous disease and, based on molecular signatures, may be classified into different subtypes with prognostic implications. In the present study, we include analysis of TUG1 expression in 796 invasive breast carcinoma and 105 normal samples of RNA sequencing (RNA-seq) datasets from The Cancer Genome Atlas (TCGA) and describe that TUG1 expression is increased in HER2-enriched and basal-like subtypes compared to luminal A. Additionally, TUG1 expression is associated with survival in HER2-enriched patients. These results reinforce the importance of TUG1 in breast cancer and outline its potential impact on specific subtypes.
Keywords: TUG1, TCGA, breast cancer, PAM50, survival, lncRNA
1. Introduction
Over the past few years, there has been increasing widespread interest in long non-coding RNAs (lncRNAs). LncRNAs are non-protein coding transcripts greater than 200 nucleotides in length and lacking in significant open reading frames [1]. These transcripts with their complex gene architecture have attracted the significant attention of researchers due to their recently discovered role in the development of some human diseases [2] as regulators of transcriptional and post-transcriptional gene expression [3]. It is estimated that the number of lncRNAs in the human genome is in the tens of thousands [4]. In addition, for the majority of these transcripts, their biological roles and molecular functions still remain unknown.
However, for some lncRNAs, their functions have been determined as involving inhibition of microRNAs (miRNAs), epigenetic regulation, chromatin state modification and chromosomal interactions, transport of proteins, and, finally, modulation of the activity or abundance of proteins or messenger RNAs (mRNAs) with which they interact [5].
Taurine upregulated 1 gene (TUG1) is a 7.1 kb lncRNA transcribed from human chromosome 22q12.2 [6]. Accumulating evidence shows that TUG1 plays an important role in several types of cancer [7].
Recently, Li et al. [8] detected hyperexpression of TUG1 in 100 samples of cancerous breast tissue and associated it to breast cancer malignancy features such as tumor size and distant metastasis. Additionally, TUG1 was associated to Ki-67 and human epidermal growth factor receptor 2 (ERBB2/HER2). Although this study evidenced the potential role of TUG1 in breast cancer, they failed to examine TUG1 expression in different subtypes, possibly due to the small number of samples analyzed [8].
Breast cancer is a heterogeneous disease at both the morphological and molecular level. Based on molecular signature Prediction Analysis on Microarrays that uses a minimal gene set (PAM50), there are five main subtypes of breast cancer: basal-like, HER2-enriched, luminal A, luminal B, and normal-like. These subtypes have specific biological features, epidemiology, and prognostic implications, and are useful in guiding patient treatment [9,10].
In the present study, we include analysis of TUG1 expression in 796 invasive breast carcinoma samples and 105 normal samples of RNA sequencing (RNA-seq) datasets from The Cancer Genome Atlas (TCGA), and describe that TUG1 expression is increased in HER2-enriched and basal-like subtypes compared to luminal A. Additionally, TUG1 expression is associated with overall survival in HER2-enriched patients. These results reinforce the importance of TUG1 in breast cancer and evidence its potential impact on specific subtypes.
2. Results
2.1. TUG1 Expression Is Increased in HER2-Enriched and Basal-Like Compared to Luminal A
The transcript level of TUG1 in breast cancer tissues was not differentially expressed when compared to adjacent non-cancerous tissue when all samples were considered (p = 0.18) nor in paired comparison analysis (p = 0.12).
