Abstract
Photosystem II (PSII) of oxygenic photosynthetic organisms normally contains exclusively chlorophyll a (Chl a) as its major light-harvesting pigment. Chl a canonically consists of the chlorin headgroup with a 20 carbon, 4 isoprene unit, phytyl tail. We have examined the 1.9 Å crystal structure of PSII from thermophilic cyanobacteria reported by Shen and coworkers in 2012 (PDB accession of 3ARC/3WU2). A newly refined electron density map from this structure, presented here, reveals that some assignments of the cofactors may be different from those modeled in the 3ARC/3WU2 structure, including a specific Chl a that appears to have a truncated tail by one isoprene unit. We provide experimental evidence using high performance liquid chromatography and mass spectrometry for a small population of Chl a esterified to a 15 carbon farnesyl tail in PSII of thermophilic cyanobacteria.
Keywords: chlorophyll a, Photosystem II, Thermosynechococcus elongatus, X-ray crystal structure
INTRODUCTION
Solar energy conversion in oxygenic photosynthesis involves light harvesting by the chlorophyll-containing complexes, Photosystem I (PSI) and Photosystem II (PSII). PSII is the primary electron donor of the photosynthetic electron-transport chain, and uses light to catalyze the oxidation of water into oxygen, protons, and free electrons that are released in the form of reduced plastoquinone molecules. PSII is a membrane-bound dimer, and each 350 kDa monomer is comprised of over 20 unique peptides and over 80 cofactors (Umena et al. 2012). The light-harvesting and water-oxidation functions of PSII are supported by chlorophyll a (Chl a) molecules and redox cofactors primarily bound to the “core” polypeptides D1, D2, CP43 and CP47. The Chl a molecules harvest light based on the UV-visible absorption spectrum shown in Figure 1, and transfer their absorbed energy to the reaction center Chl a, referred to as P680. The excited state of P680 in PSII promotes charge separation that drives plastoquinone reduction and water oxidation (McEvoy and Brudvig 2006).
Figure 1.
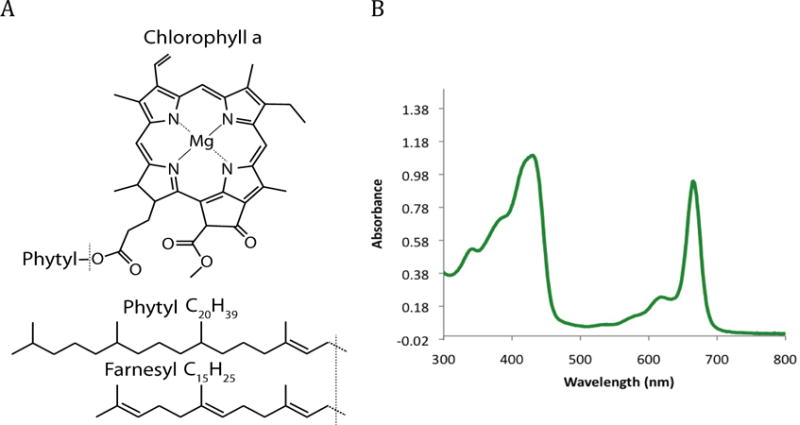
Structure of chlorophyll a and UV-visible absorption spectrum of chlorophyll a. (A) Chlorophyll a consists of a chlorophyllide a headgroup attached to a 4-isprene unit, 20 carbon, phytyl tail. This study investigates the presence of a non-canonical 3-isoprene unit, 15 carbon, farnesyl tail. (B) The UV-visible absorption spectrum of chlorophyll a.
Recently, atomic-resolution structures of PSII have been determined from thermophilic cyanobacteria Thermosynechococcus vulcanus (T. vulcanus) and Thermosynechococcus elongatus (T. elongatus). In particular, the 1.9 Å resolution structure reported by Shen and coworkers (Umena et al. 2012) has identified the coordinated structure of the PSII Mn4CaO5 cluster in the active site of the oxygen-evolving complex (OEC). Additionally, these high-resolution structure determinations give confirmation to other somewhat unexpected components or structural features in the protein that may have earlier been dismissed as artifacts of low resolution. This includes the electron density of the lipids, carotenoids, and chlorophylls that have been reported in several previous structures of PSII (Zouni et al. 2001; Kamiya and Shen 2003; Loll et al. 2005a; Guskov et al. 2009).
PSII is found in oxygenic photosynthetic organisms, plants, cyanobacteria, and green algae, and normally contains exclusively Chl a. Chl a is a magnesium-chelated chlorin tetrapyrrole that typically has a phytyl tail (Figure 1). The preference for Chl a in PSII in oxygenic photosynthetic species is in contrast to the known diversity of bacteriochlorophylls found in anoxygenic photosynthetic bacteria, which are known to have a variety of tail lengths and branched structures as evident from high performance liquid chromatography (HPLC) analysis of microbial mats (Airs and Keely 2003). In PSII, the headgroup of Chl a has been specifically evolved because of its energetic characteristics and the phytyl tail has been selected to anchor it in the highly conserved hydrophobic protein core (Björn et al. 2009).
