Abstract
Background
Dengue virus (DENV) is transmitted by mosquito and has been circulating in Guangdong, China, for over 30 years. Dengue infection causes mild to severe disease symptoms in human. Cytokine profiles were suggested to be crucial especially during the acute stage in the dengue infection.
Aim
To determine the cytokine profiles at the acute stage in patients with primary or secondary dengue infection in Guangzhou city in the 2014 outbreak.
Methods
We investigated 23 inflammatory cytokines in serum collected from dengue-infected patients and analyzed their correlations with their clinical indexes.
Results
The concentrations of CXCL9, IP-10, CXCL11, IL-8, IL-10, and CCL2 in serum were significantly higher in the groups of DENV-infected patients during the first two weeks than those of control group while CCL17 and CXCL5 showed lower expression level in the patients. Among these cytokines, CXCL9, CCL17, and CXCL5 showed statistical difference between the groups of primary and secondary infections. The platelet count and lactate dehydrogenase were correlated with the level of CCL17 and MIP-1α/CXCL5, respectively, in the group of secondary infection.
Conclusions
We determined the cytokine profiles in serum of the patients during the 2014 dengue outbreak. The expression of specific cytokines was associated with the secondary infection.
1. Introduction
Dengue virus (DENV) as a member of Flaviviridae family is an enveloped, positive-sense, single-strain RNA virus with the size of around 11 kb. It is mainly transmitted by Aedes albopictus resulting in dengue fever (DF) and, in some cases, progresses to dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS). The dengue outbreaks frequently occurred in tropical and subtropical climates areas. A report indicated that there were around 96 million with apparent dengue infections and 294 million with asymptomatic infections globally in 2010 [1]. During 2010 to 2015, around 15.7% of DENV-infected cases developed DHF and 0.7% of DENV-infected cases died globally. In endemic areas, many people have been infected by dengue virus more than once in their entire life. Indeed, around 55.9% of DENV-infected cases were known as secondary infection [2]. The nonneutralizing antibodies against heterotypic DENV in secondary infection was thought to be partially responsible for the severe dengue due to the antibody-dependent enhancement (ADE) [3]. However, many cases of primary infection could also progress to severe dengue fever.
Cytokines are a group of protein which serve as signaling transducer and modulator to regulate the host immunity especially during infection. Numbers of studies have investigated the cytokine profiles in the serum samples of the dengue-infected patients in order to understand the immune responses against this pathogen. Importantly, some studies have figured that induction of several cytokines was associated with severe dengue infection [4–7].
In mainland China, outbreaks of dengue fever mainly occurred in the southern area like Guangdong or Yunnan [8, 9]. Epidemiological studies showed that dengue 1 was the major serotype circulating in Guangdong province although small percentage of patients were infected by other three serotypes over the last 20 years which were suggested as imported incidents [10]. The most recent outbreak in Guangdong province occurred in 2014 in which around fifty thousand of dengue fever cases were reported and six patients died [11]. While majority of the patients were infected by dengue 1 viruses in this outbreak, we and others previously showed that the condition of the patients were generally mild compared to the previous large-scale outbreaks [12]. However, little was known about the cytokine profiles in these patients during the acute stage of the dengue infection. In this study, we investigated the levels of 23 inflammatory cytokines in serum from patients with either primary or secondary dengue infection during the acute phase in the 2014 dengue outbreak. In addition, their correlations with the routine clinical indexes were also analyzed.
2. Materials and Methods
2.1. Study Population and Sample Collection
During the dengue epidemics period from August to December 2014, hospitalized patients who were laboratory confirmed by either (i) DENV RNA real-time PCR (DAAN, China), (ii) DENV antigen NS1 ELISA (KeShun, China), or (iii) DENV IgM ELISA (Panbio, Australia) at the First Affiliated Hospital of Guangzhou University of Chinese Medicine were enrolled. Patients were categorized as primary infections or secondary infections according to the IgM/IgG ratio and the patients were defined as DF or severe DF according to the guideline by WHO in 2009 [13]. A total of 297 DENV-infected patients and 20 healthy controls were included in this study. Those with any disease symptom (such as fever, cough, obstruction, or other unwellness) that indicated the infection or those with any immunocompromised disease were excluded from the control group. Serum and peripheral blood samples were obtained from patients at different day after the onset day of dengue fever (day 1). Samples were further grouped to those collected in the first week (day 1 to day 7) or second week (day 8 to day 14). The patient identities were delinked from the samples during analysis. Approval for the study was obtained from the ethics committee of the First Affiliated Hospital of Guangzhou University of Chinese Medicine, and informed consent was waived by the committee.
