Abstract
High-mobility group box 1 (HMGB1) is a potential therapeutic target and novel biomarker in a variety of malignant tumors, including hepatocellular carcinoma (HCC). More recently, a number of microRNAs (miRNAs) are identified as a class of regulators for broad control of HMGB1-mediated biological actions in eukaryotic cells. In this review article we will describe representative miRNAs involved in regulating the HMGB1 signaling pathways in HCC cell lines and/or animal models. We also propose the possible mechanisms underlying the miRNA/HMGB1 axis and discuss the future clinical significance of miRNAs targeting HMGB1 molecule for HCC therapy.
1. Introduction
Hepatocellular carcinoma (HCC) is the sixth most common malignant tumor and the third leading cause of cancer-related death. In 2013, 792,000 new cases of HCC occurred globally, and 818,000 of these cases resulted in death [1]. Although treatment of HCC has changed over the last 10 years and even though patients are able to choose different therapies that improve survival, effective therapeutic strategies are still severely needed. Particularly, the identification of novel biomarkers and molecular targets would provide HCC patients with better classification schemes and treatment options [2].
MicroRNAs (miRNAs) are tiny noncoding RNAs that are 20–22 nucleotides in length. The functions of miRNAs are associated with several crucial biological processes, such as cell proliferation, differentiation, and apoptosis [3]. The 5′ end of the miRNA (“seed” region) targets the coding and 3′ untranslated regions (3′ UTRs) of the messenger RNA (mRNA) through specific paired interactions. Thus, miRNAs regulate the assembly of the miRNA-containing ribonucleoprotein (miRNP) complex and the Argonaute/EIF2C (AGO) proteins, which eventually leads to mRNA degradation [3]. Dysregulation of miRNAs can drive tumorigenesis, and, thus, identification of miRNAs as serum biomarkers may serve as a useful diagnostic approach to cancer [4]. Several studies have reported the use of miRNAs as biomarkers and their impact on the development of therapeutic strategies in breast cancer [5], colorectal cancer [6], cervical cancer [7], lung cancer [8], thyroid cancer [9], soft tissue sarcoma [10], prostate cancer [11], and pancreatic cancer [12]. The roles of miRNAs in HCC especially are being heavily studied [13]. For instance, Tsang et al. have reported that miR-125b functions as a tumor suppressor by inhibiting eukaryotic translation initiation factor 5A2 (eIF5A2) in HCC cell lines, indicating the negative regulatory impact of miR-125b on HCC [14]. Wang et al. have suggested that overexpression of miR-215 in HCC cells is correlated with upregulated chemoresistance, via targeting of dihydrofolate reductase (DHFR) and thymidylate synthase, and proliferation, via effects on P21 expression [15].
The high-mobility group box 1 protein (HMGB1) is a chromatin-binding factor that targets DNA and facilitates the process of transcriptional protein assembly. First discovered in 1973, HMGB1 is presumably released from dead cells and secreted by inflammatory cells, and HMGB1 plays central roles in various disease states [16], including autoimmune disease [17], ischemia reperfusion injury [18], inflammation, and cancer [19, 20]. By acting as a damage-associated molecular pattern (DAMP), HMGB1 binds with high affinity to several receptors, such as the receptor for advanced glycation end products (RAGE) and Toll-like receptor- (TLR-) 2, TLR-4, and TLR-9, mediating the immune response to necrosis and immune cell invasion to trauma, pathogens, and sepsis [21]. Additionally, overexpression of HMGB1 is associated with several cancer characteristics. Binding to RAGE, HMGB1 enhances cell migration and tumor metastasis, thereby promoting cancer development [22, 23]. Several studies have focused on the association between HMGB1 expression and HCC. Serum HMGB1 can be used as a marker to evaluate tumor stage and predict HCC prognosis in patients [24, 25]. Additionally, HMGB1 regulates cell proliferation [26], cell differentiation [27], and tumor metastasis via various signaling pathways [28–30] in HCC. Since HMGB1 has potential to function as a diagnostic biomarker or curative target, the function of HMGB1 in HCC has been a hot research topic in recent years.
2. Crucial Roles of HMGB1 in HCC
The roles of HMGB1 probably are of crucial importance in HCC tumorigenesis and tumor development. In fact, HCC is usually characterized as a typical inflammation-related carcinoma. Proinflammatory mediator HMGB1 is currently believed to be secreted actively by hepatitis viruses-infected hepatocytes in the chronic inflammatory status of the liver [31]. There is increasing evidence that HMGB1 is involved in the process of HCC formation and progression, such as cell proliferation, differentiation, angiogenesis, metastasis, inflammation, and immune function in in vitro and in vivo HCC models [31, 32]. The interaction of extracellular HMGB1 with its receptors RAGE/TLRs on cell surface may activate intracellular NF-κB pathways, inducing the elevation of leukocyte adhesion molecules and proinflammatory cytokines, thereby promoting inflammation and creating a sustained tumor microenvironment, and ultimately resulting in HCC [33]. It was also reported that HMGB1 promotes the growth of HCC cell line in vitro. Inhibition of HMGB1 by RNA interference not only represses the ability of cell proliferation, migration, and invasion, but also enhances apoptosis through NF-κB pathways [34]. Furthermore, previous studies indicated a close correlation between p53 and HMGB1 which plays an important role in HCC tumorigenesis. Of note, p53 was originally considered to be a tumor suppressor. However, it was demonstrated that sustained p53 activation could accelerate protumorigenic liver inflammation in rat liver by inducing release of HMGB1 [35]. More interestingly, HMGB1 and p53 also form a complex which regulates the balance between tumor cell death and survival [36]. In addition, the effect of HMGB1 is also thought to be associated with metastasis of HCC. HMGB1 activates caspase-1 through RAGE/TLR4 signaling pathway in hypoxic HCC cells, leading to cleavage and release of proinflammatory cytokines such as IL-1β and IL-18, which in turn promotes the invasiveness and metastases of HCC [28]. Altogether, these findings suggest that HMGB1 is a powerful carcinogenic mediator and serves as potential therapeutic target for HCC.
