Abstract
Purpose of Review
Clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated 9 (Cas9) has recently emerged as a top genome editing technology and has afforded investigators the ability to more easily study a number of diseases. This review discusses CRISPR/Cas9’s advantages and limitations and highlights a few recent reports on genome editing applications for alleviating dyslipidemia through disruption of proprotein convertase subtilisin/kexin type 9 (PCSK9).
Recent Findings
Targeting of mouse Pcsk9 using CRISPR/Cas9 technology has yielded promising results for lowering total cholesterol levels, and several recent findings are highlighted in this review. Reported on-target mutagenesis efficiency is as high as 90% with a subsequent 40% reduction of blood cholesterol levels in mice, highlighting the potential for use as a therapeutic in human patients.
Summary
The ability to characterize and treat diseases is becoming easier with the recent advances in genome editing technologies. In this review, we discuss how genome editing strategies can be of use for potential therapeutic applications.
Keywords: Genome editing, Dyslipidemia, CRISPR/Cas9, PCSK9, Cardiovascular disease
Introduction
Hundreds of billions of dollars are spent annually on direct and indirect costs of atherosclerotic cardiovascular disease, the leading cause of death worldwide [1]. Although this disease is an extensive and complex process involving numerous genes, proteins, and cell types, a large contributor to its progression is having pro-atherogenic lipid profiles, classified as dyslipidemia. While lifestyle changes such as diet and exercise generally help, they are often insufficient to attain healthy lipid profiles, necessitating the use of therapeutic treatments such as low-density lipoprotein cholesterol (LDL-C)-lowering drugs. Statin use has been exceedingly successful in lowering LDL-C levels, but like any medication, not all patients tolerate and respond to statins well enough and need either a different or a combined therapeutic approach to manage lipid levels.
Genome-sequencing data have now afforded the ability to understand how genetic variation influences dyslipidemia, and functional follow-up in the laboratory using cellular and animal models will be useful for developing methods to treat the condition. In this review, we describe the use of recent advances in genome editing technologies for understanding and characterizing factors associated with various diseases, with a particular emphasis on dyslipidemia. We also describe the potential use of genome editing in the treatment of this condition through the targeting of the proprotein convertase subtilisin/kexin type 9 (PCSK9) gene.
Genome Editing Using Clustered Regularly Interspaced Short Palindromic Repeat/CRISPR-Associated 9
The field of genome editing has drastically expanded in the last few years, and the application of genome editing techniques in the study of cardiovascular disorders has been expansively reviewed [2–5]. Much of the recent focus has been on harnessing clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-Associated 9 (Cas9), a system originally identified as an endogenous mechanism for bacterial adaptive immunity, for targeted genome editing [6, 7]. Potential uses for this technology include the development of cell and animal models to study specific diseases, functional analyses of how specific genetic variants contribute to diseases, and permanent disruption of a gene or correction of a harmful genetic mutation for therapeutic benefit.
In the CRISPR/Cas9 system, the Cas9 nuclease can target a specific genomic site when complexed with a synthetic, approximately 100-nucleotide “guide” RNA. The first 20 nucleotides of the synthetic guide RNA will recognize and hybridize to a complementary sequence of genomic DNA, termed the protospacer, which is situated directly next to a protospacer-adjacent motif (PAM) [6–8]. The Cas9 nuclease then introduces a double-stranded break in the genomic DNA, and the endogenous DNA repair machinery repairs the break through either non-homologous end-joining (NHEJ) or homology-directed repair (HDR) [9]. Double-stranded breaks are most often repaired by NHEJ, whereby the ends are directly rejoined together; however, this error-prone process can insert and/or delete DNA base pairs (indels) at the site of the break. If indels occur in the protein-coding sequence of a gene, they can cause frameshifts that truncate the protein, likely rendering it nonfunctional. Investigators can take advantage of this endogenous repair pathway when specifically seeking to knock out a targeted gene in cell or animal models. If introduction of a mutation into a target genomic sequence is desired, an exogenous DNA repair template such as a single-strand DNA oligonucleotide or double-strand DNA vector can be used to incorporate specific changes in the genomic DNA by way of the HDR repair pathway. However, this is a rarer event because HDR generally occurs much less frequently than NHEJ as its activity is limited to proliferating cells in the S and G2 phases of the cell cycle.