The Chl a biosynthetic pathway has been described in detail in recent reviews (Chew and Bryant 2007; Masuda and Fujita 2008). The phytyl tail is a branched 20-carbon chain alcohol that has been esterified to the chlorin ring at C17 (Figure 1). It is the product of the four isoprene-unit geranylgeranyl that has undergone three reduction events catalyzed by geranylgeraniol reductase (ChlP) to saturate three of the four double bonds (Shpilyov et al. 2005). Although fully assembled Chl a is made exclusively with the phytyl tail, small populations of Chl a with partially or fully unsaturated tails, named geranylgeranyl (GG), dihydrogeranylgeranyl (DHGG), and tetrahydrogeranylgeranyl (THGG), have been identified in PSII during the greening processes in plants (Nakamura and Watanabe 1998), indicating the possibility of minor variations in tail properties but not in length.
The final stages of biosynthesis of Chl a involve chlorophyll synthase (ChlG), which catalyzes the attachment of the isoprene tail, in its diphosphate form, to the C17 –COOH of a fully assembled chlorophyllide ring. In vitro studies of ChlG have demonstrated that, although this reaction prefers to use phytyldiphosphate or geranylgeranyldiphosphate as a substrate (both 20C and part of the canonical pathway), it can also attach farnesyl diphosphate (FPP, 15C), albeit at lower efficiency, producing Chl a with a farnesyl tail (Chl aF) (Rüdiger et al. 1980). In mesophilic cyanobacteria, FPP is not available within the cell to be used as a substrate by ChlG. However, specifically in the thermophilic cyanobacterium, T. elongatus, studies have previously identified a novel gene (named SelFPS) in the isoprenyl transferase family that specifically produces FPP optimally at 50–60 °C which is the optimal growth temperature range of T. elongates (Ohto et al. 1999). We hypothesize that the presence of this SelFPS would enrich the cell in this precursor FPP and allow FPP to be utilized by ChlG as a substrate during Chl a biosynthesis. If this is true, it would be interesting to study if this gene could aid in creating synthetic strains of thermophilic cyanobacteria, although that would be outside the scope of this study.
Upon careful examination of the chlorophylls in the reported X-ray structures of PSII, a number of the phytyl tails fail to display clear electron density for the tail regions most distal to the chlorin ring. This has been observed in previous structures and attributed to the tails being in regions of the structure that exhibit high B-factors (Loll et al. 2005b; Müh et al. 2008). However, a number of the Chl a molecules with incomplete electron densities for the tails are buried and shielded from the lipid interface; their tail shortness cannot be explained by increased mobility. One such shielded Chl a is poised end-to-end with a conserved, tightly bound phosphatidylglycerol molecule (PG). The electron density and orientation of the lipid and adjacent Chl a together (seen in Figure 2) would suggest the presence of a 15 carbon tail on the Chl a. We sought to investigate if a 15 carbon tail on Chl a could indeed be found in PSII from T. elongatus; we support this for the first time with biochemical characterization to complement the structural observation.
Figure 2.
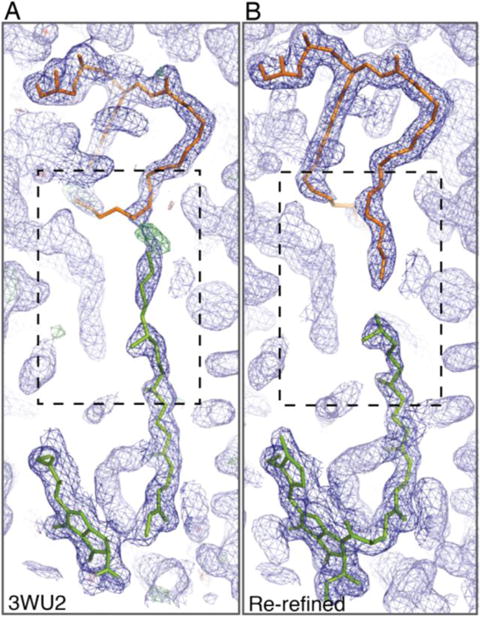
Electron density for Chl a (c-631 in the 3ARC numbering, green) adjacent to the hydrophobic tail of PG (d-714, orange) identified in the 1.9 Å PSII crystal structure (PDB 3WU2). Electron density and fitting is shown: (A) from the originally reported 1.9 Å structure with Chl c-631 having a full length 20 carbon phytyl tail extending through unresolved space into adjacent electron density and (B) from the re-refined electron density map showing Chl c-631 with a 15 carbon farnesyl tail that is truncated when the electron density of the tail stops. The σA-weighted 2Fo-Fc electron density maps are contoured at 1.0σ (Dark Blue), and the σA-weighted Fo-Fc electron density map contoured at 3.0σ (A, green). Images were generated using COOT and PyMol.
In this study, we first examine the electron density of the Chl a-PG pair in a newly re-refined map from the 1.9 Å PSII crystal structure (PDB 3ARC/3WU2) to indeed show an unambiguous truncation of electron density on the tail of Chl a. We then use HPLC coupled with mass spectrometry (MS) to identify the presence of Chl a with a farnesyl tail (Chl aF) in PSII from the thermophilic cyanobacterium T. elongatus. To our knowledge, this is the first report of Chl a having a novel tail length in cyanobacteria and, more importantly, it supports the findings of ambiguous chlorophyll tail length reported in previous crystal structures of PSII (Loll et al. 2005b; Müh et al. 2008).