2.2. Detection of Clinical Routine Indexes
Clinical routine indexes including count of white blood cells (WBC), platelet (PLT), and hematocrit (HCT) were determined from the peripheral blood. Glutamic-pyruvic transaminase (ALT), aspartate transaminase (AST), lactate dehydrogenase (LDH), creatine kinase (CK), creatinine (Cr), and urea were detected from the serum samples which were isolated from clotted blood by centrifugation.
2.3. Quantification of Inflammatory Cytokines
Inflammatory cytokines were identified and quantified using the flow cytometry cytokine bead array purchased from LEGENDplex (BioLegend, CA, USA) according to the manufacturer's instruction. Twenty-three cytokines were included: IFN-α, IFN-γ, TNF-α, IL-6, IL-8, IL-10, IL-1β, IL-18, IL-23, IL-33, IL-17A, IL-12, CXCL9, CXCL11, CXCL5, CCL2, CCL11, CCL17, CCL20, MIP-1α, MIP-1β, IP-10, and RANTES. Briefly, the cytokines were captured by beads which were conjugated with specific biotinylated antibodies. The streptavidin-phycoerythrin was subsequently added and bonded to the biotinylated antibodies, providing fluorescent signal intensities in proportion to the amount of bound analytes. The cytokines were identified by the size of beads and PE fluorescent signal and quantified according to a standard curve on BD FACSVerse flow cytometer (BD, USA).
3. Statistical Analysis
Statistical analysis was performed using SPSS 23.0 and GraphPad Prism v7.0. The mean values of clinical routine indexes and cytokines in each cohort were compared using Mann–Whitney U test or t-test. The correlations between routine indexes and cytokines were analyzed based on the Spearman rank correlation coefficient (R value). A P value <0.05 was considered as statistically significant.
4. Results
4.1. General Background and Clinical Routine Indexes
In 297 laboratory-confirmed DENV-infected patients, 234 patients were classified as primary infection while 63 patients were classified as secondary infection, without any cases developed severe dengue fever. The mean age of the patients with secondary infection was higher than those with primary infection (58.37 ± 18.874 versus 45.99 ± 18.369, P < 0.001) as presented in Table 1. The laboratory diagnosis results showed that the levels of WBC, PLT, HCT, and UREA were lower in all DENV-infected patients while ALT, AST, LDH, and CK were higher compared to the control group. However, no statistical difference was observed in the clinical routine indexes between the groups of primary infection and secondary infection (Table 2).
Table 1.
General background.
| Primary infections (A) | Secondary infections (B) | Controls | P value | |
|---|---|---|---|---|
| A versus B | ||||
| Age (mean ± SD) | 45.99 ± 18.369 | 58.37 ± 18.874 | 38.25 ± 7.468 | <0.001∗∗ |
| Gender | ||||
| Male | 112 | 31 | 9 | 0.888 |
| Female | 122 | 32 | 11 |
∗∗ P < 0.05.
Table 2.
Clinical routine indexes in infections and controls.
| Primary infections (mean ± SD) | P value | Secondary infections (mean ± SD) | P value | Controls (E) (mean ± SD) |