3. MicroRNAs Regulate HMGB1 Signaling Pathways
The crucial roles of HMGB1 signaling in tumor development and the progression of various processes are now well established. However, the mechanisms and extent to which miRNAs contribute to alterations in HMGB1 expression and/or activities in pathological conditions remain poorly understood in HCC. Representative miRNAs and their effects on HMGB1 in HCC are outlined in Table 1. In this section, we describe the main findings from relevant studies regarding miRNAs, which regulate the HMGB1 signaling pathways. These findings may provide a better understanding of the biological mechanisms and potential clinical significance of miRNAs in HCC.
Table 1.
List of microRNAs involved in HMGB1 signaling pathways.
| miRNA | Binding site | Biological effects on HMGB1 | Biological effects on phenotype | Possible involved signaling pathway | Clinical significance | Cell lines | Animal model | Ref |
|---|---|---|---|---|---|---|---|---|
| miR-200a | lncRNA-TP73-AS1 3′UTR of HMGB1 mRNA (position 2724–2730) | miR-200a negatively regulates HMGB1 expression. TP73-AS1 competes with HMGB1 for miR-200a binding. |
Inhibition of cell proliferation | HMGB1/RAGE and NF-кB targets cytokines | High lncRNA-TP73- AS1 expression in HCC is correlated with poorer prognosis. | HCCLM3, MHCC97L, SMMC772, Hep3B, HepG2 | - | [37] |
|
| ||||||||
| miR-320a | 3′UTR of HMGB1 mRNA | miR-320a suppresses the expression of HMGB1. | Suppressing the invasion and metastasis of cells | HMGB1-RAGE-MMP2/MMP9 signaling | The decrease of miR-320a is associated with the invasion and metastasis. | HepG2, SK-hep-1 | - | [38] |
|
| ||||||||
| miR-505 | 3′UTR of HMGB1 mRNA (positions 417–424) | miR-505 negatively regulates HMGB1 expression in cells. | Inhibition of cell proliferation, invasion, and EMT | TGF-β-induced EMT | - | QGY7703, SMMC7721, MHCC97 | - | [39] |
|
| ||||||||
| miR-129-2 | 3′UTR of HMGB1 mRNA (positions 387–394) | HMGB1 is a downstream mediator of the biological function of miR-129-2. | Suppressing HCC migration and invasion | HMGB1-pAKT-MMP2 /MMP9 signaling |
miR-129-2 is inversely correlated with venous infiltration, high Edmondson-Steiner grading, and advanced TNM stage. An independent prognostic factor for OS/DFS. |
HepB3, Huh7 | BALB/c nude mice | [40] |
|
| ||||||||
| miR-325 | 3′UTR of HMGB1 mRNA | miR-325 negatively regulates HMGB1 expression. | Inhibition of cell invasion and proliferation | - | miR-325 is highly associated with HCC tumor size, TNM stage, and metastasis of patients. An independent prognostic factor for OS. |
SMMC-7721, Hep3B, HepG2, Huh7, Bel7404 | - | [41] |
|
| ||||||||
| miR-21 | 3′-UTRs of RECK and TIMP3 mRNA | HMGB1 signaling increases miR-21 expression to mediate the enhanced activity of MMPs through RECK/TIMP3. | Promoting cell invasion, migration, and proliferation | HMGB1-IL-6/Stat3 signalingRECK/TIMP3-MMP | - | Primary HCC cells, Huh7, HepG2, HepB3 | Athymic nude mice | [30] |
3′UTR: 3′ untranslated region; RAGE: receptor for advanced glycation end-products; MMP: matrix metalloproteinases; EMT: epithelial-mesenchymal transition; TNM: tumor-node-metastasis; OS: overall survival; DFS: disease-free survival; RECK: reversion-inducing cysteine-rich protein with Kazal motifs; TIMP3: metalloproteinase inhibitor 3. Ref: references.
3.1. miR-200a
miR-200a belongs to the miRNA-200 (miR-200) family, which contains five members that compose two clusters [42]. Cluster I miR-200s, which include miR-200b, miR-200a, and miR-429, are located in chromosome 1 [43]. As a member of the miR-200 family, miR-200a influences the epithelial-to-mesenchymal transition (EMT) and metastasis of various tumor types [44]. A negative correlation reportedly exists between the miR-200a level and tumorigenesis, angiogenesis, and metastasis through cell signaling pathways in breast cancer [45, 46], ovarian cancer [47, 48], endometrial cancer [49], meningioma [50], pituitary cancer [51], renal cancer [52], prostate cancer [53], oral cancer [54], nasopharyngeal carcinoma [55], and HCC [37].
