Abstract
Although human genetics has resulted in the identification of novel lipid-related genes that can be targeted for the prevention of atherosclerotic vascular disease, medications targeting these genes or their protein products have short-term effects and require frequent administration during the course of the lifetime for maximal benefit. Genome-editing technologies, such as CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats-CRISPR–associated 9) have the potential to permanently alter genes in the body and produce long-term and even lifelong protection against atherosclerosis. In this review, we discuss recent advances in genome-editing technologies and early proof-of-concept studies of somatic in vivo genome editing in mice that highlight the potential of genome editing to target disease-related genes in patients, which would establish a novel therapeutic paradigm for atherosclerosis.
Keywords: animals, atherosclerosis, gene editing, lipids, mice
With atherosclerotic vascular disease being the leading cause of death in the United States and worldwide,1 enormous effort has been focused on understanding the development and progression of disease, with an eye toward improving the prevention and treatment of disease.2 In just the last decade, genotyping and sequencing studies have considerably expanded our knowledge of how genetic variation influences the pathology of atherosclerotic vascular disease and pointed to novel therapeutic targets.3 The discovery of PCSK9 (proprotein convertase subtilisin/kexin type 9) as a positive regulator of blood low-density lipoprotein (LDL) cholesterol levels via antagonism of the LDL receptor has resulted in the development of antibody-based, PCSK9-targeting therapeutics that have been demonstrated to substantially reduce blood LDL cholesterol levels in patients and even reduce the risk of a combined end point of cardiovascular death, myocardial infarction, and stroke.4
Recently developed genome-editing technologies have for the first time made it feasible to permanently alter target genes in living animals. As such, 1-time genome editing in human patients offers a compelling potential alternative to current treatment options (statins, ezetimibe, PCSK9 antibodies, etc) that entail repeated daily or monthly dosing during the course of a lifetime. In this focused review, we discuss recent advances in genome-editing technologies, such as CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats-CRISPR–associated 9) and a new generation of base editors, describe early findings with respect to efficacy and safety of genome-editing tools in preclinical models, and highlight how genome editing of atherogenic genes, such as PCSK9, might prove useful for clinical protection against atherosclerotic vascular disease. More comprehensive reviews of the use of genome editing in cardiology and the cardiovascular sciences are available elsewhere.5–9
Genome-Editing Technologies
Tools such as zinc-finger nucleases,10 transcription activator-like effector nucleases,11,12 meganucleases,13 adenoviruses,14 and adeno-associated viruses (AAVs)15 have been successfully used to introduce directed changes in genomes. During the past few years, CRISPR-Cas9 systems developed from naturally occurring adaptive immunity mechanisms in various bacterial species, such as Streptococcus pyogenes (S. pyogenes) and Staphylococcus aureus (S. aureus), have been widely applied in mammalian cells and organisms and are generally acknowledged as a game-changing technology.16–23 CRISPR-Cas9 systems comprise a Cas9 nuclease and a ≈100-nucleotide synthetic guide RNA that complex together to target and bind a specific genomic site. The synthetic guide includes around 20 nucleotides that can hybridize to a complementary sequence on either genomic DNA strand, called the protospacer, which directly borders a protospacer-adjacent motif. On hybridization of the guide RNA to genomic DNA, Cas9 induces a double-strand break in the genomic DNA.