P value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1st week (A) | 2nd week (B) | A versus E | B versus E | 1st week (C) | 2nd week (D) | C versus E | D versus E | A versus C | B versus D | ||
| Number of case | 151 | 82 | — | — | 44 | 19 | — | — | 20 | — | — |
| WBC (×109/L) | 4.02 ± 2.57 | 3.79 ± 2.58 | <0.001∗∗ | <0.001∗∗ | 3.80 ± 1.83 | 4.00 ± 1.65 | <0.001∗∗ | <0.001∗∗ | 7.30 ± 1.51 | 0.840 | 0.242 |
| PLT (×109/L) | 104.07 ± 61.90 | 100.46 ± 59.68 | <0.001∗∗ | <0.001∗∗ | 105.23 ± 65.66 | 112.79 ± 74.43 | <0.001∗∗ | <0.001∗∗ | 248.70 ± 63.40 | 0.983 | 0.599 |
| HCT | 0.39 ± 0.05 | 0.39 ± 0.04 | <0.001∗∗ | <0.001∗∗ | 0.40 ± 0.04 | 0.39 ± 0.05 | <0.001∗∗ | <0.001∗∗ | 0.44 ± 0.04 | 0.366 | 0.562 |
| ALT (U/L) | 62.54 ± 79.63 | 62.72 ± 63.17 | <0.001∗∗ | <0.001∗∗ | 54.57 ± 71.40 | 74.22 ± 73.40 | <0.001∗∗ | <0.001∗∗ | 19.70 ± 8.25 | 0.679 | 0.316 |
| AST (U/L) | 76.60 ± 64.87 | 76.48 ± 63.68 | <0.001∗∗ | <0.001∗∗ | 77.16 ± 57.99 | 109.78 ± 148.19 | <0.001∗∗ | <0.001∗∗ | 18.50 ± 3.59 | 0.799 | 0.618 |
| LDH (U/L) | 297.16 ± 154.33 | 309.37 ± 111.52 | <0.001∗∗ | <0.001∗∗ | 322.97 ± 170.13 | 535.00 ± 502.20 | <0.001∗∗ | <0.001∗∗ | 151.40 ± 15.41 | 0.303 | 0.467 |
| CK (U/L) | 235.27 ± 262.68 | 273.98 ± 378.99 | <0.001∗∗ | <0.001∗∗ | 370.50 ± 500.50 | 3651.00 ± 10569.53 | <0.001∗∗ | 0.009∗∗ | 74.55 ± 18.19 | 0.233 | 0.867 |
| Cr (μmol/L) | 84.38 ± 50.42 | 86.96 ± 60.76 | 0.056 | 0.068 | 82.68 ± 26.19 | 78.53 ± 11.85 | 0.072 | 0.094 | 71.45 ± 12.95 | 0.897 | 0.868 |
| UREA (mmol/L) | 3.71 ± 2.20 | 3.84 ± 1.92 | 0.001∗∗ | 0.003∗∗ | 3.48 ± 1.83 | 3.25 ± 1.38 | 0.001∗∗ | 0.002∗∗ | 4.64 ± 1.18 | 0.318 | 0.178 |
Mean ± SD of clinical routine indexes were presented and performed analysis using Mann–Whitney U test or t-test between two cohorts. The statistical significance is indicated by asterisks: ∗∗P < 0.05.
4.2. Inflammatory Cytokines in DENV Primary Infections and Secondary Infections
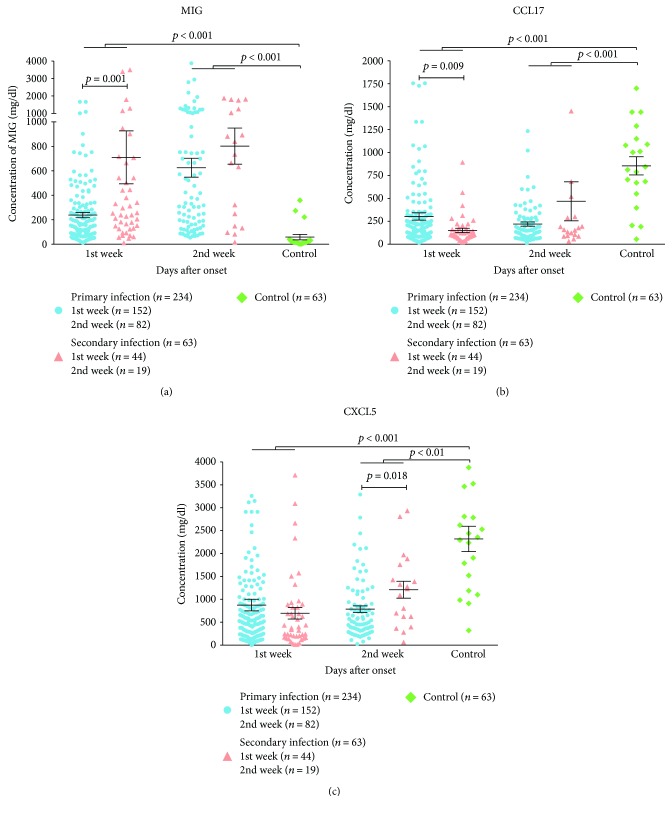
Among 23 inflammatory cytokines detected, the concentrations of IL-6, IL-1β, IL-33, IL-23, IL-17A, IL-12, IFN-α, and TNF-α were lower than the detection limit that we could not conclude their expression. The remaining 15 inflammatory cytokines were further quantified and shown in Table 3. The concentrations of CXCL9, IP-10, CXCL11, IL-8, IL-10, and CCL2 in the serum were significantly higher in the groups of DENV-infected patients during the first two weeks than the control group (P < 0.05). On the contrary, the levels of CCL17 and CXCL5 were lower in the DENV-infected patients compared to the controls. Among these cytokines, we found that CXCL9, CCL17, and CXCL5 showed statistical difference between the groups of primary and secondary infections (Figure 1). During the 1st week, the concentration of CXCL9 was higher in secondary infections than that in primary infections (710.20 ± 1435.59 versus 238.61 ± 265.29), while CCL17 was lower in secondary infections than that in primary infections (151.09 ± 155.53 versus 303.26 ± 490.43). During the 2nd week, the concentration of CXCL5 was higher in secondary infections than those in primary infections (CXCL5: 1209.47 ± 804.15 versus 786.72 ± 649.12).