miR-200a may regulate the neoplastic transition of the hepatic oval cell by directly targeting the β-catenin (CTNNB1) pathway [56]. A recent study has described the role of miR-200a in regulating HCC cell growth via direct targeting of HMGB1 [37], and the authors have reported the interaction between long noncoding RNA (lncRNA) TP73-AS1 and miR-200a during the process of HCC cell proliferation [37]. According to their findings, miR-200a inhibits HMGB1 expression. Additionally, high lncRNA-TP73-AS1 expression in patients is correlated with a worse prognosis. The protein levels of HMGB1, RAGE, and NF-κB are significantly decreased after TP73-AS1 knockdown. More detailed studies have shown that TP73-AS1 competes with HMGB1 for miR-200a binding. The interaction between miR-200a and TP73-AS1 is consequently regulated by HMGB1/RAGE expression and downstream NF-κB targets, such as inflammatory cytokines. Reduced binding between TP73-AS1 and miR-200a promotes the inhibitory actions of miR-200a toward HMGB1 and ultimately delays HCC progression [37].
3.2. miR-320a
miR-320a is downregulated in various malignant diseases. One study has demonstrated that miR-320a represses proliferation and metastasis and enhances irradiation-induced apoptosis by directly targeting STAT3 signals in lung adenocarcinoma cells [57]. Several researchers had reported that, by targeting β-catenin, miR-320 suppresses the proliferation of colon cancer cells [58] and has potential as a prognostic biomarker for colorectal cancer [59]. Other studies have also suggested that miR-320a can inhibit tumor growth in breast cancer [60], glioblastoma [61], gastric cancer [62], and acute lymphoblastic leukemia [63].
The roles of miR-320a in HCC are also under discussion. Yao et al. have reported that miR-320a deregulates guanine nucleotide-binding protein G(i) α-1 subunit (GNAI1) and facilitates the migration and invasion of HCC [64]. Yuan et al. have reported that miR-320a has a suppressive effect on the stemness features of HCC cells by repressing CTNNB1 signals and that association of lncRNA-DANCR with CTNNB1 decreases the inhibitory actions of miR-320a on CTNNB1 [65].
In an investigation of the interaction between miR-320a and HMGB1, one research group demonstrated a remarkable decline in miR-320a in HCC patients [38]. Because HMGB1 is a known direct target of miR-320a, the expression level of miR-320a might regulate the invasion and metastasis of HCC cells by targeting the HMGB1 pathway, partly indicating the potential antimetastasis role of miR-320a in HCC [38].
3.3. miR-505
Several studies discuss the role of miR-505 in different cancers. miR-505 is reportedly associated with apoptosis, and studies of drug-resistant human breast cancer cell lines have shown that miR-505 inhibits proliferation by inducing apoptosis [66]. However, high expression of miR-505 is strongly associated with a poor prognosis in lymph node-negative breast cancer patients [67]. Additionally, several reports have indicated that miR-505 plays a suppressor role in cervical carcinoma by targeting the Frizzled-4 gene (FZD4) [68] and in endometrial cancer by targeting transforming growth factor-α (TGF-α) [69]. miR-505 expression is correlated with the potential of establishing a biomarker of the imatinib therapeutic response in chronic myeloid leukemia patients [70]. Interestingly, Qin et al. have reported that miR-505 is upregulated in a nasopharyngeal cell line but that miR-505 surprisingly still inhibits metastasis and suppresses the expression of EMT markers, which are the genes associated with cell migration and invasion [71].
Similarly, the results of studies in HCC have also revealed several latent functions for miR-505. Dysregulation of miR-505 might be associated with HCC [72]. A recent article has presented several studies showing that miR-505 is decreased in various HCC cell lines. miR-505 attenuates HCC cell proliferation and invasion by inhibiting HMGB1, suggesting potential antitumor roles for miR-505 in HCC [39].
3.4. miR-129-2
miR-129-2 belongs to the miR-129 family, which presumably functions negatively in different cancers. miR-129-2 suppresses proliferation, migration, and invasion of renal carcinoma [73], esophageal carcinoma [74], breast cancer [75, 76], and lung cancer [77]. Furthermore, the miR-129-2 gene is reportedly hypermethylated in endometrial cancer [78], gastric carcinoma [79], osteosarcoma [80], and glioma [81]. miR-129-2 downregulates the expression of SOX4 [74], an oncogene of the SRY-related HMGB family, and epigenetic deregulation of miR-129-2 results in overexpression of SOX4 in endometrial cancer [78] and gastric carcinoma [79]. Lu et al. have found frequent DNA methylation of miR-129-2 in HCC and have suggested the potential clinical utility of miR-129-2 as a diagnostic biomarker for HCC [82]. Chen et al. have reported that methylation-mediated repression of miR-129-2 might promote SOX4 expression and HCC progression [83].
The relationship between miR-129-2 and HMGB1 is also under exploration. Yang et al. have reported that the expression of miR-129-2 in glioma cells is regulated by DNA methylation and that miR-129-2 inhibits glioma cell growth and promotes apoptosis by directly targeting HMGB1 [81]. Another group from China have reported that miR-129-2 is remarkably declined in HCC cells [40]. The miR-129-2 level is correlated with venous infiltration, a high Edmondson-Steiner grade, and an advanced tumor-node-metastasis (TNM) stage and serves as an independent prognostic factor to indicate overall survival and disease-free survival. Further investigations have indicated that miR-129-2 suppresses AKT phosphorylation and downregulates matrix metalloproteinase 2/9 (MMP2/9) expression. Suppression of p-AKT significantly represses HMGB1-mediated HCC cell migration and invasion. Additionally, miR-129-2 is regulated by DNA methylation. Demethylation of miR-129-2 has an inhibitory impact on cell growth, partly though inhibition [40].