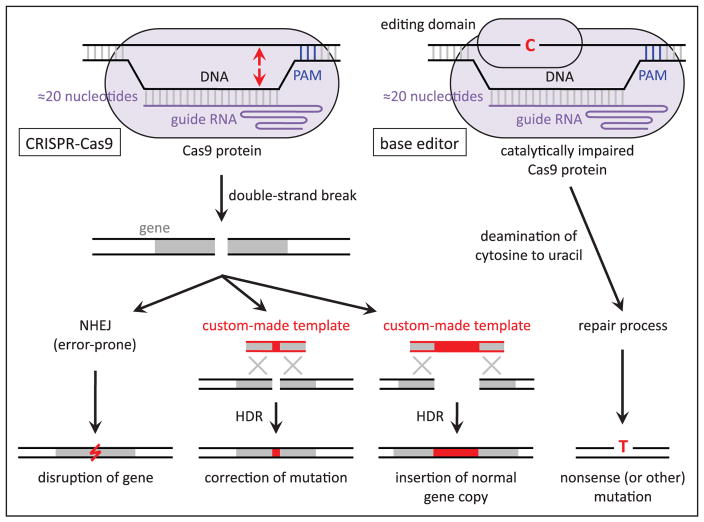
On introduction of a double-strand break, either nonhomologous end-joining (NHEJ) or homology-directed repair (HDR) is initiated by the cell (Figure). Double-stranded breaks are most commonly repaired through NHEJ without the use of a repair template, but NHEJ can result in semirandom insertion or deletion (or both) of DNA basepairs (indels) at the site of repair that can cause frameshift mutations, likely rendering the protein product of a targeted gene nonfunctional. This error-prone cellular repair process has, thus, proven useful for the intentional disruption of a gene in cellular and animal models. Through the less frequently used HDR pathway, precise introduction of a specific mutation can be achieved with a synthetic, custom-made repair template that has homology to the target site, either a single-strand DNA oligonucleotide or double-strand DNA vector. In contrast to NHEJ, which is active in all cells at all times, HDR is limited to proliferating cells in the S and G2 phases of the cell cycle. Because of this, precise genome editing by HDR is typically much less efficient than gene disruption by NHEJ, especially in living animals. This distinction has important therapeutic implications, as described below.
Figure.
Schematic of possible outcomes of genome editing that are relevant to the prevention and treatment of atherosclerosis. On the left, standard CRISPR-Cas9 is directed to a genomic site by the protospacer-adjacent motif (PAM) in the DNA and protospacer sequence in the guide RNA, whereupon it generates a double-strand break (location indicated by arrows pointing to DNA strands). The break is repaired by either nonhomologous end-joining (NHEJ) or homology-directed repair (HDR), which yield different outcomes. On the right, the base editor does not generate a double-strand break but rather converts a cytosine base (or bases) into a uracil base, which is ultimately replaced with a thymine base (with a complementary change of guanine-to-adenine on the opposite strand).
Recently, CRISPR-Cas9 has been adapted so that it cannot directly generate double-strand breaks in DNA but can nevertheless alter specific nucleotides in the DNA sequence—a phenomenon known as base editing (Figure).24–28 Addition of a cytosine deaminase domain adapted from an RNA-editing or DNA-editing enzyme to a catalytically impaired Cas9 protein confers the ability to convert cytosine bases near the CRISPR-Cas9 target site (as determined by the guide RNA) into uracil bases, which can then be converted into thymine bases by exploiting endogenous DNA repair machinery (eg, nicking just the opposite strand with Cas9, causing an adenine base to be placed opposite the uracil base during nick repair, and subsequently allowing base excision repair to replace the uracil base with thymine). In principle, base editing allows for specific alterations—such as the correction of disease-causing mutations—to be made with high efficiency and without the limitations of HDR genome editing (restriction to proliferating cells, the need for a repair template). At present, base editing is limited to cytosine-to-thymine changes or (if the base editor is targeted to the opposite strand) guanine-to-adenine changes. Nonetheless, base editing can be useful if any of the following has a desirable therapeutic effect: (1) alteration of a wild-type codon to a stop codon (nonsense mutation); (2) alteration of a wild-type codon so that the encoded amino acid disrupts the function of the protein (missense mutation); (3) direct reversal of a pathogenic thymine-to-cytosine or adenine-to-guanine mutation; or (4) because of wobble in the genetic code, alteration of a pathogenic mutant codon with a cytosine-to-thymine or guanine-to-adenine change such that the edited codon encodes the wild-type amino acid, even though the original mutation is not directly reversed (codon editing).