Table 3.
Concentration of inflammatory cytokines in infections and controls.
| Primary infection (mean ± SD) | P value | Secondary infection (mean ± SD) | P value | Control (E) (mean ± SD) |
P value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1st week (A) | 2nd week (B) | A versus E | B versus E | 1st week (C) | 2nd week (D) | C versus E | D versus E | A versus C | B versus D | ||
| Number of case | 152 | 82 | — | — | 44 | 19 | 20 | ||||
| CXCL9 (MIG) | 238.61 ± 265.29 | 625.52 ± 700.20 | <0.001∗∗ | <0.001∗∗ | 710.20 ± 1435.59 | 802.32 ± 644.12 | <0.001∗∗ | <0.001∗∗ | 58.35 ± 100.68 | 0.001∗∗ | 0.225 |
| CCL17 (TARC) | 303.26 ± 490.43 | 220.40 ± 209.15 | <0.001∗∗ | <0.001∗∗ | 151.09 ± 155.53 | 468.16 ± 926.50 | <0.001∗∗ | 0.001∗∗ | 855.40 ± 442.74 | 0.009∗∗ | 0.537 |
| CXCL5 (ENA-78) | 875.13 ± 1553.74 | 786.72 ± 649.12 | <0.001∗∗ | <0.001∗∗ | 695.59 ± 833.55 | 1209.47 ± 804.15 | <0.001∗∗ | 0.002∗∗ | 2320.05 ± 1237.33 | 0.229 | 0.018∗ |
| CCL11 (eotaxin) | 341.20 ± 209.82 | 362.48 ± 233.38 | 0.385 | 0.604 | 307.23 ± 142.02 | 468.58 ± 163.38 | 0.389 | 0.032∗ | 375.65 ± 215.28 | 0.841 | 0.002∗∗ |
| IP-10 | 7694.73 ± 5035.07 | 6382.49 ± 4539.37 | <0.001∗∗ | <0.001∗∗ | 7387.52 ± 4874.71 | 7335.05 ± 6475.22 | <0.001∗∗ | <0.001∗∗ | 367.60 ± 151.43 | 0.672 | 0.962 |
| CXCL11 (I-TAC) | 4116.99 ± 2659.76 | 1424.44 ± 1184.22 | <0.001∗∗ | <0.001∗∗ | 3725.09 ± 3105.49 | 1214.58 ± 1088.54 | <0.001∗∗ | <0.001∗∗ | 254.50 ± 531.58 | 0.199 | 0.326 |
| IL-8 | 1911.43 ± 4409.65 | 1226.95 ± 2478.91 | <0.001∗∗ | 0.004∗∗ | 2407.39 ± 5175.86 | 1793.00 ± 2336.76 | <0.001∗∗ | 0.002∗∗ | 958.70 ± 2112.73 | 0.616 | 0.098 |
| IL-10 | 45.88 ± 61.41 | 11.16 ± 11.80 | <0.001∗∗ | <0.001∗∗ | 60.11 ± 81.01 | 7.58 ± 5.07 | <0.001∗∗ | <0.001∗∗ | 1.50 ± 0.61 | 0.240 | 0.280 |
| CCL2 (MCP-1) | 9768.83 ± 9708.82 | 5807.54 ± 3414.33 | <0.001∗∗ | <0.001∗∗ | 8611.84 ± 6343.42 | 8009.00 ± 10517.43 | <0.001∗∗ | <0.001∗∗ | 2427.95 ± 845.28 | 0.809 | 0.339 |
| IFN-γ | 251.03 ± 917.31 | 15.05 ± 36.22 | <0.001∗∗ | <0.001∗∗ | 93.39 ± 152.62 | 7.79 ± 15.62 | <0.001∗∗ | 0.054 | 0.35 ± 1.57 | 0.226 | 0.136 |
| IL-18 | 205.80 ± 221.93 | 198.98 ± 211.30 | 0.005∗∗ | 0.047∗ | 286.20 ± 397.11 | 141.05 ± 115.95 | 0.029∗ | 0.325 | 105.45 ± 55.26 | 0.834 | 0.364 |
| MIP-1α | 244.68 ± 398.14 | 253.67 ± 431.30 | 0.006∗∗ | 0.003∗∗ | 223.41 ± 262.85 | 210.26 ± 157.82 | 0.156 | 0.002∗∗ | 127.20 ± 142.06 | 0.392 | 0.633 |