Although many different CRISPR/Cas9 systems have been adapted since the advent of this technology’s use for genome editing [6–8, 10–13], the originally identified system from Streptococcus pyogenes is still the most commonly used CRISPR-Cas9 system today. Different genomic sites can be targeted by simply changing the protospacer in the guide RNA, making CRISPR/Cas9 exceptionally easy to design and use compared to other genome editing technologies such as transcription activator-like effector nucleases (TALENs) or zinc finger nucleases (ZFNs). Further, multiple guide RNAs targeting numerous sites can be introduced into a single vector for potentially improved editing efficiency. A large number of potential target sites can be screened for editing efficiency rather quickly and with low cost, usually resulting in identification of at least one suitable guide RNA for the desired application.
While there are many benefits to using CRISPR/Cas9, it still has several serious limitations to overcome before it can be routinely used for therapeutic purposes in human patients. First and foremost, the potential for “off-target” mutagenesis elsewhere in the genome from the intended target is a substantial concern; disruption of a different gene, initiation of oncogenesis, and altered cell survival/proliferation could all potentially cause considerable harm when using genome editing as a therapy. A large amount of work has aimed at improving on-target efficiency while reducing off-target effects, including alteration of the guide RNA [14, 15], use of a nickase or catalytically dead version of Cas9 [16–18], “high-fidelity” mutations in Cas9 [19, 20], and fusion of base-editing domains to Cas9 for specific base pair changes [21–24].
A second drawback to the CRISPR/Cas9 system is the necessity for a PAM to be situated particularly close to the intended target site. For example, the S. pyogenes Cas9 PAM sequence, NGG, occurs on average every eight base pairs but in some regions of the genome much less frequently than that, which can be problematic if a specific on-target site has no nearby PAM. Most other Cas9 proteins require more complex PAM sequences (examples include Staphylococcus aureus with a PAM of NNGRR [12••, 25], Streptococcus thermophilus with a PAM of NNAGAAW [6], and Neisseria meningitidis with a PAM of NNNNGATT [10, 11]), which present even more restrictions. Fortunately, this limitation has been partially resolved with the classification and use of a diverse variety of Cas9 proteins from different bacterial species and, further, with structure-guided mutagenesis that can alter the PAM sequence specificity to expand the number of targetable sites [25, 26].
A third drawback is that even if a desired target site has conveniently located PAMs, it does not necessarily mean that on-target mutagenesis efficiency will be high. Extensive locus-to-locus variability has been observed. Further, there appears to be substantial variability in the degree to which genome editing can be achieved in different tissues. Although this may not be a serious limitation for the treatment of dyslipidemia, due to the relative efficiency of genome editing in both mouse and human hepatocytes (see below), the reported low efficiencies of genome editing in other tissues such as cardiac muscle may limit its therapeutic potential for diseases arising in those tissues.
Genome Editing for Disease Modeling
Recent advances in genome editing are allowing investigators to more easily study the effects of genetic variants on disease processes, which can inform the development of therapeutics in the future. Human induced pluripotent stem cell (iPSC) lines are one system for determining the functional consequences of a mutation and how it may contribute to certain diseases. Cells collected from patients can be reprogrammed into iPSCs, which can in turn be differentiated into specific cell types for downstream functional study. Although it may be beneficial to use a cell line derived directly from the patient to characterize how a specific genetic mutation contributes to a disease phenotype, difficulties arise when comparing lines between different patients that vary in genetic and epigenetic backgrounds. Therefore, use of genome editing to create matched, isogenic cell lines that differ only with respect to specific mutations allows a more rigorous characterization of the functional consequences of the mutations. Additionally, genome editing to introduce specific mutations in iPSCs affords the opportunity to screen for therapies to treat conditions caused by the mutations. A number of examples of the use of iPSCs for the study and treatment of lipid metabolism disorders have recently been reported [27–32]. In one notable example, iPSC-derived hepatocytes from a patient with familial hypercholesterolemia were used to identify cardiac glycosides as a potential treatment for dyslipidemia [31].