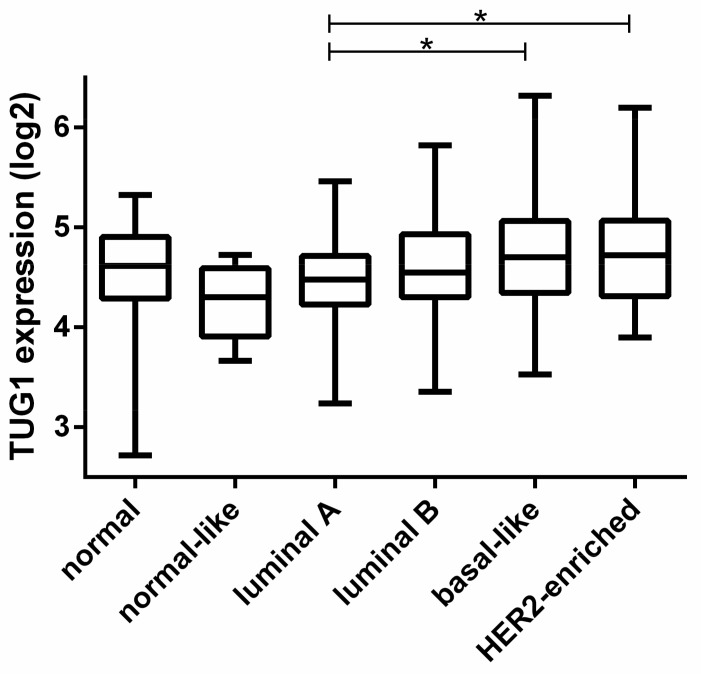
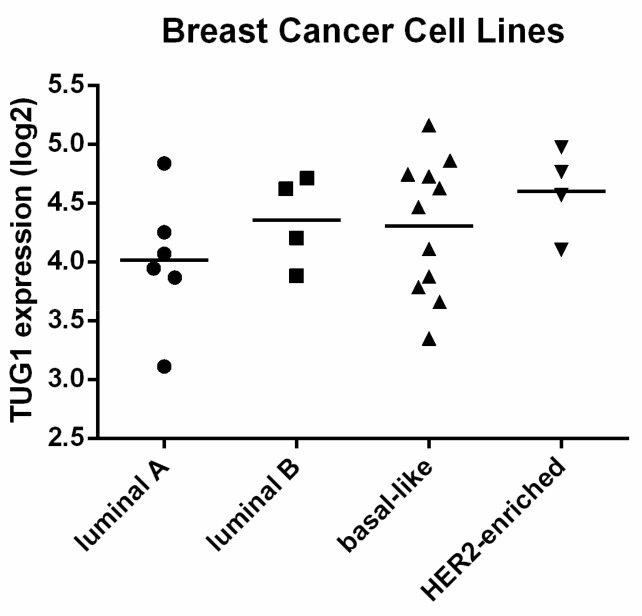
Related to subtype PAM50 classification, TUG1 was found to be hypoexpressed in patients classified as having the luminal A subtype when compared to HER2-enriched and basal-like subtypes (p < 0.001) (Figure 1). In breast cancer cell lines, although TUG1 expression profile were similar to that seen in patients, the difference was not significant (p = 0.39) (Figure A1).
Figure 1.
Expression analysis of the taurine upregulated 1 gene (TUG1) transcript in Prediction Analysis on Microarrays that uses a minimal gene set (PAM50) breast cancer subgroups. TUG1 expression data were from “The Cancer Genome Atlas (TCGA)”, and the PAM50 gene signatures rely on 50 discriminatory genes to segregate tumors into luminal A (n = 227), luminal B (n = 122), human epidermal growth factor receptor 2 (HER2)-enriched (n = 56), basal-like (n = 94), and “normal-like” (n = 7). Samples categorized as “normal” are adjacent non-tumor breast tissue (n = 105). * p < 0.001.
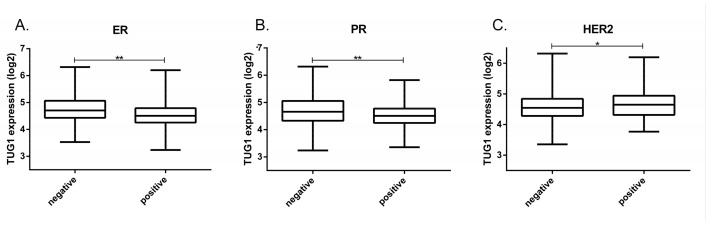
High TUG1 expression was also associated with positive HER2 and negative status for estrogen and progesterone receptors (ER/PR) (p ≤ 0.001) (Figure 2). Association with the neoplasm disease stage was not found (p = 0.43) (data available at TANRIC platform)
Figure 2.