| RANTES | 11621.99 ± 4007.26 | 16017.28 ± 3794.66 | <0.001∗∗ | 0.214 | 11632.41 ± 4207.56 | 17477.47 ± 3340.15 | <0.001∗∗ | 0.933 | 17165.85 ± 2359.73 | 0.819 | 0.200 |
| MIP-1β | 196.97 ± 180.47 | 145.11 ± 188.44 | 0.237 | 0.511 | 165.84 ± 128.60 | 128.05 ± 100.70 | 0.845 | 0.491 | 146.85 ± 97.90 | 0.214 | 0.677 |
| CCL20 (MIP-3α) | 46.79 ± 198.24 | 21.38 ± 63.88 | 0.318 | 0.517 | 19.66 ± 27.61 | 15.84 ± 18.98 | 0.976 | 0.942 | 56.05 ± 178.53 | 0.212 | 0.401 |
Mean ± SD of cytokines were presented and performed analysis using Mann–Whitney U test or t-test between two cohorts. Statistical significance is indicated by asterisks: ∗∗P < 0.01 or ∗P < 0.05.
Figure 1.
Inflammatory cytokines with statistical differences between primary infections and secondary infections were showed. P values obtained from Mann–Whitney U test were annotated.
4.3. Correlation between Clinical Routine Indexes and Inflammatory Cytokines
We next tested the correlation between the routine clinical indexes and inflammatory cytokines in DENV infection. We found that several cytokines showed statistically significant correlation with clinical routine indexes (Table 4). In the group of all dengue-infected patients or primary infection, there were only weak correlation in specific cytokines with urea, AST, or LDH. However, PLT was found to be strongly negative correlated with the level of CCL17 (R = −0.31, P = 0.012) in the group of secondary infection. In addition, the level of LDH was also significantly associated with the expression of MIP-1α (R = 0.49, P = 0.001) and CXCL5 (R = 0.39, P = 0.015).
Table 4.
Correlation between clinical routine indexes and cytokines.
| R | P | |
|---|---|---|
| Primary infection | ||
| UREA with | ||
| RANTES | −0.131 | 0.049 |
| CCL17 | −0.196 | 0.003 |
| CXCL5 | −0.147 | 0.027 |
| IL-8 | −0.149 | 0.025 |
| IL-10 | 0.132 | 0.048 |
| CCL2 | −0.151 | 0.023 |
| Secondary infection | ||
| PLT with | ||
| CCL17 | −0.314 | 0.012 |
| LDH with | ||
| MIP-1α | 0.493 | 0.001 |
| CXCL5 | 0.388 | 0.015 |
| All infected patients | ||
| UREA with | ||
| RANTES | −0.126 | 0.033 |
| CCL11 | −0.137 | 0.020 |
| CCL17 | −0.167 | 0.005 |
| CXCL5 | −0.143 | 0.015 |
| IL-8 | −0.128 | 0.030 |
| IL-10 | −0.127 | 0.031 |
| CCL2 | −0.146 | 0.013 |
| AST with | ||
| CCL17 | 0.125 | 0.031 |
| LDH with | ||
| MIP-1α | 0.186 | 0.006 |
Correlations between clinical routine indexes and cytokines were analyzed based on the Spearman rank correlation coefficient and P value. Only those with a P value <0.05 are presented in the table.