The production of in vivo model systems is also being greatly facilitated with the use of genome editing tools. Mouse models can be rapidly created with the use of CRISPR/Cas9 injected directly into single-cell mouse embryos [33, 34] to knock out genes or knock in specific mutations. The same technique can be used to genetically modify animals that were previously not amenable to modification. The modeling of cardiovascular disorders in mice [12••, 35••, 36••], rats [37–42], rabbits [43–45], zebrafish [46], and pigs [47, 48] using genome editing technologies has recently been reported.
Disruption of PCSK9 Using CRISPR/Cas9 In Vivo
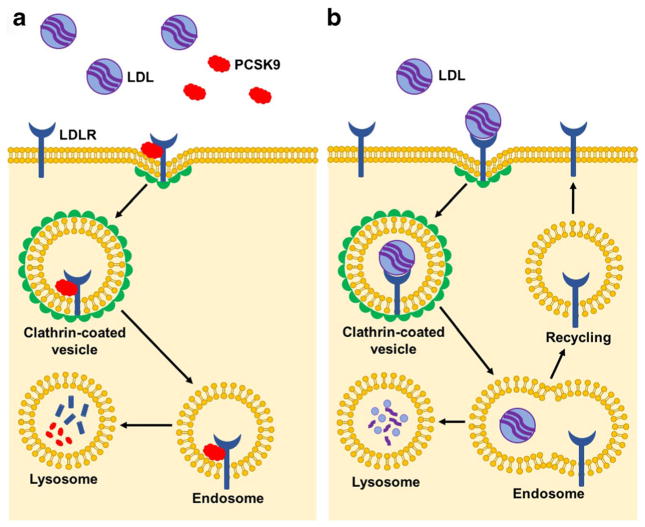
PCSK9, a protein that increases plasma LDL-C levels via inhibition of LDL clearance, has recently emerged as a promising protein and genetic target. Naturally occurring PCSK9 loss-of-function mutations not only reduce blood LDL-C levels but also reduce coronary heart disease risk by as much as 88% [49]. Mechanistic studies have shown that circulating PCSK9 binds to the LDL receptor (LDLR) and, upon endocytosis of the PCSK9/LDLR complex, promotes lysosomal degradation of LDLR instead of LDLR trafficking back to the plasma membrane (Fig. 1). When PCSK9 binding to LDLR is disrupted by either small molecule targeting of the PCSK9 protein or genetic silencing of the PCSK9 gene, endocytosed LDLR is not degraded and is instead recycled back to the plasma membrane where it is available for LDL binding and clearance (Fig. 1). Multiple pharmaceutical companies have developed inhibitors of PCSK9 through antibody targeting or siRNA or antisense oligonucleotide silencing in order to reduce blood LDL-C levels and, ultimately, atherosclerotic vascular disease. These therapeutic approaches have started to yield clinical success [50], but as with statin drugs, they have the substantial limitation of needing to be taken at repeated intervals over the course of a patient’s lifetime to obtain the desired therapeutic benefit. In light of these observations, genome editing of PCSK9 offers a highly attractive alternative approach as it could significantly reduce coronary heart disease risk while also offering a lifelong therapeutic effect with only a single treatment.
Fig. 1.