TUG1 transcript expression associated with breast cancer relevant receptors; (A) TUG1 expression in positive (n = 385) or negative (n = 114) estrogen receptor (ER); (B) TUG1 expression in positive (n = 328) or negative (n = 170) progesterone receptor (PR); (C) TUG1 expression in positive (n = 74) or negative (n = 413) HER2. TUG1 expression data were from TCGA. * p = 0.001; ** p < 0.0001. ER, estrogen receptor; PR, progesterone receptor.
2.2. TUG1 Has a Different Impact on Survival Depending on Molecular Subtype
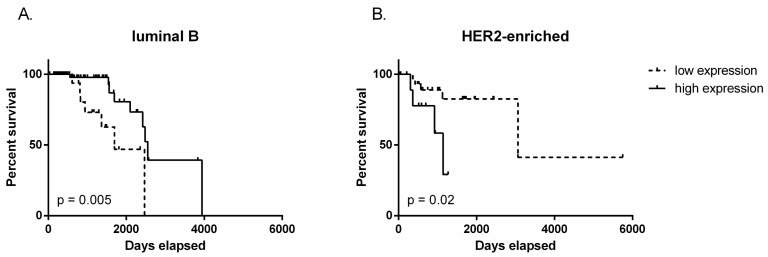
Considering the differences in TUG1 in molecular subgroups, we investigated the prognostic significance of expression in each subtype. We found that decreased expression of TUG1 is associated with improved disease-free survival in HER2-enriched patients (the lowest 75% expression -P75, p = 0.02) but high expression of TUG1 is associated with increased disease-free survival in luminal B patients (the lowest 25% expression - P25, p = 0.005) (Figure 3), suggesting that the prognostic value of TUG1 could be different in specific subtypes.
Figure 3.
Disease-free survival determined by Kaplan-Meier plots and the log-rank test according to TUG1 expression. (A) luminal B: 25% of patients with lower and 75% of patients with higher TUG1 expression; (B) HER2-enriched: 75% of patients with lower and 25% of patients with higher TUG1 expression. TUG1 disease-free survival information of breast cancer patients was downloaded from the TCGA portal.
The classification of mammary tumors by the TNM system is based on the main morphological attributes of malignant tumors, which are clinically and histopathologically determined. The parameters evaluated are: primary tumor extension (T), presence and extent of regional lymph node metastases (N), and the presence of distant metastases (M) [11,12]. For subtypes evidenced in univariate analysis of survival, multivariate Cox regression was performed. Age at initial pathologic diagnosis, tumor size (T1 to T4), involvement of lymph nodes (N0 to N3), and presence of metastatic disease (M0 or M1) were considered as covariates, along with TUG1 expression. In luminal B samples (Table 1), all the covariates tested failed to show significance. However, in HER2-enriched samples (Table 2), the analysis concluded that TUG1 expression, age at initial pathologic diagnosis, and positive metastasis are associated with prognosis.
Table 1.
Multivariate Cox analysis of luminal B samples.
| Multivariate Cox Analysis | ||
|---|---|---|
| Covariate | Hazard Ratio (95% CI) | p-Value |
| Age at initial diagnosis | 1.0038 (0.9480–1.063) | 0.897 |
| TUG1 Expression | 0.5427 (0.1539–1.914) | 0.342 |
| Tumor Size (T1, n = 17; T2, n = 84; T3, n = 15; T4, n = 6) |
1.4296 (0.4925–4.150) | 0.511 |
| Node Metastasis (N0, n = 52; N1, n = 46; N2, n = 16; N3, n = 8) |
0.8022 (0.2955–2.178) | 0.665 |
| Distant Metastasis (M0, n = 114; M1, n = 5) |
3.3331 (0.2284–48.651) | 0.379 |
CI, confidence interval. T1–T4, tumor size; N0–N3, involvement of lymph nodes; M0–M1, absence/presence of metastatic disease
Table 2.