3.5. miR-325
A few studies have reported functions for miR-325 in cancer. Wong et al. have shown that miR-325 is upregulated in squamous cell carcinoma of the tongue [84]. Yao et al. have reported that in non-small-cell lung cancer, HMGB1 is negatively regulated by miR-325 and that an aberrant expression level of miR-325 affects cell invasion and proliferation [85]. One study that investigated the expression of miR-325 and its impact via HMGB1 targeting in HCC has shown that the expression level of miR-325 is negatively correlated with HMGB1 in HCC. miR-325 functions as a tumor suppressor by targeting HMGB1 and might serve as a potential prognostic marker for HCC [41].
3.6. miR-21
In contrast to the miRNAs described above, miR-21 is frequently overexpressed in different malignant tumor cell lines and tissues, such as glioblastoma [86], breast cancer [87, 88], myeloma [89], colon cancer [90], prostate cancer [91], B-cell lymphoma [92], and lung cancer [93]. Further functional studies have indicated that miR-21 plays a key role in tumor progression through its association with antiapoptotic effects, radical invasion, and metastasis [86, 88]. miR-21 is distinguished from the other miRNAs mentioned in this review because miR-21 has been described as an oncomiR due to its facilitative impact on tumorigenesis [94].
The link between miR-21 and HCC has also has been thoroughly studied. Wang et al. have reported that the serum miR-21 level is associated with HCC prognosis and can even be an earlier and more precise biomarker than AFP [95]. Huang et al. have indicated that elevated expression of miR-21 is associated with HCC progression [96]. Guo et al. have shown that, combined with AFP, circulating miR-21 might enable an HCC diagnosis, particularly in AFP-negative patients [97]. Additionally, Hu et al. have shown that lncRNA growth arrest-specific transcript 5 (GAS5) moderates the expression of miR-21 and ultimately attenuates the migration and invasion of HCC cells [98]. Different mechanisms or pathways of miR-21 that promote the malignancy of HCC have also been identified. By using locked nucleic acids, Najafi et al. have generated a model of miR-21 degradation in an HCC cell line to assess the effect of miR-21 degradation on cell viability and apoptosis and ultimately have found that downregulation of miR-21 decreases HCC cell viability by inducing apoptosis and necrosis [99]. Jiang et al. have reported that the liver cancer stem cells are enhanced by overexpression of miR-21, which then leads to high invasion and migration of HCC [100]. Li et al. have confirmed that miR-21 and miR-183 negatively regulate cytokine signaling 6 (SOCS6), a negative regulator of the cytokine receptor pathway, and subsequently modulate tumor growth and invasion [101]. Moreover, several studies have revealed the connection between hepatitis B virus (HBV) X protein (HBx), miR-21, and HCC. Qiu et al. have indicated that HBx downregulates the expression of programmed cell death 4 (PDCD4), whose expression level is low in HCC patients and which can attenuate carcinogenesis, partially via miR-21 [102]. Li et al. have observed that upregulation of miR-21 is induced by the HBx-interleukin-6- (IL-6-) STAT3 pathway and that miR-21 is a critical factor in the process of HBx-induced transformation, suggesting the promoting effect of miR-21 in early carcinogenesis [103].
One research group from China has focused on the link between liver inflammation and HCC under the effects of HMGB1 and dysregulation of miR-21 [30]. The researchers have identified a balanced level of expression between HMGB1 and miR-21 in HCC cells and patients and have verified that these levels are regulated by the IL-6-STAT3 signaling axis. Further studies have shown that, through the IL-6-STAT3 pathway, HMGB1 signaling elevates the level of miR-21 expression and subsequently inhibits posttranscriptional processing of the MMP inhibitors RECK and TIMP3. By attenuating RECK and TIMP3 which could repress the malignant characteristics of HCC, miR-21 enhances the activity of MMPs and ultimately promotes HCC progression [30].
4. Biological Mechanisms of the MicroRNA/HMGB1 Axis
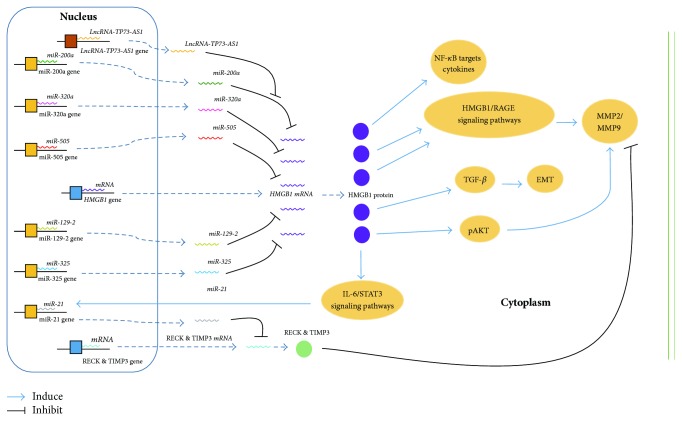
HCC releases DAMP proteins, such as HMGB1, which interact with their receptors, triggering a proinflammatory signaling cascade that contributes to tumor progression [104, 105]. HMGB1 can be recognized by siRNA and activate downstream signaling molecules. Although the linkages between miRNAs and HMGB1 in HCC have been discussed thoroughly above, a universal conclusion of the molecular mechanisms of the interactions between miRNAs and HMGB1 is required. Based on miRNAs described in Section 3, we propose a hypothesis that the mechanism of biological mechanisms of the miRNA/HMGB1 axis as shown in Figure 1.