5. Discussion
Proinflammatory cytokines were secreted to initiate the inflammation and to control the DENV replication especially at the early stage of infection. However, dysregulation of these cytokines was also considered as important reason in dengue pathogenesis especially in DHF and DSS [14, 15]. In this study, we have focused to investigate the cytokine induction profiles from the patients infected with dengue viruses during the Guangdong outbreak in 2014. We also compared their levels between the patients with primary and secondary infections as well as the clinical implications. Interestingly, we found that while several proinflammatory cytokines such as CXCL9, IP-10, CXCL11, IL-8, IL-10, and so on were highly upregulated in the patients after dengue infection, the levels of CCL17 and CXCL5 were significantly lower than the controls. These results provided information for us to understand the interplay between the virus and the host responses during the acute stage of dengue infection. CXCL9, IP-10 (CXCL10), and CXCL11 are the ligands of CXCR3. They are chemoattractants of T lymphocytes and NK cells and make contribution in terms of antiviral and antimicrobial [16–18]. Specifically, CXCR3-ligand interaction attracts Th1 cells and promotes Th1 cell maturation. Recent study demonstrated that there was an increase of T cell activation in asymptomatic viremia compared to those with clinical dengue [19]. In addition, CXCL10 (IP-10) was shown to compete with dengue virus for binding to cell surface heparan sulfate and led to a reduction of dengue infection to cells [20–22]. Our results indicated that the CXCR3-related pathways were crucial at the acute phase DENV infection, and T cell responses may be one of the key immune component to regulate the dengue infection. However, dysregulation of these CXCR ligands was suggested to associate with the disease severity. For example, CXCL9 was identified as a risk factor of increasing vascular permeability in DENV infection [14]. It has been also demonstrated that IP-10 was more elevated in patients who developed dengue hemorrhagic fever or dengue shock syndrome compared to the less severe controls [23]. Although the IP-10 and CXCL9 are considered associated with increase in vascular permeability which leads to severe dengue fever, they are only consider as the risk factors contributed to the severity of the dengue infection. The entire immune environment which contained additional factors in regulation of other cytokines may also play a crucial role in progression of diseases. Indeed, we do not know if the concentrations of the two cytokines were high enough in the patients to lead to the severe outcome as we did not include the patients with severe dengue fever in our study. On the contrary, CCL17 is constitutively expressed in thymus and plays similar role as CXCL9 in inflammation. However, the concentration of CCL17 in primary and secondary infections of this study were lower than that in controls. Wong et al. figured that CCL17 deficiency in mice could be a protective factor to prevent severe colitis by reducing production of other proinflammatory cytokines [21]. Moreover, CCR4, as the only known receptor for CCL17, was also found to markedly decreased the tissue injury in DENV infected mice when it was blocked [24]. These results suggested that reduction of CCL17 may benefit to the host during dengue infection. The functions of CXCL5 have been well studied in cancer and bacterial infection but rarely and ambiguous in dengue. The downregulated CXCL5 may play similar role with CCL17 in DENV infection.
Previous studies have found a significant increase in IFN-γ and TNF-α in the patients with dengue infection [4, 7]. Both of them are important proinflammatory cytokines and have been shown to associate with the disease severity. While the upregulation of the IFN-γ can induce plasma leakage, TNF-α may indicate for the activation of vascular endothelial cell and may increase vascular permeability which lead to hemorrhagic fever [25–27]. In our study, we did not observe a detectable level of TNF-α from the serum of our patients collected from the first two weeks of infection. In addition, highly varied level of IFN-γ in the serum was also found at the first week. It is likely that the mild condition of the patients does not necessary trigger these immune responses given the fact that only very small proportion of patients developed severe forms of dengue fever in this outbreak. In addition, it has been noted that the levels of IFN-γ and TNF-α varied among different studies. Comparable levels of these two cytokines between patients and healthy controls have also been reported in some studies [28, 29]. It has been known that CXCL9, IP-10 (CXCL10), and CXCL11 are all IFN-γ-induced chemokines. The observation of lacking IFN-γ induction could not explain why there were still high level of CXCR3 ligands expressed in the serum. These CXCR3 ligands may be regulated by other stimuli such as IFN-β which was not included in this study.
One of the limitation in our study was that we did not know the serotype of the dengue virus in the patients. Also, we could not identify the previous infection history of those patients with secondary infection. Thus, the impact of the serotype on the cytokine profiles was not clear. However, epidemiological data suggested that serotype 1 was the major subtype circulated in Guangdong province over the last 20 years and large proportion of patients were also infected by dengue 1 in the 2014 outbreak [10]. It is thus logical to postulate that the results we obtained should mainly reflect the impact of dengue 1 infection. Furthermore, we could not compare the cytokine profile between mild and severe dengue patients as there were no severe patients admitted to our hospital. Despite of these limitations and there are some similar studies that reveal the cytokine profile in dengue-infected patients, our study has done a more comprehensive detection of 23 cytokines in up to 297 infected patients. Our results not only extended our understanding on the immunological features of the patients in the Guangzhou dengue outbreak in 2014 but also determined the association of clinical routine indexes and these inflammatory cytokines which provide new insight to the pathogenesis of dengue fever in human.