Multivariate Cox analysis of HER2-enriched samples.
| Multivariate Cox Analysis | ||
|---|---|---|
| Covariate | Hazard Ratio (95% CI) | p-Value |
| Age at initial diagnosis | 1.130 (1.0110–1.264) | 0.0314 |
| TUG1 Expression | 37.262 (2.5634–541.647) | 0.00807 |
| Tumor Size (T1, n = 8; T2, n = 36; T3, n = 8; T4, n = 3) |
2.374 (0.5688–9.904) | 0.23564 |
| Node Metastasis (N0, n = 21; N1, n = 17; N2, n = 11; N3, n = 6) |
1.757 (0.6721–4.593) | 0.25039 |
| Distant Metastasis (M0, n = 53; M1, n = 2) |
27.257 (1.2025–617.812) | 0.03791 |
CI, confidence interval. T1–T4, tumor size; N0–N3, involvement of lymph nodes; M0–M1, absence/presence of metastatic disease. Bold entries denote statistically significant values.
3. Discussion
The lncRNA TUG1 was initially identified in developing murine retinal cells, upregulated by taurine, and necessary for the proper formation of photoreceptors [6]. Associated with chromatin-modifying complexes, TUG1 plays a role in gene regulation, including repression of genes involved in cell cycle regulation [13], and it has been extensively associated with diverse tumor types.
For example, TUG1 is highly expressed and associated with poor prognostic markers in bladder, hepatic, colorectal, gastric, renal, and ovarian cancer. Additionally, TUG1 silencing inhibits proliferation and induces apoptosis in cells derived from bladder cancer, osteosarcoma, hepatic, renal, colon, and gastric cancer; it is also associated with migration, invasion, tumorigenicity, the cell cycle, and angiogenesis in many other tumor types [7,14,15,16]. Although most of these works presented TUG1 as having an oncogenetic role, it is downregulated and associated with better survival in glioblastoma and non-small cell lung cancer, and TUG1 may function as a tumor suppressor in these cancer cell models [17,18]. Recent papers have reviewed TUG1 expression in different kinds of cancer [19,20]. Through meta-analysis studies, they investigated the relationship of TUG1 with clinicopathological features in cancer to delineate the potential clinical applications of this lncRNA on prognosis. Although some conflicting results were found, both studies indicated that increased lncRNA TUG1 is an independent prognostic biomarker for unfavorable overall survival in cancer patients.
The involvement of TUG1 in breast cancer is still controversial. High expression of TUG1 was found in 100 cancerous tissues and associated with poor features such as tumor size, distant metastasis, and TNM (Tumor, Node, Metastasis) staging [8]. High expression was also observed in 24 breast cancer samples when compared to adjacent tissues and potential oncogenic functioning of TUG1 in breast cancer cell lines was evidenced by retardation of cell proliferation, cell migration, and invasion; suppressed cell cycle progression; and increased apoptosis after knockdown of TUG1. By another side, Fan and colleagues [21], studying 58 matched pairs of breast cancer and normal tissue samples, observed that TUG1 overexpression promoted cell migration and invasion while TUG1 knockdown had the opposite effects, suggesting that TUG1 is a tumor suppressor in breast cancer.
Technological advances are generating enormous amount of data, and the public accessibility of this information is accelerating the connection between scientific research and clinical application. Cancer genome data from large-scale projects such as TCGA is being used to discover new relevant aspects of cancer genomics such as the development of therapeutic strategies and identification of specific biomarkers.
We included expression analysis of TUG1 in 796 tumor samples’ and 105 normal breast tissue samples’ RNA-seq datasets from TCGA. This large number of samples enables broad analyses, allowing, for example, the possibility of focusing on the specific nuances of each subtype.