With respect to ease of use, CRISPR-Cas9 and its derivative base editors are unrivaled when compared with other genome-editing technologies; with a change of only the ≈20-nucleotide protospacer sequence in the guide RNA, Cas9 can be redirected to any genomic site, in contrast to other technologies where whole proteins or viral vectors have to be re-engineered. However, CRISPR-Cas9 systems have several important limitations (Table). First, the desired target genomic site has to be close to a naturally occurring protospacer-adjacent motif sequence. This can sometimes be remedied by the use of different Cas9 proteins that offer an expanded set of protospacer-adjacent motif sequences.20,21,29,30 Second, the repair of double-strand DNA breaks at the desired target site can have unintended consequences, such as insertion of undesired sequences, large deletions that impinge on the target gene in undesired ways or affect surrounding genes, and even chromosomal rearrangements. This can be remedied by the use of base editors that do not require the introduction of double-strand DNA breaks to make the desired alterations, although the base editors are themselves limited by their restriction to specific kinds of basepair changes and the need for appropriately juxtaposed protospacer-adjacent motif sequences.24–28 Third, CRISPR-Cas9 can cause off-target mutagenesis at unintended sites in the genome, potentially leading to undesired gene disruption, alteration of cell survival or proliferation, or promotion of oncogenesis. This limitation has been addressed in a substantial body of literature since the introduction of CRISPR-Cas9, including but not limited to use of truncated guide RNAs or mutated Cas9 nucleases to reduce off-target mutagenesis.31–37 Fourth, from a therapeutic perspective, CRISPR-Cas9 genome editing would cause durable changes that with the current state of the technology could not be easily reversed, unlike with most medications (although this presumes that genome-editing technology will not advance considerably in the coming years). This could prove problematic if unintended negative consequences of a therapeutic intervention were to emerge later in life.
Table.
Current Limitations of CRISPR-Cas9 for Therapeutic Use
| Precise genome editing with HDR is inefficient, especially in nonproliferating cells. |
| Restricted to genomic sites with a PAM. |
| Potential for unintended consequences at the target site, eg, insertion of undesired sequences, large deletions, and chromosomal rearrangements. |
| Off-target mutagenesis can occur at unintended sites in the genome but is difficult to detect with current technology. |
| Durable changes to the genome that are currently prohibitive to reverse. |
| Too large to easily accommodate in AAV vector—the preferred viral vector for human therapeutics. |
| Requires testing in human or humanized models to accurately assess efficacy and safety. |
| Immunologic responses against Cas9-expressing cells, especially if AAV is used. |
AAV indicates adeno-associated virus; HDR, homology-directed repair; and PAM, protospacer-adjacent motif.
Disruption of Atherogenic Genes by Genome Editing
Naturally occurring loss-of-function mutations that are protective against atherosclerotic vascular disease but that do not cause serious adverse health consequences, even in the homozygous or compound heterozygous state (ie, full knockout of gene function), have been described in several genes, most notably PCSK9, ANGPTL3, and APOC3.38–44 Each of these genes is preferentially expressed in hepatocytes, with the protein product secreted into the bloodstream. These findings showcase these genes as highly promising therapeutic targets. PCSK9 has garnered much attention since its discovery in 2003 as a regulator of blood LDL cholesterol levels. Dysregulation of the PCSK9 protein through gain-of-function mutations impairs LDL clearance via antagonism of LDL receptor and results in autosomal dominant hypercholesterolemia.45–47 Conversely, disruption of PCSK9 activity through either naturally occurring loss-of-function mutations, antibody-based therapeutic inhibition of the PCSK9 protein, or siRNA gene silencing can substantially reduce circulating LDL-C levels and even decrease coronary heart disease risk.4,38,48,49