Figure 1.
Schematic representation of microRNAs regulating HMGB1-mediated mechanisms.
To our knowledge, gene expression in eukaryotic cells is regulated at multiple levels, including transcriptionally, posttranscriptionally, translationally, posttranslationally, and epigenetically. Furthermore, miRNAs mainly participate in regulating progression at the posttranscriptional level. Similar to the biological behaviors of these tiny noncoding RNAs, several specific miRNAs bind to the HMGB1 mRNA, via the particular targeting effect of the “seed” region of miRNAs in the coding region and 3′ UTR of the mRNA, thus guiding the process of HMGB1 mRNA degradation, influencing downstream signal pathways, such as HMGB1-RAGE-MMP2/MMP9 signaling [40], and ultimately altering the malignant tumor characteristics of HCC (Figure 1). Additionally, several other molecules take part in the miRNA-HMGB1 axis. Multiple lncRNAs [37], while competing with HMGB1 mRNAs, interact with several peculiar miRNAs, which explains the miRNA posttranscriptional regulation of HMGB1. Furthermore, miRNAs may also regulate the production of downstream molecules. The HMGB1 protein stimulates HCC cells to generate oncomiRs, which inhibit tumor suppressors such as RECK and TIMP3 [30], and promote HCC malignancy.
Altogether, the miRNA-HMGB1 axis probably is an important mechanism that is active during the posttranscriptional regulatory processes that target HMGB1 in HCC.
5. Future Perspective Regarding Clinical Significance
Several recent studies about the linkages between miRNAs and HMGB1 have highlighted the availability of miRNAs that can serve as prognostic predictors of HCC in patients. For example, Liu et al. have noted that the relationship between the miR-129-2 level and advanced malignant features is reversely correlated, which might provide hints toward patient prognosis [40]. Additionally, Li et al. have shown that the negative correlation between miR-325 and tumor size, TNM stage, and metastasis in HCC highlights miR-325 as an independent prognostic factor for HCC patients [41]. Therefore, these findings suggest that the miRNA expression profile will serve as a credible tool for diagnostics, tumor staging, and prognostic predictions of HCC in the future.
Moreover, some studies, which have confirmed the functions of miRNAs on HMGB1 in different signaling pathways during the regulation of tumor progression, have suggested several novel available therapies that are based on the miRNA-HMGB1 axis to combat HCC. Overexpression of miRNAs that target HMGB1 mRNA or silence oncomiRs that inhibit tumor suppressor genes might reduce the production of HMGB1, block downstream signaling pathways, and consequently downregulate HCC malignancy. Combined with current methods of treatment, including surgery, chemotherapy, radiotherapy, radiofrequency ablation, hepatic arterial chemoembolization (TACE), and immunotherapy, the regulatory effects of miRNA-HMGB1 in HCC may play important roles in targeted therapy and precision medicine in the future.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (81502794), Zhejiang Provincial Natural Science Foundation of China (LY16H240001), Science and Technology Department of Zhejiang Province (2014C03030, 2017F10003), the Qianjiang Talents Project of Zhejiang Province (QJD1602027), and the Health and Family Planning Commission of Zhejiang Province (2017PY015, 2018KY038, and 11-ZC02).
Conflicts of Interest
The authors declare no conflicts of interest.
Authors' Contributions
Jianing Yan and Shibo Ying contributed equally to this work.
References
- 1.Fitzmaurice C., Dicker D., Pain A., et al. The global burden of cancer 2013. JAMA Oncology. 2015;1(4):505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forner A., Llovet J. M., Bruix J. Hepatocellular carcinoma. The Lancet. 2012;379(9822):1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 3.Calin G. A., Croce C. M. MicroRNA signatures in human cancers. Nature Reviews Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 4.Chen X., Ba Y., Ma L., et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Research. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 5.Graveel C. R., Calderone H. M., Westerhuis J. J., Winn M. E., Sempere L. F. Critical analysis of the potential for microRNA biomarkers in breast cancer management. Breast Cancer : Targets and Therapy. 2015;7:59–79. doi: 10.2147/BCTT.S43799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stiegelbauer V., Perakis S., Deutsch A., et al MicroRNAs as novel predictive biomarkers and therapeutic targets in colorectal cancer. World Journal of Gastroenterology. 2014;20(33):11727–11735. doi: 10.3748/wjg.v20.i33.11727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ribeiro J., Sousa H. MicroRNAs as biomarkers of cervical cancer development: A literature review on miR-125b and miR-34a. Molecular Biology Reports. 2014;41(3):1525–1531. doi: 10.1007/s11033-013-2998-0. [DOI] [PubMed] [Google Scholar]
- 8.Wang R. J., Zheng Y. H., Wang P., Zhang J. Z. Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in non-small cell lung cancer. International Journal of Clinical and Experimental Pathology. 2015;8(1):765–771. [PMC free article] [PubMed] [Google Scholar]