MATERIALS AND METHODS
Cell culture and purification of PSII core complexes
Thermosynechococcus elongatus BP-1 cells were grown in a 13 L cylindrical bioreactor cultured at 55 °C, bubbled with 2% air-balanced CO2, and illuminated with four 15 W Cool White fluorescent light bulbs. Photosystem II core complexes were purified from T. elongatus according to (Shen et al. 2010) with modifications. All procedures performed under dim green light.
Cells were harvested at an OD730 of 1.2 and pelleted at 25 °C. Cell walls were disrupted by bead beating in buffer T1 (30 mM Hepes-NaOH, pH 7.0, 10 mM MgCl2, 5 mM CaCl2, and 25% glycerol). Broken cells were collected in a small volume of buffer T2 (30 mM Hepes-NaOH, pH 7.0, 10 mM MgCl2) and centrifuged at 4400g for 5 min at 4 °C to remove unbroken cells and residual beads. Thylakoid membranes were pelleted and resuspended in a small volume of buffer T1 to a Chl concentration of 1 mg/mL, solubilized with LDAO (30% N,N-dimethyldodecylamine N-oxide, Sigma) to a final concentration of 0.27% v/v and incubated for 5 min while stirring on ice. Solubilized thylakoids were added to an equal volume of buffer T1, mixed gently, and ultracentrifuged at 125,000g for 30 min at 4 °C. Crude purified PSII is found in the supernatant and pelleted using 13% w/v PEG 1,450 by centrifuging at 125,000g for 30 min at 4 °C. The pellet containing crude PSII was resuspended in a small volume of buffer T2 at 1 mg/mL Chl concentration and solubilized for 30 min with 1% β-DDM (n-dodecyl-beta-maltoside, Avanti Polar Lipids) by stirring on ice. Solubilized PSII was passed through a 0.45 μm filter and loaded onto a Q-sepharose HP anion exchange column (GE) that was equilibrated at 4 °C with 100% column buffer A (30 mM Mes-NaOH, pH 6.0, 3 mM CaCl2. 0.03% β-DDM), 0% column buffer B (30 mM Mes-NaOH, pH 6.0, 3 mM CaCl2, 1 M NaCl, 0.03% β-DDM). After washing with 5 column volumes of 170 mM NaCl, a linear gradient was applied from 170 mM - 400 mM NaCl. The PSII fraction was concentrated with an Amicon 100 kDa cutoff protein concentrator. Pure PSII was resuspended in a small volume of buffer T3 (3 mM Mes-NaOH, pH 6.0, 20 mM NaCl, 3 mM CaCl2 and 25% glycerol) and stored under liquid nitrogen until further use.
Pure PSII particles displayed maximal oxygen-evolution activity of up to ~3500 μmol O2/mg Chl/h in the presence of electron acceptors 0.25 mM phenyl-p-benzoquinone (PPBQ) and 1 mM K3Fe(CN)6 in O2-assay buffer (20 mM Mes-NaOH, pH 6.0, 20 mM NaCl, 3 mM CaCl2) with constant stirring at 30 °C. Oxygen-evolution activity was measured using a Clarke-type platinum oxygen electrode.
Extraction of chlorophylls
Chlorophylls were extracted by mixing <100 μL of cell pellet or concentrated purified PSII complexes in 1–2 mL of methanol. Extraneous debris was separated by centrifugation at 13,800g using a Fisher Scientific microcentrifuge (model 235C). The dark green supernatant containing the extracted pigments was collected for immediate analysis.
Separation by HPLC
The supernatant containing the Chl extract was chromatographed using a Waters Atlantis T3 analytical column (4.6 × 250 mm, 5 μm particle size) and a Waters 600 HPLC system with a 2996 photodiode array detector. The column temperature was maintained at 30 °C using a Phenomenex Thermosphere TS-130 column heater. The mobile phase was programmed as follows: the initial solvent composition, which was 6:4 v/v acetonitrile/methanol (hereafter referred to as solvent A) was adjusted by linear gradient to 80% solvent A, 20% ethyl acetate over a period of 20 min; this composition was then delivered isocratically for 10 min, after which time the linear gradient continued to a final concentration of 60% solvent A, 40% ethyl acetate at 50 min. The flow rate was 1.5 mL/min.
Fractions that displayed absorption spectra consistent with that of Chl were collected in amber glass vials containing 2 drops of concentrated (12 M) hydrochloric acid, which removed the central magnesium. This was found to help stabilize the Chls during preparation for mass spectrometry. The samples contained in the individual vials were then evaporated to dryness using a gentle stream of nitrogen gas.
Analysis by mass spectrometry
The samples were redissolved in methanol and analyzed using an Applied Biosystems API2000 mass spectrometer equipped with an electrospray ionization (ESI) probe. The mobile phase was acetonitrile delivered at 50 μL/min, and the instrument parameters were as follows: curtain gas, 20 psi; ion spray voltage, 5500 V; nebulizer gas, 20 psi; declustering potential, 20 V; focusing potential, 200 V; entrance potential, 10 V. Product ion analyses were conducted with the collision gas set to 50 psi.