PCSK9 inhibition of LDLR recycling. a Circulating proprotein convertase subtilisin/kexin 9 (PCSK9) binds to the low-density lipoprotein receptor (LDLR) and, upon internalization of the complex by endocytosis, promotes the degradation of LDLR in the lysosomal compartment. Consequently, lower levels of LDLR are available at the plasma membrane for LDL binding and clearance. b Upon inhibition of PCSK9, whether through antibody targeting of circulating PCSK9 or disruption of the PCSK9 gene, endocytosed LDLR is not readily degraded and recycles back to the plasma membrane. As more receptor is available to bind to LDL, there is increased LDL clearance and lower circulating LDL levels
Recent work has described the ability to permanently disrupt PCSK9 with high efficiency using the CRISPR/Cas9 system in vivo. In the first published study, Ding et al. [35••] injected mice with an adenovirus containing the S. pyogenes Cas9 and a guide RNA targeting exon 1 of Pcsk9. Substantial NHEJ-mediated indel mutagenesis in mouse livers was observed 3–4 days post-injection and, importantly, resulted in a highly significant ~90% reduction in blood PCSK9 protein levels with a concomitant 35–40% decrease in total blood cholesterol levels. Ten off-target sites that most closely matched the on-target site with respect to sequence similarity and were deemed the most likely candidates to harbor off-target mutations were assessed, and no evidence of mutagenesis was detected. In another set of experiments, Wang et al. [36••] tested the ability to permanently disrupt human PCSK9 using CRISPR/Cas9 in a humanized-liver mouse model in vivo; the authors reasoned that this approach would allow preclinical assessment of in vivo genome editing for the use of CRISPR/Cas9 therapy in human patients. Similar to the first study, the authors injected an adenovirus containing the S. pyogenes Cas9 and a guide RNA targeting exon 1 in human PCSK9 in Fah−/−Rag2−/−Il2rg−/− (FRG KO) mice carrying transplanted, engrafted human hepatocytes. Substantial NHEJ-mediated indel mutagenesis of the human PCSK9 gene was observed 3–4 days post injection, resulting in >50% reduction of the blood levels of secreted human PCSK9 protein. No off-target mutagenesis was detected at any of the eight top off-target sites.
Although these two studies used adenovirus as a delivery method, which is not optimal for use in human patients [51, 52], the studies represent proof-of-principle examples of the highly efficient use of CRISPR/Cas9 technology to alter PCSK9 levels in vivo. Adenovirus was used because S. pyogenes Cas9 is too large to be accommodated in a single adeno-associated viral (AAV) vector, which is more suitable for clinical use. Recent work by Ran et al. [12••] demonstrated the NHEJ-mediated disruption of Pcsk9 in mouse liver using S. aureus Cas9, which is small enough to be delivered by AAV. In accordance with the original results of Ding et al., Ran et al. observed >90% decrease of blood PCSK9 protein levels, with a resultant 40% decrease in blood cholesterol levels and no evidence of off-target mutagenesis.
These three studies highlight the ability to alter the genome in order to permanently decrease cholesterol levels and thus potentially treat dyslipidemia in human patients. Other genome editing strategies, such as targeted disruption of lipoprotein production or gene replacement therapy for enhanced cholesterol clearance, may also be feasible for treating dyslipidemia. As genome editing technology advances, along with our knowledge of genetic contributors to cardiovascular disease, we can expect more studies showcasing ways to definitively treat disease.
Conclusion
Although some treatment options are available for individuals suffering from atherosclerotic cardiovascular disease, development of a safe, one-time genome editing treatment would provide a potent new therapeutic option. However, the current limitations of genome editing technology need to be overcome before it can routinely be used in patients. Dyslipidemia may be a particularly suitable condition to address with genome editing, as preclinical studies have already established that efficient targeting of genes associated with cholesterol levels is feasible. Undoubtedly, genome editing will be a useful tool in the coming years for both understanding and preventing cardiovascular disease.
Footnotes
Compliance with Ethical Standards
Conflict of Interest Alexandra Chadwick and Kiran Musunuru declare they have no conflicts of interest.
Human and Animal Rights and Informed Consent All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted.