Acknowledgments
The authors are grateful to their laboratory colleagues for the specimens they provided. Research funding was supported by Science Research Project of the Guangdong Province (Grant no. 2016A050503047).
Conflicts of Interest
The authors have no conflict of interest to declare.
Authors' Contributions
Yingchun Zhou and Chris Ka Pun Mok conceived the study design. Junyuan Huang and Weiwen Liang designed the methods and experiments, carried out the laboratory experiments, analyzed the data, interpreted the results, and wrote the paper. Chris Ka Pun Mok made the statistical analysis of the databases. Shaoyan Chen analyzed and helped with the interpretation of the data of the work. Ye Zhu and Haiming Chen collected the specimen and experimental date. Yingchun Zhou revised the manuscript critically for important intellectual content. All authors read and approved the final manuscript.
References
- 1.Bhatt S., Gething P. W., Brady O. J., et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo C., Zhou Z., Wen Z., et al. Global epidemiology of dengue outbreaks in 1990–2015: a systematic review and meta-analysis. Frontiers in Cellular and Infection Microbiology. 2017;7:7–317. doi: 10.3389/fcimb.2017.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guzman M. G., Alvarez M., Halstead S. B. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: an historical perspective and role of antibody-dependent enhancement of infection. Archives of Virology. 2013;158(7):1445–1459. doi: 10.1007/s00705-013-1645-3. [DOI] [PubMed] [Google Scholar]
- 4.Zhao L., Huang X., Hong W., et al. Slow resolution of inflammation in severe adult dengue patients. BMC Infectious Diseases. 2016;16(1, article 291) doi: 10.1186/s12879-016-1596-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang Y., Kou Z., Zhang F., et al. Both viremia and cytokine levels associate with the lack of severe disease in secondary dengue 1 infection among adult Chinese patients. PLoS One. 2010;5(12, article e15631) doi: 10.1371/journal.pone.0015631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliveira R. A. D. S., Cordeiro M. T., Moura P. M. M. F., et al. Serum cytokine/chemokine profiles in patients with dengue fever (DF) and dengue hemorrhagic fever (FHD) by using protein array. Journal of Clinical Virology. 2017;89:39–45. doi: 10.1016/j.jcv.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Pandey N., Jain A., Garg R. K., Kumar R., Agrawal O. P., Lakshmana Rao P. V. Serum levels of IL-8, IFNγ, IL-10, and TGF β and their gene expression levels in severe and non-severe cases of dengue virus infection. Archives of Virology. 2015;160(6):1463–1475. doi: 10.1007/s00705-015-2410-6. [DOI] [PubMed] [Google Scholar]
- 8.Guo R., Lin J., Li L., et al. The prevalence and endemic nature of dengue infections in Guangdong, South China: an epidemiological, serological, and etiological study from 2005–2011. PLoS One. 2014;9(1, article e85596) doi: 10.1371/journal.pone.0085596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.James A. A., Lun Z.-R., Wu J.-Y., Chen X.-G. Dengue fever in mainland China. The American Journal of Tropical Medicine and Hygiene. 2010;83(3):664–671. doi: 10.4269/ajtmh.2010.09-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai S., Huang Z., Zhou H., et al. The changing epidemiology of dengue in China, 1990–2014: a descriptive analysis of 25 years of nationwide surveillance data. BMC Medicine. 2015;13(1, article 100) doi: 10.1186/s12916-015-0336-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang L., Luo X., Shao J., et al. Epidemiology and characteristics of the dengue outbreak in Guangdong, Southern China, in 2014. European Journal of Clinical Microbiology & Infectious Diseases. 2016;35(2):269–277. doi: 10.1007/s10096-015-2540-5. [DOI] [PubMed] [Google Scholar]
- 12.Lin Y. P., Luo Y., Chen Y., et al. Clinical and epidemiological features of the 2014 large-scale dengue outbreak in Guangzhou city, China. BMC Infectious Diseases. 2016;16(1):p. 102. doi: 10.1186/s12879-016-1379-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Health Organization and the Special Programme for Research and Training in Tropical Diseases (TDR) Dengue guidelines for diagnosis, treatment, prevention and control: new edition. http://www.who.int/rpc/guidelines/9789241547871/en/ [PubMed]