Structural examination and structure re-refinement of the 3ARC/3WU2 PSII model
We began model re-refinement of 3ARC prior to the release of the updated 3WU2 coordinates because the 3ARC model had several large residual peaks inside the OEC, causing a concern about the validity of the OEC model in the given S1 state. The experimental structure factors reported for 3ARC were retrieved from the PDB followed by anisotropy correction using the program PHASER in suite CCP4 for this re-refinement. (Read 2001; Winn et al. 2011). Atomic model refinement started with the 3ARC model (Umena et al. 2012) after deleting all multiple conformations of Chl a, including the Chl a-PG pair, and any unknown lipids of unknown lengths for model rebuilding, and then followed by inspection of the descending heights of both positive and negative peaks in the residual electron density maps to about 4.5σ in each iteration as described elsewhere (Wang 2017). The model was re-refined using Refmac5 (Murshudov et al. 1997). Some of the unknown lipid molecules previously identified were eventually replaced with low-molecular weight polyethylene glycol (LMW-PEG) molecules with various unit lengths, in view of the fact that PEG was included in the original crystallization. The ideal geometry (bond lengths and bond angles) of the OEC was obtained from the quantum-mechanics/molecular modeling of the S1 state (Luber et al. 2011) but with a 5 times reduced weighting relative to the ideal protein geometry. There are three variants of chlorophylls and two different variants of heme groups in the re-refined model.
RESULTS
The re-refined PSII model (PDB accession code 5V2C) has improved working and free R-factors of 9.6% and 16.3% in comparison to the values of 15.6% and 19.4% previously reported for 3WU2, an updated version of 3ARC, respectively (Table 1). As a consequence of model re-refinement, we observe an apparent truncation of tail length in a number of Chls, which is similar to that described in earlier reports of PSII crystal structures (Loll et al. 2005b; Müh et al. 2008), although this was not reported for the 1.9 Å structure. The discrepancy in reporting often implies that earlier low-resolution structures might have mistakes. Our model re-refinement combined with biochemical analysis suggests otherwise. Of most interest is a Chl a (Chl-631 in 3ARC, and Chl-31 in 3WU2) that can be found to bind to PsbC, its headgroup ligated to a water network near Trp63 of the peptide PsbC.
Table 1.
A summary of crystallographic statistics of 5V2C PSII model re-refinement.
| PDB accession | 5V2C |
| Space group | P212121 |
| Unit cell parameters | a = 122.19 Å, b = 228.51 Å, c = 286.40 Å |
| Resolution range (highest shell) (Å) | 178.62–1.90 (1.95–1.90) |
| Number of reflections | 591,665 (45,752) |
| Working R-factor (%) | 9.16 (22.8) |
| Free R-factor (%) | 16.28 (28.0) |
| Mean atomic B-factor (Å2) | 39.48 |
| Number of atoms | 57,587 |
| Reflection-to-atom Ratio | 10.27 |
| Estimated standard uncertainties (from R-factor) (Å) | 0.157 |
| Estimated standard uncertainties (free R-factor) (Å) | 0.098 |
| Estimated standard uncertainties (maximum-likelihood) (Å) | 0.157 |
| Root mean squares bond-length deviations (Å) | 0.021 |
| Root mean squares bond-angle deviations (°) | 3.27 |
Figure 2 displays the electron density for the Chl a-PG pair of interest for the second monomer of the reported 1.9 Å model (Figure 2A) and the re-refined electron density (Figure 2B). The tail of this Chl a interacts end-to-end with the hydrophobic tail of the most tightly bound PG molecule (code LHG-d710 in 3ARC and LHG-410 in 3WU2), and shows clear gaps in the electron density after the third isoprene unit of the tail of the Chl a (Figure 2). The tail of the PG is also extending out of the electron density to compensate for the extended phytyl tail from the Chl a (Figure 2A). With the abrupt truncation of a full isoprene unit, the electron density for this Chl a is unlikely to be the product of a gradually increasing B-factor along the tail due to its atomic mobility; rather it reveals a possible physiological relevance. As described above, a close examination of the literature regarding chlorophyll synthase activity (Rüdiger et al. 1980) and FPP production in T. elongatus (Ohto et al. 1999) reveals a plausible correlation between the biosynthetic pathway for chlorophyll a with a 15 carbon farnesyl tail and what has been observed here.
In light of model re-refinement, we sought to biochemically identify varying tail Chl a species using HPLC, UV-visible spectroscopy, and mass spectrometry. Figure 3 shows the HPLC chromatograph of methanol-extracted samples, monitored at 663 nm to identify any Chl species. In addition to the major Chl a and pheophytin peaks, we were able to resolve a number of unidentified minor peaks that eluted very early from the column (Figure 3, peak 1) with UV-visible spectra consistent with Chl a. All peaks 1–6 shown in Figure 3 had a QY absorption peak at 663 nm.
Figure 3.