High expression of TUG1 in breast cancer tissues compared to adjacent tissues was not confirmed in the present study; herein TUG1 expression was quite similar in tumor and control samples.
Li et al. [8] found TUG1 was upregulated in invasive breast cancer, more specifically in Ki67 and HER2-positive patients, however, they failed to examine differences between subtypes, a limitation of their paper. The present study, based on TCGA analysis, described high TUG1 expression in basal-like and HER-enriched patients compared to luminal A patients. Additionally, high expression of TUG1 was confirmed in HER2-positive patients, and we also found high expression in ER- and PR-negative patients.
PAM50 classification provided some additional biological insights into molecular subtypes. These subtype groupings have important implications in clinical practice, mainly for the identification of patients for whom chemotherapy could be avoided, such as low risk endocrine-positive patients, usually HER2-negative [10]. Furthermore, there are many molecular mechanisms to be better understood in these subtypes considering, for example, identification of novel therapeutic targets, mainly in high-risk patient groups in which chemotherapy has only modest benefits.
High expression of TUG1 was found in HER2-enriched and basal-like patients in comparison to luminal A, usually low risk, endocrine-positive patients. HER2-enriched and basal-like patients have a higher risk of relapse and may not be susceptible to the effects of anti-HER2 target treatment (mainly in the basal-like group) [22]. TUG1 expression in these subtypes may be acting in the molecular pathways but additional investigation is needed to understand the specific networks.
Survival analysis considering all patients with breast cancer did not evidence differences but we found correlations between TUG1 expression and overall-survival in patients classified as luminal B or HER2-enriched: patients with the lowest TUG1 expression had lower survival in luminal B patients, while the highest TUG1 expression was associated with poor overall survival in HER2-enriched patients.
In multivariate Cox regression including TUG1 expression and other clinical factors (age at initial pathologic diagnosis and TNM classification), the results from the luminal B samples failed to demonstrate an association. There were a great number of censored samples at the beginning of survival analysis, so these results should be viewed with caution. The multivariate results from HER2-enriched samples showed that TUG1 expression, age at initial pathologic diagnosis, and distant metastasis were significantly correlated with prognosis, however, it is important to take into account that the number HER2-enriched samples presenting with distant metastasis (n = 2) was too small to draw any reliable conclusions.
TUG1 mechanisms of action and regulation is not completely known. Like its complex clinical associations, its mechanism of cellular action appears to be diverse.
The first mechanism described shows that TUG1 is induced in p53-wild type cells, but not in p53-mutant cells, and binds to polycomb repressive complex 2 (PRC2) to silence genes involved in cell-cycle regulation [13].
TUG1 was found to decrease miR-145 expression; more specifically, there was a reciprocal repression between TUG1 and miR-145. This regulation may induce expression of many miR-145 targets, for example, the zinc finger E-box binding homeobox 2 (ZEB2) [23]. Additionally, TUG1 may be induced by nuclear transcription factor SP1 and repress, by polycomb repressive complex 2 (PRC2), Kruppel-like factor 2 (KLF2) transcription [16].
Notch1 activation may induce TUG1 expression [24], and this lncRNA possibly interacts with Wnt/β-catenin signaling. On the other hand, the potential tumor suppression function of TUG1 may be considered, for example, by its regulation of PTEN expression [25].
As well as TUG1 mechanisms seeming to be complex and associated with multiple signaling pathways, its clinical relevance seems to depend on the context of each subtype.
Additional studies are needed to better understand how TUG1 is included in molecular pathways in each subtype. The recognition that lncRNAs such as TUG1 are differentially expressed in different subtypes, and recognition of their relationships to known molecular pathways for a subtype, opens important perspectives toward this aim.
In conclusion, TUG1 expression was found to be higher in patients classified as HER2-enriched and basal-like, compared to luminal A. Additionally, TUG1 expression was associated with survival in HER2-enriched patients. These findings suggest a potential role of this lncRNA in specific subtypes of breast cancer and in the development of different phenotypes. Further studies are needed to corroborate and extend our results.