- 14.Dalrymple N. A., Mackow E. R. Endothelial cells elicit immune-enhancing responses to dengue virus infection. Journal of Virology. 2012;86(12):6408–6415. doi: 10.1128/JVI.00213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothman A. L. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nature Reviews Immunology. 2011;11(8):532–543. doi: 10.1038/nri3014. [DOI] [PubMed] [Google Scholar]
- 16.Egesten A., Eliasson M., Johansson H. M., et al. The CXC chemokine MIG/CXCL9 is important in innate immunity against Streptococcus pyogenes. The Journal of Infectious Diseases. 2007;195(5):684–693. doi: 10.1086/510857. [DOI] [PubMed] [Google Scholar]
- 17.Reid-Yu S. A., Tuinema B. R., Small C. N., Xing L., Coombes B. K. CXCL9 contributes to antimicrobial protection of the gut during Citrobacter rodentium infection independent of chemokine-receptor signaling. PLoS Pathogens. 2015;11(2, article e1004648) doi: 10.1371/journal.ppat.1004648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang W., Hu K., Luo S., et al. Herpes simplex virus type 2 infection of human epithelial cells induces CXCL9 expression and CD4+ T cell migration via activation of p38-CCAAT/enhancer-binding protein-β pathway. The Journal of Immunology. 2012;188(12):6247–6257. doi: 10.4049/jimmunol.1103706. [DOI] [PubMed] [Google Scholar]
- 19.Simon-Lorière E., Duong V., Tawfik A., et al. Increased adaptive immune responses and proper feedback regulation protect against clinical dengue. Science Translational Medicine. 2017;9(405, article eaal5088) doi: 10.1126/scitranslmed.aal5088. [DOI] [PubMed] [Google Scholar]
- 20.Chen J.-P., Lu H.-L., Lai S.-L., et al. Dengue virus induces expression of CXC chemokine ligand 10/IFN-γ-inducible protein 10, which competitively inhibits viral binding to cell surface heparan sulfate. The Journal of Immunology. 2006;177(5):3185–3192. doi: 10.4049/jimmunol.177.5.3185. [DOI] [PubMed] [Google Scholar]
- 21.Wong K. L., Chen W., Balakrishnan T., Toh Y. X., Fink K., Wong S.-C. Susceptibility and response of human blood monocyte subsets to primary dengue virus infection. PLoS One. 2012;7(5, article e36435) doi: 10.1371/journal.pone.0036435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ip P. P., Liao F. Resistance to dengue virus infection in mice is potentiated by CXCL10 and is independent of CXCL10-mediated leukocyte recruitment. The Journal of Immunology. 2010;184(10):5705–5714. doi: 10.4049/jimmunol.0903484. [DOI] [PubMed] [Google Scholar]
- 23.Fallahi P., Elia G. Interferon-γ-induced protein 10 in dengue virus infection. La Clinica Terapeutica. 2016;167(6):e186–e191. doi: 10.7417/CT.2016.1966. [DOI] [PubMed] [Google Scholar]
- 24.Guabiraba R., Marques R. E., Besnard A.-G., et al. Role of the chemokine receptors CCR1, CCR2 and CCR4 in the pathogenesis of experimental dengue infection in mice. PLoS One. 2010;5(12, article e15680) doi: 10.1371/journal.pone.0015680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurane I. Dengue hemorrhagic fever with special emphasis on immunopathogenesis. Comparative Immunology, Microbiology and Infectious Diseases. 2007;30(5-6):329–340. doi: 10.1016/j.cimid.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 26.Cardier J. E., Mariño E., Romano E., et al. Proinflammatory factors present in sera from patients with acute dengue infection induce activation and apoptosis of human microvascular endothelial cells: possible role of TNF-α in endothelial cell damage in dengue. Cytokine. 2005;30(6):359–365. doi: 10.1016/j.cyto.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 27.Mangada M. M., Rothman A. L. Altered cytokine responses of dengue-specific CD4+ T cells to heterologous serotypes. The Journal of Immunology. 2005;175(4):2676–2683. doi: 10.4049/jimmunol.175.4.2676. [DOI] [PubMed] [Google Scholar]
- 28.Rathakrishnan A., Wang S. M., Hu Y., et al. Cytokine expression profile of dengue patients at different phases of illness. PLoS One. 2012;7(12, article e52215) doi: 10.1371/journal.pone.0052215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de la Cruz Hernández S. I., Puerta-Guardo H. N., Flores Aguilar H., et al. Primary dengue virus infections induce differential cytokine production in Mexican patients. Memórias do Instituto Oswaldo Cruz. 2016;111(3):161–167. doi: 10.1590/0074-02760150359. [DOI] [PMC free article] [PubMed] [Google Scholar]