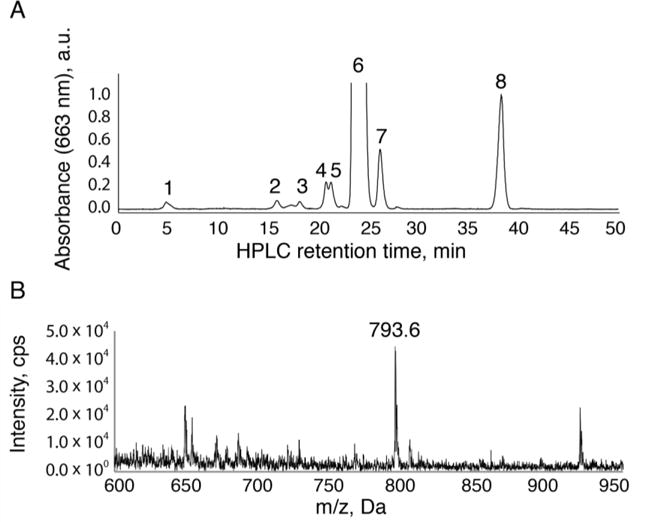
HPLC elution profile of methanol extracted pigments from PSII purified from T. elongatus in normal growth conditions (55 °C). The HPLC chromatogram in (A) represents a single representative experiment from over 3 repetitions. Peak 1 is the previously unidentified Chl a with a farnesyl tail with a demetallated mass of 793.6 m/z and its mass spectrometry analysis can be seen in (B). Peaks 2-5 represent Chl a with the phytyl tail that has not been fully reduced from geranylgeranyl and Chl a with oxidized headgroups. Peak 6 is Chl a. Peak 7 is Chl a′ (demonstrating a small contamination of PSI in this sample). Peak 8 is pheophytin. All peaks 1-6 have a QY absorption maximum at 663 nm, consistent with Chl a headgroup in methanol.
To identify the mass of the esterified tail associated with the early-eluting Chl a peaks, the fractions were collected and analyzed with ESI-MS. The dominant mass of the early eluting Chl a-like peaks is 793 m/z (Figure 3B). This mass indicates the difference of a single isoprene unit (−68 m/z) and also 8 hydrogens relative to the mass of demetallated Chl a with a phytyl tail (871 m/z). Our samples have been demetallated with HCl after purification by column chromatography to avoid oxidation of the Chl a headgroup. The difference of 8 hydrogens can be attributed to additional double bonds in the tail, which has ruled out the possibility of shortening the tail by some chemical truncation of the longer tail during the analysis. The short elution time of peak 1 relative to Chl a is consistent with a Chl a esterified to a farnesyl tail (Rüdiger et al. 1980). Taken together, the mass, UV-visible spectrum, and retention time of peak 1 indicates a structure of Chl a with a 15 carbon farnesyl tail, Chl aF.
In addition to the newly identified Chl aF, we easily resolved the Chl a peak (peak 6, 893 m/z) that frequently saturated the detector, as well as other minor chlorophyll peaks representing additional bonds of the phytyl tail and oxidized chlorophylls (peaks 2-5) (Nakamura et al. 2001). We also observed the Chl a′ epimer native to PSI (peak 7, 893 m/z) and the naturally occurring pheophytin (peak 8, 871 m/z).
To confirm that the Chl aF compound could indeed be produced from the biochemical pathway described above, in which the tail precursor FPP is synthesized by a FPP synthase unique to T. elongatus with an optimal activity at 60 °C (Ohto et al. 1999), we examined the correlation of Chl aF with growth temperature. We examined growth temperatures from 45 °C to 60 °C by studying pigment extracts from whole cells and purified PSII from T. elongatus (Figure 4). For this study, we used samples that were less concentrated so as not to saturate the detector in the HPLC. We used the software Origin to determine the area under the Chl a peak as well as the peaks eluting between 4‐14 minutes relating to Chl aF (HPLC chromatograms can be found in the SI). We observed an increase in the production of Chl aF that correlates with increasing temperature. The highest ratio between Chl aF and Chl a was at 60 °C in both whole cells and purified PSII. We also observe a modest enrichment of Chl aF in purified PSII samples versus whole cell extracts. Based on the correlation observed here, PSII appears to be a major target for Chl aF localization.
Figure 4.
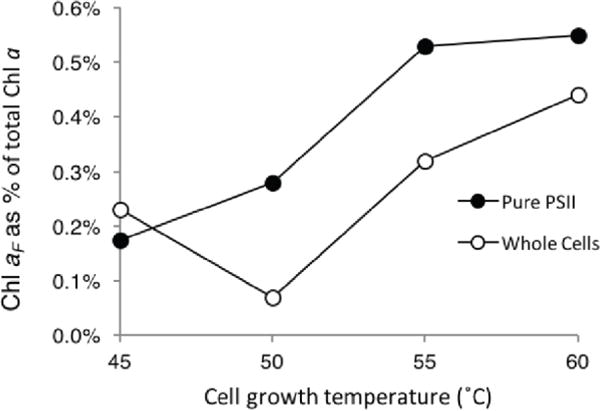
Growth temperature dependence of the normalized abundance of Chl aF versus total chlorophyll a. Abundance was determined by calculating the area under the curve using the software Origin for all peaks from 4–14 minutes for Chl aF and the dominant peak eluting at 24 minutes for Chl a. Closed symbols represent abundance of Chl aF in the purified PSII sample. Open symbols represent the abundance of Chl aF in the whole cell pellet used for PSII purification.