Genome editing to introduce loss-of-function frameshift mutations via NHEJ into any of these genes within the hepatocytes of a patient should, in principle, mimic the atheroprotective effects of naturally occurring loss-of-function variants in the gene. Permanent disruption of PCSK9 with genome editing in vivo has been recently reported in several proof-of-principle studies in mice. In the first such study, an adenoviral vector containing S. pyogenes Cas9 and a guide RNA targeting the first exon of Pcsk9 was injected into mice, resulting in a high rate of NHEJ-mediated indel mutagenesis—the majority of the Pcsk9 alleles in the liver—that reduced circulating PCSK9 protein levels by >90% and reduced total cholesterol levels by ≈40%.50 An important limitation of the S. pyogenes CRISPR-Cas9 system is that it is too large to fit within an AAV vector—the preferred viral vector for gene therapy applications because of its favorable safety profile compared with adenoviral vectors. A subsequent study used AAV to deliver the smaller Cas9 from S. aureus and a guide RNA targeting Pcsk9 into mouse liver; this study also demonstrated efficient disruption of Pcsk9 alleles in the liver, reduction of circulating PCSK9 protein levels by >90%, and reduction of total cholesterol levels by ≈40%.20 Two studies have explored the possibility of targeting Apob in mice to reduce the production and secretion of lipoprotein particles into the bloodstream, thereby reducing blood cholesterol levels by a different mechanism than PCSK9 inhibition20,51; however, both studies documented substantial hepatic steatosis, consistent with observations in people with naturally occurring loss-of-function mutations in APOB.52
Although these studies are encouraging, the information they yielded is not directly relevant to human therapeutics. Murine and human gene orthologs differ substantially in their primary nucleotide sequences (relevant to efficiency, ie, on-target mutagenesis), as do the mouse and human genomes (relevant to safety, ie, off-target mutagenesis). In a follow-up study, the ability of CRISPR-Cas9 to disrupt the human PCSK9 gene in human hepatocytes in vivo was assessed using a liver-humanized mouse model (based on the Fah−/− Rag2−/− Il2rg−/− mouse, which permits the controlled ablation of endogenous mouse hepatocytes) in which primary human hepatocytes had been transplanted and engrafted in the liver in large proportions. S. pyogenes CRISPR-Cas9 delivered by an adenoviral vector efficiently disrupted the human PCSK9 gene and significantly reduced circulating human PCSK9 protein levels.53 Notably, S. aureus CRISPR-Cas9 demonstrated no appreciable activity against the human PCSK9 gene in human hepatocytes in the same liver-humanized model (unpublished observations) in contrast to the aforementioned study in which efficient activity was seen with S. aureus CRISPR-Cas9 in wild-type mice.20 This suggests that different CRISPR-Cas9 systems might have different degrees of activity in different species and emphasizes the importance of testing CRISPR-Cas9–based therapeutics directly in authentic human cells rather than extrapolating entirely from observations in nonhuman model systems.
As alluded to above, unintended on-target and off-target mutagenesis is the most worrisome concern on the safety of CRISPR-Cas9 genome editing. In the aforementioned Pcsk9/PCSK9 and Apob studies, no off-target mutagenesis was observed at a limited number of genomic sites predicted to be most likely to experience mutations, with the caveat that next-generation DNA sequencing is not sensitive enough to recognize mutagenesis occurring at rates much <0.1%. However, in one of the Apob studies, integration of AAV vector sequences at the Apob target site was observed at rates as high as 26%.51 This observation underscores the fact that NHEJ repair is unpredictable in nature, potentially leading to unintended consequences even at the desired target site.
In principle, base editors are a safer alternative to standard CRISPR-Cas9 because the lack of need for double-strand DNA breaks means there would be greatly reduced on-target indel mutagenesis, and any off-target mutagenesis would be largely limited to cytosine-to-thymine substitutions. In general, cytosine-to-thymine changes would be more benign than indels or chromosomal rearrangements resulting from double-strand breaks. This is true even in coding sequences because of wobble in the genetic code rendering many cytosine-to-thymine changes into synonymous variants, whereas most frameshifts and in-frame insertions or deletions would be deleterious. Furthermore, the indels can be as large as kilobases and affect multiple genes.
What is less clear is whether base editors can match standard CRISPR-Cas9 with respect to efficacy, although in vitro studies suggest that this is in fact the case.54 In a recent proof-of-principle study, an adenoviral vector containing the base editor called BE3 and a guide RNA targeting Pcsk9 was injected into mice, resulting in the introduction of site-specific nonsense mutations (altering one or both guanine bases in a tryptophan TGG (thymine-guanine-guanine) codon into adenine bases, yielding stop codons) in ≤31% of the Pcsk9 alleles in the liver.55 The base editing reduced circulating PCSK9 protein levels by >50% and reduced total cholesterol levels by ≈30%, establishing base editing as a viable therapeutic approach. No evidence of off-target mutagenesis, either indels or base edits, was observed. A major limitation of this work is that base editors are larger than standard Cas9 proteins, making them even more challenging to deliver with AAV.