4. Patients and Methods
Information from 796 invasive breast carcinoma and 105 non-neoplastic cancer-adjacent breast tissues was collected. All RNA-seq data and clinical information is available in the TCGA database following ethics, laws, and policies from the program. Clinicopathological characteristics of the studied population is presented in Table 3.
Table 3.
Clinicopatholigical Features.
| Characteristics | ||
|---|---|---|
| Gender | Male (n = 8) | Female (n = 784) |
| Age at diagnosis | ≤35 (n = 27) | >35 (n = 765) |
| ER status | Negative (n = 173) | Positive (n = 582) |
| PR status | Negative (n = 247) | Positive (n = 506) |
| HER2 status | Negative (n = 628) | Positive (n = 113) |
| Tumor Size | ≤2cm (n = 202) | ≥2cm (n = 560) |
| Node Metastasis | Negative (n = 369) | Positive (n = 396) |
| Distant Metastasis | Negative (n = 745) | Positive (n = 14) |
Patient information from TCGA. Data only for those with TUG1 expression and clinical data available (n = 796).
Clinical guidelines for determining estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (ERBB2/HER2) status for breast cancers have been established in TCGA samples. According to the current clinical guidelines jointly issued by the American Society of Clinical Oncology (ASCO) and the College of American Pathology (CAP), in use since January 2010, a breast tumor is considered ER- and PR-positive if the immunohistochemistry (IHC) value of the corresponding nuclear staining is ≥1%. Before 2010 there was no universal standard for ER and PR status, so local hospitals used their own thresholds for their clinical practices. Considering that the year of diagnosis of all breast cancer cases collected in this study ranged from 1988 to 2011, determining the clinical status for ER and PR followed a mixture of thresholds. For HER2, following the current ASCO/CAP guideline, a breast tumor with IHC values of 0 or 1+ is called “Negative”, level 2+ is “Equivocal”, and level 3+ is “Positive”. The equivocal cases are then analyzed by Fluorescence In Situ Hybridization (FISH), where a case is called “Positive” if the FISH ratio is ≥2.2, and “Negative” if the FISH ratio is ≤1.8 [26]. Subtype stratification was based on the identification of gene expression by the 50-gene PAM50 predictor [27].
Large-scale RNA-seq datasets from The Cancer Genome Atlas (TCGA) were assessed and analyzed by the open-access web resource “The Atlas of Noncoding RNAs in Cancer” (TANRIC) [28]
On the TANRIC website, RNA-seq datasets for breast cancer cell lines are also available. We included TUG1 expression information from 25 cell lines, classified as luminal A (n = 6), luminal B (n = 4), HER2-enriched (n = 4), or basal (n = 11) subtypes [29,30].
Analysis of variance (ANOVA) with post-hoc Tukey testing was used to examine TUG1 expression across tumor subtypes and disease stages. Student’s t-test assessed statistical differences between positive and negative ER, PR, and HER2 status. The homogeneity among the variances was tested by Bartlett’s test. More details of the algorithm for expression quantification and statistics are better described by Li et al. [28].
For survival analysis, patients were stratified into values below and above the median and the quartiles (P25 and P75). The Kaplan-Meier curves and log rank tests were used to estimate overall survival, which was calculated to the last follow-up, or death event. Additionally, multivariate Cox proportional hazards regression analysis included TUG1 expression, age, tumor size, involvement of lymph nodes, and presence of metastatic disease, with calculation of the hazard ratio (HR) and 95% confidence interval (95% CI).
Data were analysed statistically using the Statistical Package for the Social Sciences (SPSS) software for Windows, version 15.0 (SPSS Inc., Chicago, IL, USA). Cox proportional hazards regression analysis was performed using the R package [31].