References
Papers of particular interest, published recently, have been highlighted as:
•• Of major importance
- 1.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, et al. Heart disease and stroke statistics—2013 update: a report from the American Heart Association. Circulation. 2013;127(1):e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chadwick AC, Musunuru K. Genome editing for the study of cardiovascular diseases. Curr Cardiol Rep. 2017;19(3):22. doi: 10.1007/s11886-017-0830-5. doi:10. 1007/s11886-017-0830-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strong A, Musunuru K. Genome editing in cardiovascular diseases. Nat Rev Cardiol. 2017;14(1):11–20. doi: 10.1038/nrcardio.2016.139. [DOI] [PubMed] [Google Scholar]
- 4.Seeger T, Porteus M, Wu JC. Genome editing in cardiovascular biology. Circ Res. 2017;120(5):778–80. doi: 10.1161/CIRCRESAHA.116.310197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Termglinchan V, Karakikes I, Seeger T, Wu JC. Current status of genome editing in cardiovascular medicine. In: Turksen K, editor. Genome editing. Cham: Springer International Publishing; 2016. pp. 107–26. [Google Scholar]
- 6.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–2. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 9.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4(6):435–45. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 10.Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A. 2013;110(39):15644–9. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10(11):1116–21. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–91. doi: 10.1038/nature14299. This study demonstrated the high efficacy of CRISPR/Cas9 in the mouse liver using Staphylococcus aureus Cas9 delivered by AAV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirano H, Gootenberg JS, Horii T, Abudayyeh OO, Kimura M, Hsu PD, et al. Structure and engineering of Francisella novicida Cas9. Cell. 2016;164(5):950–61. doi: 10.1016/j.cell.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32(3):279–84. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–32. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–9. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31(9):833–8. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32(6):577–82. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351(6268):84–8. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–5. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–4. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma Y, Zhang J, Yin W, Zhang Z, Song Y, Chang X. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods. 2016;13(12):1029–35. doi: 10.1038/nmeth.4027. [DOI] [PubMed] [Google Scholar]
- 23.Hess GT, Fresard L, Han K, Lee CH, Li A, Cimprich KA, et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods. 2016;13(12):1036–42. doi: 10.1038/nmeth.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353(6305) doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 25.Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523(7561):481–5. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Ge X, Yang F, Zhang L, Zheng J, Tan X, et al. Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci Rep. 2014;4:5405. doi: 10.1038/srep05405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12(2):238–51. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta RM, Meissner TB, Cowan CA, Musunuru K. Genome-edited human pluripotent stem cell-derived macrophages as a model of reverse cholesterol transport—brief report. Arterioscler Thromb Vasc Biol. 2016;36(1):15–8. doi: 10.1161/ATVBAHA.115.305956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pashos EE, Park Y, Wang X, Raghavan A, Yang W, Abbey D, et al. Large, diverse population cohorts of hiPSCs and derived hepatocyte-like cells reveal functional genetic variation at blood lipid-associated loci. Cell Stem Cell. 2017;20(4):558–570e10. doi: 10.1016/j.stem.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maetzel D, Sarkar S, Wang H, Abi-Mosleh L, Xu P, Cheng AW, et al. Genetic and chemical correction of cholesterol accumulation and impaired autophagy in hepatic and neural cells derived from Niemann-Pick Type C patient-specific iPS cells. Stem Cell Reports. 2014;2(6):866–80. doi: 10.1016/j.stemcr.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cayo MA, Mallanna SK, Di Furio F, Jing R, Tolliver LB, Bures M, et al. A drug screen using human iPSC-derived hepatocyte-like cells reveals cardiac glycosides as a potential treatment for hypercholesterolemia. Cell Stem Cell. 2017;20(4):478–489. e5. doi: 10.1016/j.stem.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warren CR, O’Sullivan JF, Friesen M, Becker CE, Zhang X, Liu P, et al. Induced pluripotent stem cell differentiation enables functional validation of GWAS variants in metabolic disease. Cell Stem Cell. 2017;20(4):547–557. e7. doi: 10.1016/j.stem.