Correction of Atherogenic Gene Mutations or Insertion of Protective Genes
Some patients experience premature atherosclerosis that is largely attributable to a single mutation that disrupts protein function, for example, autosomal dominant hypercholesterolemia from a mutation in LDLR, APOB, or PCSK9.45,56,57 In principle, genome editing could address such a mutation by several different strategies: (1) the use of HDR with a synthetic repair template or if the mutation is either a thymine-to-cytosine change or an adenine-to-guanine change, through the use of base editing whereby the mutant allele is corrected, (2) impairment of the mutation’s dominant effect by NHEJ-mediated disruption of the mutant allele (while sparing the wild-type allele), or (3) HDR-mediated insertion of additional wild-type copies of the affected gene to dilute the mutant allele’s effect.
Depending on the strategy used, the on-target editing efficiency of currently available CRISPR-Cas9 technology might not be sufficient to achieve the desired goal. This is particularly true for the use of HDR to either correct a mutation or insert wild-type gene copies. Although the latter has been successfully undertaken with hemophilia in a mouse model,58 that disease can be treated if only a small percentage of cells express clotting factor because achieving only 5% of the normal factor levels is sufficient to eliminate the most serious aspects of the disease. The same is not true of hypercholesterolemia and its attendant risk of atherosclerotic vascular disease. To date, the most successful execution of HDR in a mouse model (ornithine transcarbamylase deficiency) resulted in 10% to 20% allele correction; notably this level of correction was observed in newborn mice with rapidly proliferating hepatocytes but not in adult mice with relatively static hepatocytes in which only 1% to 2% allele correction was observed.59 For these reasons, NHEJ and base editing are the more likely avenues for success in the prevention of atherosclerosis for the foreseeable future.
Next Steps
Generally speaking, the next steps in translating antiatherogenic genome-editing approaches into the clinic will entail optimizing their efficacy and safety. For both, an important consideration is the delivery route. As adenoviral vectors are disfavored for use in humans because of safety concerns, AAVs are the preferred viral delivery vehicle.60,61 AAV has 2 shortcomings that will need to be addressed. First, the size limitation inherent in AAV vectors prevents the delivery of S. pyogenes Cas9 or base editors. Exploration of smaller Cas9 proteins from other bacterial species is needed to identify those that are effective in human hepatocytes but small enough to be accommodated by AAV. Second, AAV vectors result in prolonged expression (ie, weeks to months) of the delivered genes. Although this may be advantageous for gene therapy applications, prolonged Cas9 expression might be counterproductive in that it could increase the burden of off-target mutagenesis and could elicit immunologic responses against the Cas9-expressing cells, the same cells in which the desired genome editing would have occurred. Indeed, the development of antibodies against Cas9 has been documented in AAV-treated mice.62 One potential solution is the inclusion in the vector of an additional guide RNA that targets the Cas9 gene itself, resulting in cleavage of the vector that self-limits the duration of expression.63 An entirely different solution would be the use of lipid nanoparticles to deliver Cas9 messenger RNA into hepatocytes, relieving the size restriction imposed by AAV, as well as providing Cas9 in a form that is rapidly degraded in cells.64,65
Although the proof-of-principle preclinical studies that have been performed to date have shown little evidence of off-target mutagenesis, the limited sensitivity of next-generation DNA sequencing cannot rule out rare mutations. In the context of the billions of cells in the human liver that would need to be targeted by genome editing, rare mutations could still be problematic. Furthermore, next-generation DNA sequencing can only be applied at a discrete number of genomic sites, not genome wide, and so investigators need to decide beforehand the sites at which they think off-target mutations are most likely to occur. There are techniques to empirically determine the most likely off-target genomic sites in an unbiased way, but to date, these techniques are limited to in vitro experiments.20,66–69 Unbiased approaches that can be applied in vivo are needed to better assess the safety of potential genome-editing therapies.