- 9.Fuziwara C. S., Kimura E. T. MicroRNA deregulation in anaplastic thyroid cancer biology. International Journal of Endocrinology. 2014;2014 doi: 10.1155/2014/743450.743450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujiwara T., Kunisada T., Takeda K., et al. MicroRNAs in soft tissue sarcomas: Overview of the accumulating evidence and importance as novel biomarkers. BioMed Research International. 2014;2014 doi: 10.1155/2014/592868.592868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa-Pinheiro P., Ramalho-Carvalho J., Vieira F. Q., et al. MicroRNA-375 plays a dual role in prostate carcinogenesis. Clinical Epigenetics. 2015;7, article no. 42 doi: 10.1186/s13148-015-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halkova T., Cuperkova R., Minarik M., Benesova L. MicroRNAs in pancreatic cancer: Involvement in carcinogenesis and potential use for diagnosis and prognosis. Gastroenterology Research and Practice. 2015;2015 doi: 10.1155/2015/892903.892903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang N., Ekanem N. R., Sakyi C. A., Ray S. D. Hepatocellular carcinoma and microRNA: New perspectives on therapeutics and diagnostics. Advanced Drug Delivery Reviews. 2015;81:62–74. doi: 10.1016/j.addr.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 14.Tsang F. H., Au V., Lu W.-J., et al. Prognostic marker microRNA-125b inhibits tumorigenic properties of hepatocellular carcinoma cells via suppressing tumorigenic molecule eIF5A2. Digestive Diseases and Sciences. 2014;59(10):2477–2487. doi: 10.1007/s10620-014-3184-5. [DOI] [PubMed] [Google Scholar]
- 15.Wang L., Wang Y. M., Xu S., et al. MicroRNA-215 is upregulated by treatment with Adriamycin and leads to the chemoresistance of hepatocellular carcinoma cells and tissues. Molecular Medicine Reports. 2015;12(4):5274–5280. doi: 10.3892/mmr.2015.4012. [DOI] [PubMed] [Google Scholar]
- 16.Kang R., Chen R., Zhang Q. HMGB1 in health and disease. Molecular Aspects of Medicine. 2014 doi: 10.1016/j.mam.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magna M., Pisetsky D. S. The role of HMGB1 in the pathogenesis of inflammatory and autoimmune diseases. Molecular Medicine. 2014;20(1):138–146. doi: 10.2119/molmed.2013.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang W. J., Toledo-Pereyra L. H. The role of HMGB1 and HSP72 in ischemia and reperfusion injury. Journal of Surgical Research. 2011;166(2):219–221. doi: 10.1016/j.jss.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 19.Sims G. P., Rowe D. C., Rietdijk S. T., Herbst R., Coyle A. J. HMGB1 and RAGE in inflammation and cancer. Annual Review of Immunology. 2010;28:367–388. doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- 20.Ying S., Jiang Z., He X., et al. Serum HMGB1 as a Potential Biomarker for Patients with Asbestos-Related Diseases. Disease Markers. 2017;2017 doi: 10.1155/2017/5756102.5756102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lotze M. T., Tracey K. J. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nature Reviews Immunology. 2005;5(4):331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 22.Tang D., Kang R., Zeh H. J., III, Lotze M. T. High-mobility group box 1 and cancer. Biochimica et Biophysica Acta—Gene Regulatory Mechanisms. 2010;1799(1-2):131–140. doi: 10.1016/j.bbagrm.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ying S., Xiao X., Chen T., Lou J. PPAR ligands function as suppressors that target biological actions of HMGB1. PPAR Research. 2016;2016:10. doi: 10.1155/2016/2612743.2612743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng B. Q., Jia C. Q., Liu C. T., et al. Serum high mobility group box chromosomal protein 1 is associated with clinicopathologic features in patients with hepatocellular carcinoma. Digestive and Liver Disease. 2008;40(6):446–452. doi: 10.1016/j.dld.2007.11.024. [DOI] [PubMed] [Google Scholar]
- 25.Xiao J., Ding Y., Huang J., et al. The association of HMGB1 gene with the prognosis of HCC. PLoS ONE. 2014;9(2) doi: 10.1371/journal.pone.0089097.e89097 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Yaser A. M., Huang Y., Zhou R. R., et al. The role of receptor for advanced glycation end products (RAGE) in the proliferation of hepatocellular carcinoma. International Journal of Molecular Sciences. 2012;13(5):5982–5997. doi: 10.3390/ijms13055982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiwatashi K., Ueno S., Abeyama K., et al. A novel function of the receptor for advanced glycation end-products (RAGE) in association with tumorigenesis and tumor differentiation of HCC. Annals of Surgical Oncology. 2008;15(3):923–933. doi: 10.1245/s10434-007-9698-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan W., Chang Y., Liang X., et al. High-mobility group box 1 activates caspase-1 and promotes hepatocellular carcinoma invasiveness and metastases. Hepatology. 2012;55(6):1863–1875. doi: 10.1002/hep.25572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J. L., Wu D. W., Cheng Z. Z., Han W. Z., Xu S. W., Sun N. N. Expression of High Mobility Group Box-B1 (HMGB-1) and Matrix metalloproteinase-9 (MMP-9) in non-small cell lung cancer (NSCLC) Asian Pacific Journal of Cancer Prevention. 2014;15(12):4865–4869. doi: 10.7314/apjcp.2014.15.12.4865. [DOI] [PubMed] [Google Scholar]