DISCUSSION
In this study, we use HPLC and mass spectrometry to characterize a small population of chlorophyll a in the thermophilic cyanobacterium, Thermosynechococcus elongatus, that has a 15 carbon farnesyl tail in place of the canonical 20 carbon phytyl tail esterified to the chlorin ring of the headgroup. Experimental evidence presented here supports the presence of this Chl a variant as the fitted molecule for the electron density in the 1.9 Å PSII structure shown in Figure 2 (noted as cla 631 in PDB 3ARC, cla504 in PDB 3WU2).
The specific chlorophyll that we describe here and is shown in Figure 2 is tightly associated with the antenna protein CP43, which holds 13 of the 35 chlorophyll a pigments of PSII. Its headgroup is directed towards the luminal side of the thylakoid membrane and is ligated by waters instead of the canonical histidine making it more likely to be exchangeable. There is a nearby tryptophan to its headgroup, PsbC-W63, that is thought to contribute to its energetics (Mueh et al 2015). However it also lies very close to the internal lipid bilayer that is unique to PSII and its interactions with the lipids are thought to be more important to its energetics than its headgroup (Mueh et al 2015). The tail is pointed towards a tightly bound PG d-714 lipid that can be found in CP43. Although proximity of this PG headgroup to both quinone sites could favorably stabilize the Em of the quinones, the PG is highly shielded from the aqueous environment and thus unlikely involved in any cation-tuning effect of the Em. Additionally, treatments with lipases are unlikely to access this PG. Thus the role of the PG is conserved and protected and therefore any Chl a that is directly in contact with it must be conserved as well to maintain an optimal binding pocket.
Both the original 3ARC/3WU2 electron density in the 1.9 Å resolution PSII structure and the electron density of the re-refined model have unambiguously shown that electron density abruptly ends after the third isoprene unit of the tail of Chl 631/504 (Figure 2). Given the continuous density for the adjacent tail of PG d-714, there is no room to fit the fourth isoprenyl moiety if this Chl a indeed contains the four isoprenyl units of the normal phytyl tail. The simplest interpretation of this feature is that this Chl molecule contains a farnesyl tail instead. Both PSII monomers of the 3ARC/3WU2 structure at 1.90 Å resolution, as well as those of the recently published low-dosage PSII structures at a resolution of ~ 1.85 Å and the Sr-substituted PSII structure at 2.1 Å resolution, exhibited a similar electron density property in this regard.
The observations of Chl a tail variations are not unique to the 3ARC/3WU2 data set described here. An examination of other higher resolution data sets (Suga et al. 2015; Tanaka et al. 2017), including the low-dosage data sets of 5B5E and 5B66, shows that variations of Chl tail lengths are universally present in all PSII crystal structures. The similarity observed in both high- and low-dosage X-ray data sets has ruled out the possibility of a radiation-induced truncation of Chl a tails. It is a concern that sample heterogeneity of Chl a could pose a problem for crystallization and the quality of crystals obtained. In many cases, crystallization could be used for purification so that different tail lengths may be separated in some crystals or enriched in others. Thus, the fraction of Chl aF observed in a crystal structure derived from one particular crystal may not bear a strong correlation with the rest of crystals in the preparation, nor with the fraction observed in the entire sample used for crystallization.
Using reverse-phase HPLC, UV-visible spectroscopy and mass spectrometry, we were able to qualitatively identify the presence of chlorophyll a with a 15 carbon farnesyl tail, Chl aF, in both T. elongatus whole cells and purified PSII-enriched samples. This compound eluted early as expected from the column, had a UV-visible spectrum consistent with Chl a, but a unique mass of 793, i.e., it is Chl a with a farnesyl tail. In addition, we observed a number of other minor peaks eluting between 4 and 14 minutes that were related to the same original mass of Chl aF with an additional oxygen (Figure S1). This may make the non-oxidized Chl aF peaks appear smaller in the chromatogram. The distribution of peaks of this kind has previously been observed elsewhere for the same compound generated by exposing anaerobic heliobacteria to oxygen (Ferlez et al. 2015), in part because the double bonds in the farnesyl tail are very sensitive to oxidation (Molińska et al. 2015). Additionally, the more polar nature of Chl aF than chlorophyll a with a phytyl tail (Chl aP) can affect its solubility in organic solvents used for extraction, leading possibly to an incomplete extraction. Due to these conditions and other unknown parameters, it is likely that the apparent abundance of the Chl aF is underrepresented in our studies making quantitation unreliable. Finally, we observed that Chl aF co-elutes with a large population of carotenoids, which may explain why this compound has eluded detection in previous studies (Iwai et al. 2008), and perhaps its abundance appears to be reduced in our current studies.