Acknowledgments
We thank Clarice Groeneveld for her helpful suggestions for the statistical analysis. This collaborative study was supported by the Public Research Agencies CAPES and CNPq.
Appendix A
Figure A1.
Expression analysis of TUG1 transcript in cell lines. TUG1 expression data were from “TCGA. Symbols represents the number of cell lines analyzed: luminal A (n = 6), luminal B (n = 4), HER2-enriched (n = 4) or basal (n = 11). Analysis of variance ANOVA was used to examine TUG1 expression across cell lines (p = 0.39).
Author Contributions
D.F.G., C.M., and J.C.O. conceived and designed the experiments; C.M., I.J.C., and R.C. analyzed the data; D.F.G., E.M.S.F.R., and J.C.O. wrote the paper.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Rao A.K.D.M., Rajkumar T., Mani S. Perspectives of long non-coding RNAs in cancer. Mol. Biol. Rep. 2017;44:203–218. doi: 10.1007/s11033-017-4103-6. [DOI] [PubMed] [Google Scholar]
- 2.Fatica A., Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 3.Melissari M.T., Grote P. Roles for long non-coding RNAs in physiology and disease. Pflug. Arch. 2016;468:945–958. doi: 10.1007/s00424-016-1804-y. [DOI] [PubMed] [Google Scholar]
- 4.Hon C.C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J., Gough J., Denisenko E., Schmeier S., Poulsen T.M., Severin J., et al. An atlas of human long non-coding rnas with accurate 5′ ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dykes I.M., Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genom. Proteom. Bioinform. 2017;15:177–186. doi: 10.1016/j.gpb.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young T.L., Matsuda T., Cepko C.L. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr. Biol. 2005;15:501–512. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 7.Li Z., Shen J., Chan M.T., Wu W.K. TUG1: A pivotal oncogenic long non-coding RNA of human cancers. Cell Prolif. 2016;49:471–475. doi: 10.1111/cpr.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li T., Liu Y., Xiao H., Xu G. Long non-coding RNA TUG1 promotes cell proliferation and metastasis in human breast cancer. Breast Cancer. 2017;24:535–543. doi: 10.1007/s12282-016-0736-x. [DOI] [PubMed] [Google Scholar]
- 9.Sugita B., Gill M., Mahajan A., Duttargi A., Kirolikar S., Almeida R., Regis K., Oluwasanmi O.L., Marchi F., Marian C., et al. Differentially expressed miRNAs in triple negative breast cancer between african-american and non-hispanic white women. Oncotarget. 2016;7:79274–79291. doi: 10.18632/oncotarget.13024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nielsen T.O., Parker J.S., Leung S., Voduc D., Ebbert M., Vickery T., Davies S.R., Snider J., Stijleman I.J., Reed J., et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin. Cancer Res. 2010;16:5222–5232. doi: 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singletary S.E., Connolly J.L. Breast cancer staging: working with the sixth edition of the AJCC Cancer Staging Manual. CA Cancer J. Clin. 2006;56:37–47. doi: 10.3322/canjclin.56.1.37. [DOI] [PubMed] [Google Scholar]
- 12.Giuliano A.E., Connolly J.L, Edge S.B., Mittendorf E.A., Rugo H.S., Solin L.J., Weaver D. L., Winchester D.J., Hortobagyi G.N. Breast Cancer-Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017;67:290–303. doi: 10.3322/caac.21393. [DOI] [PubMed] [Google Scholar]
- 13.Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., Thomas K., Presser A., Bernstein B.E., van Oudenaarden A., et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma B., Li M., Zhang L., Huang M., Lei J.B., Fu G.H., Liu C.X., Lai Q.W., Chen Q.Q., Wang Y.L. Upregulation of long non-coding RNA TUG1 correlates with poor prognosis and disease status in osteosarcoma. Tumour Biol. 2016;37:4445–4455. doi: 10.1007/s13277-015-4301-6. [DOI] [PubMed] [Google Scholar]