- 30.Chen M., Liu Y., Varley P., et al. High-mobility group box 1 promotes hepatocellular carcinoma progression through MIR-21-mediated matrix metalloproteinase activity. Cancer Research. 2015;75(8):1645–1656. doi: 10.1158/0008-5472.CAN-14-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou R. R., Kuang X. Y., Huang Y., et al. Potential role of High mobility group box 1 in hepatocellular carcinoma. Cell Adhesion & Migration. 2014;8(5):493–498. doi: 10.4161/19336918.2014.969139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., Xiang L., Li H., et al. The role of HMGB1 signaling pathway in the development and progression of hepatocellular carcinoma: a review. International Journal of Molecular Sciences. 2015;16(9):22527–22540. doi: 10.3390/ijms160922527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nogueira-Machadoa J. A., de Oliveira Volpea C. M. HMGB-1 as a target for inflammation controlling. Recent Patents on Endocrine Metabolic & Immune Drug Discovery. 2012;6(3):201–209. doi: 10.2174/187221412802481784. [DOI] [PubMed] [Google Scholar]
- 34.Jiang W., Wang Z., Li X., et al. Reduced high-mobility group box 1 expression induced by RNA interference inhibits the bioactivity of hepatocellular carcinoma cell line HCCLM3. Digestive Diseases and Sciences. 2012;57(1):92–98. doi: 10.1007/s10620-011-1944-z. [DOI] [PubMed] [Google Scholar]
- 35.Yan H. X., Wu H. P., Zhang H. L., et al. P53 promotes inflammation-associated hepatocarcinogenesis by inducing HMGB1 release. Journal of Hepatology. 2013;59(4):762–768. doi: 10.1016/j.jhep.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livesey K. M., Kang R., Vernon P., et al. p53/HMGB1 complexes regulate autophagy and apoptosis. Cancer Research. 2012;72(8):1996–2005. doi: 10.1158/0008-5472.can-11-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S., Huang Y., Huang Y., et al. The long non-coding RNA TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE regulation. Journal of Experimental & Clinical Cancer Research. 2017;36(1, article no. 51) doi: 10.1186/s13046-017-0519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lv G., Wu M., Wang M., et al. miR-320a regulates high mobility group box 1 expression and inhibits invasion and metastasis in hepatocellular carcinoma. Liver International. 2017;37(9):1354–1364. doi: 10.1111/liv.13424. [DOI] [PubMed] [Google Scholar]
- 39.Lu L., Qiu C., Li D., Bai G., Liang J., Yang Q. MicroRNA-505 suppresses proliferation and invasion in hepatoma cells by directly targeting high-mobility group box 1. Life Sciences. 2016;157:12–18. doi: 10.1016/j.lfs.2016.05.039. [DOI] [PubMed] [Google Scholar]
- 40.Liu Z., Dou C., Yao B., et al. Methylation-mediated repression of microRNA-129-2 suppresses cell aggressiveness by inhibiting high mobility group box 1 in human hepatocellular carcinoma. Oncotarget . 2016;7(24):36909–36923. doi: 10.18632/oncotarget.9377. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Li H., Huang W., Luo R. The microRNA-325 inhibits hepatocellular carcinoma progression by targeting high mobility group box 1. Diagnostic Pathology. 2015;10(1, article no. 117) doi: 10.1186/s13000-015-0323-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Humphries B., Yang C. The microRNA-200 family: small molecules with novel roles in cancer development, progression and therapy. Oncotarget . 2015;6(9):6472–6498. doi: 10.18632/oncotarget.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altuvia Y., Landgraf P., Lithwick G., et al. Clustering and conservation patterns of human microRNAs. Nucleic Acids Research. 2005;33(8):2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Senfter D., Madlener S., Krupitza G., Mader R. M. The microRNA-200 family: Still much to discover. Biomolecular Concepts. 2016;7(5-6):311–319. doi: 10.1515/bmc-2016-0020. [DOI] [PubMed] [Google Scholar]
- 45.Park S. M., Gaur A. B., Lengyel E., Peter M. E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes & Development. 2008;22(7):894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu S. J., Hu J. Y., Kuang X. Y., et al. MicroRNA-200a promotes anoikis resistance and metastasis by targeting YAP1 in human breast cancer. Clinical Cancer Research. 2013;19(6):1389–1399. doi: 10.1158/1078-0432.CCR-12-1959. [DOI] [PubMed] [Google Scholar]
- 47.Mateescu B., Batista L., Cardon M., et al. MiR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nature Medicine. 2011;17(12):1627–1635. doi: 10.1038/nm.2512. [DOI] [PubMed] [Google Scholar]
- 48.Zuberi M., Mir R., Das J., et al. Expression of serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clinical and Translational Oncology. 2015;17(10):779–787. doi: 10.1007/s12094-015-1303-1. [DOI] [PubMed] [Google Scholar]
- 49.Li R., He J. L., Chen X. M., et al. MiR-200a is involved in proliferation and apoptosis in the human endometrial adenocarcinoma cell line HEC-1B by targeting the tumor suppressor PTEN. Molecular Biology Reports. 2014;41(4):1977–1984. doi: 10.1007/s11033-014-3045-5. [DOI] [PubMed] [Google Scholar]
- 50.Saydam O., Shen Y., Würdinger T., et al. Downregulated microRNA-200a in meningiomas promotes tumor growth by reducing E-cadherin and activating the Wnt/β-catenin signaling pathway. Molecular and Cellular Biology. 2009;29(21):5923–5940. doi: 10.1128/mcb.00332-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liao C., Chen W., Fan X., et al. MicroRNA-200c inhibits apoptosis in pituitary adenoma cells by targeting the PTEN/Akt signaling pathway. Oncology Research : Featuring Preclinical and Clinical Cancer Therapeutics. 2013;21(3):129–136. doi: 10.3727/096504013X13832473329999. [DOI] [PubMed] [Google Scholar]