The data we presented here were highly reproducible as indicated from numerous individual samples run during the development phases of this study. This does not exclude the possible minor contamination of PSI in our PSII samples that were prepared by using standard purification procedures. For example, peak 7 in the HPLC trace in Figure 3A represents Chl a’, an epimer of Chl a that is found exclusively in PSI (Nakamura et al. 2003). The relative abundance of this compound compared to the Pheo peak (peak 8) is very small and indicates only a very minor contamination of PSI in the sample).
By examining pigment extracts from whole cells and purified PSII over four different growth temperatures, we see a trend for Chl aF to be produced more at 60 °C. These results support the hypothesis that the FPP synthase gene unique to T. elongatus (Ohto et al. 1999) may enrich the pool of FPP enough for Chl aF synthesis in vivo. FPP would also serve as a precursor for other compounds to aid the cell in adapting to deal with higher temperatures such as by altering the rigidity of the lipid membrane. Additionally, this is important for biofuel research that aims to over-produce long hydrocarbon chains for jet fuel. Researchers have been studying how to overproduce FPP in the cell, convert it to farnesene with farnesene synthase (Halfmann et al. 2014), and examine the suitability of farnesene as a drop-in candidate for jet fuel (Hellier et al. 2013). The results presented here demonstrate that over-production of isoprene chain biofuels, or biofuel precursors, in cyanobacteria may perturb the metabolic pathway of chlorophyll biosynthesis and photosynthetic function, and should be considered during future tests.
CONCLUSION
This study has used biochemical analysis to support the long-standing observation in the reported PSII crystal structures that there is a minor population of Chl a with shortened tails in PSII from thermophilic cyanobacteria with shortened tails. We have confirmed this observation by reporting the electron density from a re-refined map based on the reported 1.9 Å structure. For the first time, we have correlated this observation with HPLC and mass spectrometry analysis to confirm the existence of a minor population of Chl a with a farnesyl tail, instead of the canonical phytyl tail. This study demonstrates the importance of pairing structural data with biochemical analysis of cofactors bound to PSII to understand the full picture of enzyme structure, function and regulation.
Supplementary Material
Acknowledgments
This work was funded by the Department of Energy Grant DE-FG02-05ER15646 (G.W.B.). Work in the laboratory of H.A.F. was supported by grants from the National Science Foundation (MCB-1243565) and the University of Connecticut Research Foundation. Crystallographic analysis of this study was in part funded by National Institutes of Health Program Grant P01 GM022779.
References
- Airs RL, Keely BJ. A high resolution study of the chlorophyll and bacteriochlorophyll pigment distributions in a calcite/gypsum microbial mat. Org Geochem. 2003;34:539–551. doi: 10.1016/S0146-6380(02)00244-9. [DOI] [Google Scholar]
- Björn LO, Papageorgiou GC, Blankenship RE, Govindjee A viewpoint: why chlorophyll a? Photosynth Res. 2009;99:85–98. doi: 10.1007/s11120-008-9395-x. [DOI] [PubMed] [Google Scholar]
- Chew AGM, Bryant DA. Chlorophyll biosynthesis in bacteria: The origins of structural and functional diversity. Annu Rev Microbiol. 2007;61:113–129. doi: 10.1146/annurev.micro.61.080706.093242. [DOI] [PubMed] [Google Scholar]
- Ferlez B, Dong W, Siavashi R, et al. The effect of bacteriochlorophyll g oxidation on energy and electron transfer in reaction centers from heliobacterium modesticaldum. J Phys Chem B. 2015;119:13714–13725. doi: 10.1021/acs.jpcb.5b03339. [DOI] [PubMed] [Google Scholar]
- Guskov A, Kern J, Gabdulkhakov A, et al. Cyanobacterial photosystem II at 2.9-Å resolution and the role of quinones, lipids, channels and chloride. Nat Struct Mol Biol. 2009;16:334–342. doi: 10.1038/nsmb.1559. [DOI] [PubMed] [Google Scholar]
- Halfmann C, et al. Genetically engineering cyanobacteria to convert CO2, water, and light into the long-chain hydrocarbon farnesene. App Microbiol Biotechnol. 2014;98:9869–9877. doi: 10.1007/s00253-014-6118-4. [DOI] [PubMed] [Google Scholar]
- Hellier P, et al. Combustion and emissions characterization of terpenes with a view to their biological production in cyanobacteria. Fuel. 2013;111:670–688. doi: 10.1016/j.fuel.2013.04.042. [DOI] [Google Scholar]
- Iwai M, Maoka T, Ikeuchi M, Takaichi S. 2,2′-beta-Hydroxylase (CrtG) is involved in carotenogenesis of both nostoxanthin and 2-hydroxymyxol 2′-fucoside in Thermosynechococcus elongatus strain BP-1. Plant Cell Physiol. 2008;49:1678–1687. doi: 10.1093/pcp/pcn142. [DOI] [PubMed] [Google Scholar]