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–8. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. Onestep generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154(6):1370–9. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35••.Ding Q, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, et al. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115(5):488–92. doi: 10.1161/CIRCRESAHA.115.304351. This study established the high efficacy of CRISPR/Cas9 targeting of Pcsk9 in the mouse liver. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Raghavan A, Chen T, Qiao L, Zhang Y, Ding Q, et al. CRISPR-Cas9 targeting of PCSK9 in human hepatocytes in vivo—brief report. Arterioscler Thromb Vasc Biol. 2016;36(5):783–6. doi: 10.1161/ATVBAHA.116.307227. doi:10.1161/ATVBAHA.116.307227. •• This study demonstrated high efficacy of CRISPR/Cas9 in a liver-humanized mouse model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacob HJ, Lazar J, Dwinell MR, Moreno C, Geurts AM. Gene targeting in the rat: advances and opportunities. Trends Genet. 2010;26(12):510–8. doi: 10.1016/j.tig.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreno C, Hoffman M, Stodola TJ, Didier DN, Lazar J, Geurts AM, et al. Creation and characterization of a renin knockout rat. Hypertension. 2011;57(3):614–9. doi: 10.1161/HYPERTENSIONAHA.110.163840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Endres BT, Priestley JR, Palygin O, Flister MJ, Hoffman MJ, Weinberg BD, et al. Mutation of Plekha7 attenuates salt-sensitive hypertension in the rat. Proc Natl Acad Sci U S A. 2014;111(35):12817–22. doi: 10.1073/pnas.1410745111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larcher T, Lafoux A, Tesson L, Remy S, Thepenier V, Francois V, et al. Characterization of dystrophin deficient rats: a new model for Duchenne muscular dystrophy. PLoS One. 2014;9(10):e110371. doi: 10.1371/journal.pone.0110371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Xie D, Huang J, Lv F, Shi D, Liu Y, et al. Cold-inducible RNA-binding protein regulates cardiac repolarization by targeting transient outward potassium channels. Circ Res. 2015;116(10):1655–9. doi: 10.1161/CIRCRESAHA.116.306287. [DOI] [PubMed] [Google Scholar]
- 42.Zhu X, Fang J, Jiang DS, Zhang P, Zhao GN, Zhu X, et al. Exacerbating pressure overload-induced cardiac hypertrophy: novel role of adaptor molecule Src homology 2-B3. Hypertension. 2015;66(3):571–81. doi: 10.1161/HYPERTENSIONAHA.115.05183. [DOI] [PubMed] [Google Scholar]
- 43.Yang D, Xu J, Zhu T, Fan J, Lai L, Zhang J, et al. Effective gene targeting in rabbits using RNA-guided Cas9 nucleases. J Mol Cell Biol. 2014;6(1):97–9. doi: 10.1093/jmcb/mjt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ji D, Zhao G, Songstad A, Cui X, Weinstein EJ. Efficient creation of an APOE knockout rabbit. Transgenic Res. 2015;24(2):227–35. doi: 10.1007/s11248-014-9834-8. [DOI] [PubMed] [Google Scholar]
- 45.Niimi M, Yang D, Kitajima S, Ning B, Wang C, Li S, et al. ApoE knockout rabbits: a novel model for the study of human hyperlipidemia. Atherosclerosis. 2016;245:187–93. doi: 10.1016/j.atherosclerosis.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Zou J, Tran D, Baalbaki M, Tang LF, Poon A, Pelonero A, et al. An internal promoter underlies the difference in disease severity between N- and C-terminal truncation mutations of Titin in zebrafish. elife. 2015;4:e09406. doi: 10.7554/eLife.09406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Du Y, Shen B, Zhou X, Li J, Liu Y, et al. Efficient generation of gene-modified pigs via injection of zygote with Cas9/sgRNA. Sci Rep. 2015;5:8256. doi: 10.1038/srep08256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Umeyama K, Watanabe K, Watanabe M, Horiuchi K, Nakano K, Kitashiro M, et al. Generation of heterozygous fibrillin-1 mutant cloned pigs from genome-edited foetal fibroblasts. Sci Rep. 2016;6:24413. doi: 10.1038/srep24413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354(12):1264–72. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 50.Sabatine MS, Giugliano RP, Keech AC, Honarpour N, Wiviott SD, Murphy SA, et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med. 2017 doi: 10.1056/NEJMoa1615664. [DOI] [PubMed] [Google Scholar]
- 51.Muruve DA. The innate immune response to adenovirus vectors. Hum Gene Ther. 2004;15(12):1157–66. doi: 10.1089/hum.2004.15.1157. [DOI] [PubMed] [Google Scholar]
- 52.Gregory SM, Nazir SA, Metcalf JP. Implications of the innate immune response to adenovirus and adenoviral vectors. Future Virol. 2011;6(3):357–74. doi: 10.2217/fvl.11.6. [DOI] [PMC free article] [PubMed] [Google Scholar]