Although there is much work still to be done in preclinical animal models, genome-editing therapies will eventually need to be tested in humans. With respect to atherosclerotic vascular disease, the best candidates for first-in-human studies would be patients for whom the potential benefits greatly outweigh the risks. They might include (1) patients with autosomal dominant hypercholesterolemia who are unable to achieve lipid goals with existing therapies and (2) elderly patients with high risk for future coronary events (eg, secondary prevention patients) who have maximal indications for lipid-lowering therapy. With respect to the latter, their life expectancy would be sufficiently limited that the most feared complication of genome editing—the development of cancer over time—might not be a serious consideration. On demonstration of efficacy paired with acceptable safety profiles in these selected patients, genome-editing therapies could then be entertained for larger segments of the patient population.
Highlights.
Genome-editing tools, especially those based on CRISPR-Cas9, provide a novel approach to the development of durable therapies for atherosclerosis.
Proof-of-principle studies in mouse models establish the feasibility of genome-editing therapies but also highlight challenges with the approach.
CRISPR-Cas9 genome editing currently has substantial limitations that need to be addressed with technology development if it is to be used in patients.
Nonstandard Abbreviations and Acronyms
- AAV
adeno-associated virus
- CRISPR-Cas9
clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR–associated 9
- HDR
homology-directed repair
- LDL
low density lipoprotein
- NHEJ
nonhomologous end-joining
- PCSK9
proprotein convertase subtilisin/kexin type 9
Footnotes
Disclosures
None.
References
- 1.Benjamin EJ, Blaha MJ, Chiuve SE, et al. Heart disease and stroke statistics–2017 update: a report from the American Heart Association. Circulation. 2017;135:e146–e603. doi: 10.1161/CIR.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tabas I, García-Cardeña G, Owens GK. Recent insights into the cellular biology of atherosclerosis. J Cell Biol. 2015;209:13–22. doi: 10.1083/jcb.201412052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khera AV, Kathiresan S. Genetics of coronary artery disease: discovery, biology and clinical translation. Nat Rev Genet. 2017;18:331–344. doi: 10.1038/nrg.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabatine MS, Giugliano RP, Keech AC, Honarpour N, Wiviott SD, Murphy SA, Kuder JF, Wang H, Liu T, Wasserman SM, Sever PS, Pedersen TR FOURIER Steering Committee and Investigators. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med. 2017;376:1713–1722. doi: 10.1056/NEJMoa1615664. [DOI] [PubMed] [Google Scholar]
- 5.Musunuru K. The hope and hype of CRISPR-Cas9 genome editing: a review. JAMA Cardiol. 2017;2:914–919. doi: 10.1001/jamacardio.2017.1713. [DOI] [PubMed] [Google Scholar]
- 6.Strong A, Musunuru K. Genome editing in cardiovascular diseases. Nat Rev Cardiol. 2017;14:11–20. doi: 10.1038/nrcardio.2016.139. [DOI] [PubMed] [Google Scholar]
- 7.Miano JM, Zhu QM, Lowenstein CJ. A CRISPR path to engineering new genetic mouse models for cardiovascular research. Arterioscler Thromb Vasc Biol. 2016;36:1058–1075. doi: 10.1161/ATVBAHA.116.304790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seeger T, Porteus M, Wu JC. Genome editing in cardiovascular biology. Circ Res. 2017;120:778–780. doi: 10.1161/CIRCRESAHA.116.310197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doetschman T, Georgieva T. Gene editing with CRISPR/Cas9 RNA-directed nuclease. Circ Res. 2017;120:876–894. doi: 10.1161/CIRCRESAHA.116.309727. [DOI] [PubMed] [Google Scholar]