- Kamiya N, Shen J-R. Crystal structure of oxygen-evolving Photosystem II from Thermosynechococcus vulcanus at 3.7 Å resolution. Proc Natl Acad Sci U S A. 2003;100:98–103. doi: 10.1073/pnas.0135651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loll B, Kern J, Saenger W, et al. Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature. 2005a;438:1040–1044. doi: 10.1038/nature04224. [DOI] [PubMed] [Google Scholar]
- Loll B, Kern J, Zouni A, et al. The antenna system of photosystem II from Thermosynechococcus elongatus at 3.2 Å resolution. Photosynth Res. 2005b;86:175–184. doi: 10.1007/s11120-005-4117-0. [DOI] [PubMed] [Google Scholar]
- Luber S, et al. S1-state model of the O2-evolving complex of photosystem II. Biochemistry. 2011;50:6308–6311. doi: 10.1021/bi200681q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T, Fujita Y. Regulation and evolution of chlorophyll metabolism. Photochem Photobiol Sci. 2008;7:1131–1149. doi: 10.1039/B807210H. [DOI] [PubMed] [Google Scholar]
- McEvoy JP, Brudvig GW. Water-splitting chemistry of photosystem II. Chem Rev. 2006;106:4455–4483. doi: 10.1021/cr0204294. [DOI] [PubMed] [Google Scholar]
- Molińska E, Klimczak U, Komaszyło J, et al. Double bond stereochemistry influences the susceptibility of short-chain isoprenoids and polyprenols to decomposition by thermo-oxidation. Lipids. 2015;50:359–370. doi: 10.1007/s11745-015-3998-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Müh F, Renger T, Zouni A. Crystal structure of cyanobacterial photosystem II at 3.0 Å resolution: A closer look at the antenna system and the small membrane-intrinsic subunits. Plant Physiol Bioch. 2008;46:238–264. doi: 10.1016/j.plaphy.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Müh F, Plöckinger M, Ortmayer H, Schmidt am Busch M, Lindorfer D, Adolphs J, Renger T. The quest for energy traps in the CP43 antenna of photosystem II. J Photochem Photobiol B: Biol. 2015;152:286–300. doi: 10.1016/j.jphotobiol.2015.05.023. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Akai M, Yoshida E, Taki T. Reversed-phase HPLC determination of chlorophyll a′ and phylloquinone in Photosystem I of oxygenic photosynthetic organisms. Eur J Biochem. 2003;270:2446–2458. doi: 10.1046/j.1432-1033.2003.03616.x. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Tanaka S, Watanabe T. Normal-phase HPLC separation of possible biosynthetic intermediates of pheophytin a and chlorophyll a′. Anal Sci. 2001;17:509–513. doi: 10.2116/analsci.17.509. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Watanabe T. HPLC determination of photosynthetic pigments during greening of etiolated barley leaves. Evidence for the biosynthesis of chlorophyll a′. FEBS Lett. 1998;426:201–204. doi: 10.1016/S0014-5793(98)00344-5. [DOI] [PubMed] [Google Scholar]
- Ohto C, Ishida C, Nakane H, et al. A thermophilic cyanobacterium Synechococcus elongatus has three different Class I prenyltransferase genes. Plant Mol Biol. 1999;40:307–321. doi: 10.1023/A:1006295705142. [DOI] [PubMed] [Google Scholar]
- Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Cryst. 2001;D57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- Rüdiger W, Benz J, Guthoff C. Detection and partial characterization of activity of chlorophyll synthetase in etioplast membranes. Eur J Biochem. 1980;109:193–200. doi: 10.1111/j.1432-1033.1980.tb04784.x. [DOI] [PubMed] [Google Scholar]
- Shen J-R, Kawakami K, Koike H. Purification and crystallization of oxygen-evolving photosystem II core complex from thermophilic cyanobacteria. In: Carpenter Robert., editor. Photosynthesis Research Protocols, Methods in Molecular Biology. Vol. 684. Humana Press; Totowa, NJ: 2010. [DOI] [PubMed] [Google Scholar]
- Shpilyov AV, Zinchenko VV, Shestakov SV, et al. Inactivation of the geranylgeranyl reductase (ChlP) gene in the cyanobacterium Synechocystis sp. PCC 6803. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2005;1706:195–203. doi: 10.1016/j.bbabio.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Suga M, Akita F, Hirata K, et al. Native structure of photosystem II at 1.95 Å resolution viewed by femtosecond X-ray pulses. Nature. 2015;517:99–103. doi: 10.1038/nature13991. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Fukushima Y, Kamiya N. Two different structures of the oxygen-evolving complex in the same polypeptide frameworks of Photosystem II. J Am Chem Soc. 2017;139:1718–1721. doi: 10.1021/jacs.6b09666. [DOI] [PubMed] [Google Scholar]
- Umena Y, Kawakami K, Shen J-R, Kamiya N. Crystal structure of oxygen-evolving Photosystem II at a resolution of 1.9 Å. Nature. 2012;473:55–60. doi: 10.1038/nature09913. [DOI] [PubMed] [Google Scholar]
- Wang J. Systematic analysis of residual density suggests that a major limitation in well-refined X-ray structures of proteins is the omission of ordered solvent. Protein Sci. 2017 doi: 10.1002/pro.3145. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Cryst. 2011;D67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouni A, Witt HT, Kern J, et al. Crystal structure of Photosystem II from Synechococcus elongatus at 3.8 Å resolution. Nature. 2001;409:739–743. doi: 10.1038/35055589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