- 15.Dong R., Liu G.B., Liu B.H., Chen G., Li K., Zheng S., Dong K.R. Targeting long non-coding RNA-TUG1 inhibits tumor growth and angiogenesis in hepatoblastoma. Cell Death Dis. 2016;7 doi: 10.1038/cddis.2016.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang M.D., Chen W.M., Qi F.Z., Sun M., Xu T.P., Ma P., Shu Y.Q. Long non-coding RNA TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Mol. Cancer. 2015;14 doi: 10.1186/s12943-015-0431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang E.B., Yin D.D., Sun M., Kong R., Liu X.H., You L.H., Han L., Xia R., Wang K.M., Yang J.S., et al. P53-regulated long non-coding rna tug1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5 doi: 10.1038/cddis.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J., An G., Zhang M., Ma Q. Long non-coding RNA TUG1 acts as a miR-26a sponge in human glioma cells. Biochem. Biophys. Res. Commun. 2016;477:743–748. doi: 10.1016/j.bbrc.2016.06.129. [DOI] [PubMed] [Google Scholar]
- 19.Li N., Shi K., Kang X., Li W. Prognostic value of long non-coding RNA TUG1 in various tumors. Oncotarget. 2017;8:65659–65667. doi: 10.18632/oncotarget.20025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J., Lin J., Li Y., Zhang Y., Chen X. Prognostic role of lncRNA TUG1 for cancer outcome: Evidence from 840 cancer patients. Oncotarget. 2017;8:50051–50060. doi: 10.18632/oncotarget.17844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan S., Yang Z., Ke Z., Huang K., Liu N., Fang X., Wang K. Downregulation of the long non-coding RNA TUG1 is associated with cell proliferation, migration, and invasion in breast cancer. Biomed. Pharm. 2017;95:1636–1643. doi: 10.1016/j.biopha.2017.09.076. [DOI] [PubMed] [Google Scholar]
- 22.Colleoni M., Munzone E. Extended adjuvant chemotherapy in endocrine non-responsive disease. Breast. 2013;22(Suppl. 2):S161–S164. doi: 10.1016/j.breast.2013.07.031. [DOI] [PubMed] [Google Scholar]
- 23.Tan J., Qiu K., Li M., Liang Y. Double-negative feedback loop between long non-coding RNA TUG1 and miR-145 promotes epithelial to mesenchymal transition and radioresistance in human bladder cancer cells. FEBS Lett. 2015;589:3175–3181. doi: 10.1016/j.febslet.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 24.Katsushima K., Natsume A., Ohka F., Shinjo K., Hatanaka A., Ichimura N., Sato S., Takahashi S., Kimura H., Totoki Y., et al. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat. Commun. 2016;7 doi: 10.1038/ncomms13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du Z., Sun T., Hacisuleyman E., Fei T., Wang X., Brown M., Rinn J.L., Lee M.G., Chen Y., Kantoff P.W., et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat. Commun. 2016;7 doi: 10.1038/ncomms10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Network C.G.A. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker J.S., Mullins M., Cheang M.C., Leung S., Voduc D., Vickery T., Davies S., Fauron C., He X., Hu Z., et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J., Han L., Roebuck P., Diao L., Liu L., Yuan Y., Weinstein J.N., Liang H. TANRIC: An Interactive Open Platform to Explore the Function of lncRNAs in Cancer. Cancer Res. 2015;75:3728–3737. doi: 10.1158/0008-5472.CAN-15-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai X., Cheng H., Bai Z., Li J. Breast cancer cell line classification and its relevance with breast tumor subtyping. J. Cancer. 2017;8:3131–3141. doi: 10.7150/jca.18457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neve R.M., Chin K., Fridlyand J., Yeh J., Baehner F.L., Fevr T., Clark L., Bayani N., Coppe J.P., Tong F., et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.R Development Core Team . A Language and Environment for Statistical Computing 2017. R Foundation for Statistical Computing; Vienna, Austria: 2011. [Google Scholar]