- 10.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 11.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller JC, Tan S, Qiao G, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 13.Grizot S, Smith J, Daboussi F, Prieto J, Redondo P, Merino N, Villate M, Thomas S, Lemaire L, Montoya G, Blanco FJ, Pâques F, Duchateau P. Efficient targeting of a SCID gene by an engineered single-chain homing endonuclease. Nucleic Acids Res. 2009;37:5405–5419. doi: 10.1093/nar/gkp548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki K, Mitsui K, Aizawa E, Hasegawa K, Kawase E, Yamagishi T, Shimizu Y, Suemori H, Nakatsuji N, Mitani K. Highly efficient transient gene expression and gene targeting in primate embryonic stem cells with helper-dependent adenoviral vectors. Proc Natl Acad Sci USA. 2008;105:13781–13786. doi: 10.1073/pnas.0806976105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan IF, Hirata RK, Wang PR, Li Y, Kho J, Nelson A, Huo Y, Zavaljevski M, Ware C, Russell DW. Engineering of human pluripotent stem cells by AAV-mediated gene targeting. Mol Ther. 2010;18:1192–1199. doi: 10.1038/mt.2010.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 20.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR, Aryee MJ, Joung JK. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding Q, Regan SN, Xia Y, Oostrom LA, Cowan CA, Musunuru K. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12:393–394. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, Shimatani Z, Kondo A. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353 doi: 10.1126/science.aaf8729. pii:aaf8729. [DOI] [PubMed] [Google Scholar]
- 26.Ma Y, Zhang J, Yin W, Zhang Z, Song Y, Chang X. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods. 2016;13:1029–1035. doi: 10.1038/nmeth.4027. [DOI] [PubMed] [Google Scholar]
- 27.Hess GT, Frésard L, Han K, Lee CH, Li A, Cimprich KA, Montgomery SB, Bassik MC. Directed evolution using dCas9-targeted somatic hyper-mutation in mammalian cells. Nat Methods. 2016;13:1036–1042. doi: 10.1038/nmeth.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim YB, Komor AC, Levy JM, Packer MS, Zhao KT, Liu DR. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci USA. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 39.Zhao Z, Tuakli-Wosornu Y, Lagace TA, Kinch L, Grishin NV, Horton JD, Cohen JC, Hobbs HH. Molecular characterization of loss-of-function mutations in PCSK9 and identification of a compound heterozygote. Am J Hum Genet. 2006;79:514–523. doi: 10.1086/507488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Musunuru K, Pirruccello JP, Do R, et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 2010;363:2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stitziel NO, Khera AV, Wang X, et al. PROMIS and Myocardial Infarction Genetics Consortium Investigators. ANGPTL3 deficiency and protection against coronary artery disease. J Am Coll Cardiol. 2017;69:2054–2063. doi: 10.1016/j.jacc.2017.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crosby J, Peloso GM, Auer PL, et al. TG and HDL Working Group of the Exome Sequencing Project, National Heart, Lung, and Blood Institute. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N Engl J Med. 2014;371:22–31. doi: 10.1056/NEJMoa1307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jørgensen AB, Frikke-Schmidt R, Nordestgaard BG, Tybjærg-Hansen A. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N Engl J Med. 2014;371:32–41. doi: 10.1056/NEJMoa1308027. [DOI] [PubMed] [Google Scholar]
- 44.Saleheen D, Natarajan P, Armean IM, et al. Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature. 2017;544:235–239. doi: 10.1038/nature22034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abifadel M, Varret M, Rabès JP, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 46.Benjannet S, Hamelin J, Chrétien M, Seidah NG. Loss- and gain-of-function PCSK9 variants: cleavage specificity, dominant negative effects, and low density lipoprotein receptor (LDLR) degradation. J Biol Chem. 2012;287:33745–33755. doi: 10.1074/jbc.M112.399725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maxwell KN, Breslow JL. Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc Natl Acad Sci USA. 2004;101:7100–7105. doi: 10.1073/pnas.0402133101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson JG, Farnier M, Krempf M, et al. ODYSSEY LONG TERM Investigators. Efficacy and safety of alirocumab in reducing lipids and cardiovascular events. N Engl J Med. 2015;372:1489–1499. doi: 10.1056/NEJMoa1501031. [DOI] [PubMed] [Google Scholar]
- 49.Fitzgerald K, White S, Borodovsky A, Bettencourt BR, Strahs A, Clausen V, Wijngaard P, Horton JD, Taubel J, Brooks A, Fernando C, Kauffman RS, Kallend D, Vaishnaw A, Simon A. A highly durable RNAi therapeutic inhibitor of PCSK9. N Engl J Med. 2017;376:41–51. doi: 10.1056/NEJMoa1609243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding Q, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, Cowan CA, Rader DJ, Musunuru K. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jarrett KE, Lee CM, Yeh YH, Hsu RH, Gupta R, Zhang M, Rodriguez PJ, Lee CS, Gillard BK, Bissig KD, Pownall HJ, Martin JF, Bao G, Lagor WR. Somatic genome editing with CRISPR/Cas9 generates and corrects a metabolic disease. Sci Rep. 2017;7:44624. doi: 10.1038/srep44624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Young SG, Northey ST, McCarthy BJ. Low plasma cholesterol levels caused by a short deletion in the apolipoprotein B gene. Science. 1988;241:591–593. doi: 10.1126/science.3399894. [DOI] [PubMed] [Google Scholar]
- 53.Wang X, Raghavan A, Chen T, Qiao L, Zhang Y, Ding Q, Musunuru K. CRISPR-Cas9 targeting of PCSK9 in human hepatocytes in vivo-brief report. Arterioscler Thromb Vasc Biol. 2016;36:783–786. doi: 10.1161/ATVBAHA.116.307227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim D, Lim K, Kim ST, Yoon SH, Kim K, Ryu SM, Kim JS. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat Biotechnol. 2017;35:475–480. doi: 10.1038/nbt.3852. [DOI] [PubMed] [Google Scholar]
- 55.Chadwick AC, Wang X, Musunuru K. In vivo base editing of PCSK9 as a therapeutic alternative to genome editing. Arterioscler Thromb Vasc Biol. 2017;37:1741–1747. doi: 10.1161/ATVBAHA.117.309881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lehrman MA, Schneider WJ, Südhof TC, Brown MS, Goldstein JL, Russell DW. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985;227:140–146. doi: 10.1126/science.3155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soria LF, Ludwig EH, Clarke HR, Vega GL, Grundy SM, McCarthy BJ. Association between a specific apolipoprotein B mutation and familial defective apolipoprotein B-100. Proc Natl Acad Sci USA. 1989;86:587–591. doi: 10.1073/pnas.86.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Haurigot V, Doyon Y, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475:217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang Y, Wang L, Bell P, McMenamin D, He Z, White J, Yu H, Xu C, Morizono H, Musunuru K, Batshaw ML, Wilson JM. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muruve DA. The innate immune response to adenovirus vectors. Hum Gene Ther. 2004;15:1157–1166. doi: 10.1089/hum.2004.15.1157. [DOI] [PubMed] [Google Scholar]
- 61.Gregory SM, Nazir SA, Metcalf JP. Implications of the innate immune response to adenovirus and adenoviral vectors. Future Virol. 2011;6:357–374. doi: 10.2217/fvl.11.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chew WL, Tabebordbar M, Cheng JK, Mali P, Wu EY, Ng AH, Zhu K, Wagers AJ, Church GM. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat Methods. 2016;13:868–874. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Y, Liu X, Zhang Y, Wang H, Ying H, Liu M, Li D, Lui KO, Ding Q. A self-restricted CRISPR system to reduce off-target effects. Mol Ther. 2016;24:1508–1510. doi: 10.1038/mt.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yin H, Song CQ, Dorkin JR, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang M, Zuris JA, Meng F, Rees H, Sun S, Deng P, Han Y, Gao X, Pouli D, Wu Q, Georgakoudi I, Liu DR, Xu Q. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc Natl Acad Sci USA. 2016;113:2868–2873. doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, Alt FW. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, Aryee MJ, Joung JK. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, Hwang J, Kim JI, Kim JS. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12:237–243. 1–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 69.Tsai SQ, Nguyen NT, Malagon-Lopez J, Topkar VV, Aryee MJ, Joung JK. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat Methods. 2017;14:607–614. doi: 10.1038/nmeth.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]