- 52.Pecot C. V., Rupaimoole R., Yang D., et al. Tumour angiogenesis regulation by the miR-200 family. Nature Communications. 2013;4, article 2427 doi: 10.1038/ncomms3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barron N., Keenan J., Gammell P., et al. Biochemical relapse following radical prostatectomy and miR-200a levels in prostate cancer. The Prostate. 2012;72(11):1193–1199. doi: 10.1002/pros.22469. [DOI] [PubMed] [Google Scholar]
- 54.Park N. J., Zhou H., Elashoff D., et al. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clinical Cancer Research. 2009;15(17):5473–5477. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia H., Ng S. S., Jiang S., et al. miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochemical and Biophysical Research Communications. 2010;391(1):535–541. doi: 10.1016/j.bbrc.2009.11.093. [DOI] [PubMed] [Google Scholar]
- 56.Liu J., Ruan B., You N., et al. Downregulation of miR-200a induces EMT phenotypes and CSC-like signatures through targeting the β-catenin pathway in hepatic oval cells. PLoS ONE. 2013;8(11) doi: 10.1371/journal.pone.0079409.e79409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lv Q., Hu J. X., Li Y. J., et al. MiR-320a effectively suppresses lung adenocarcinoma cell proliferation and metastasis by regulating STAT3 signals. Cancer Biology & Therapy. 2017;18(3):142–151. doi: 10.1080/15384047.2017.1281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun J. Y., Huang Y., Li J. P., et al. MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting β-catenin. Biochemical and Biophysical Research Communications. 2012;420(4):787–792. doi: 10.1016/j.bbrc.2012.03.075. [DOI] [PubMed] [Google Scholar]
- 59.Fang Z., Tang J., Bai Y., et al. Plasma levels of microRNA-24, microRNA-320a, and microRNA-423-5p are potential biomarkers for colorectal carcinoma. Journal of Experimental & Clinical Cancer Research. 2015;34, article 86 doi: 10.1186/s13046-015-0198-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bronisz A., Godlewski J., Wallace J. A., et al. Reprogramming of the tumour microenvironment by stromal PTEN-regulated miR-320. Nature Cell Biology. 2012;14(2):159–167. doi: 10.1038/ncb2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo T., Feng Y., Liu Q., et al. MicroRNA-320a suppresses in GBM patients and modulates glioma cell functions by targeting IGF-1R. Tumor Biology. 2014;35(11):11269–11275. doi: 10.1007/s13277-014-2283-4. [DOI] [PubMed] [Google Scholar]
- 62.Zhu Y., Zhang Y., Sui Z., Zhang Y., Liu M., Tang H. USP14 de-ubiquitinates vimentin and miR-320a modulates USP14 and vimentin to contribute to malignancy in gastric cancer cells. Oncotarget . 2017;8(30):48725–48736. doi: 10.18632/oncotarget.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Diakos C., Zhong S., Xiao Y., et al. TEL-AML1 regulation of survivin and apoptosis via miRNA-494 and miRNA-320a. Blood. 2010;116(23):4885–4893. doi: 10.1182/blood-2009-02-206706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yao J., Liang L. H., Zhang Y., et al. GNAI1 Suppresses Tumor Cell Migration and Invasion and is Post-Transcriptionally Regulated by Mir-320a/c/d in Hepatocellular Carcinoma, Cancer Biol Med. GNAI1 Suppresses Tumor Cell Migration and Invasion and is Post-Transcriptionally Regulated by Mir-320a/c/d in Hepatocellular Carcinoma, Cancer Biol Med. 2012;9(4):234–41. doi: 10.7497/j.issn.2095-3941.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuan S. X., Wang J., Yang F., et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63(2):499–511. doi: 10.1002/hep.27893. [DOI] [PubMed] [Google Scholar]
- 66.Yoshioka Y., Minoura K., Takahashi R.-U., et al. An integrative genomic analysis revealed the relevance of microRNA and gene expression for drug-resistance in human breast cancer cells. Molecular Cancer. 2011;10, article no. 135 doi: 10.1186/1476-4598-10-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jonsdottir K., Janssen S. R., Da Rosa F. C., et al. Validation of expression patterns for nine miRNAs in 204 lymph-node negative breast cancers. PLoS ONE. 2012;7(11) doi: 10.1371/journal.pone.0048692.e48692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma C., Xu B., Husaiyin S., et al. MicroRNA-505 predicts prognosis and acts as tumor inhibitor in cervical carcinoma with inverse association with FZD4. Biomedicine & Pharmacotherapy. 2017;92:586–594. doi: 10.1016/j.biopha.2017.04.028. [DOI] [PubMed] [Google Scholar]
- 69.Chen S., Sun K. X., Liu B. L., Zong Z. H., Zhao Y. MicroRNA-505 functions as a tumor suppressor in endometrial cancer by targeting TGF-aα. Molecular Cancer. 2016;15(1, article no. 11) doi: 10.1186/s12943-016-0496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramachandran S. S., Muiwo P., Ahmad H. M., et al. miR-505-5p and miR-193b-3p: potential biomarkers of imatinib response in patients with chronic myeloid leukemia. Leukemia & Lymphoma. 2017;58(8):1981–1984. doi: 10.1080/10428194.2016.1272681. [DOI] [PubMed] [Google Scholar]
- 71.Qin Z., He W., Tang J., et al. MicroRNAs Provide Feedback Regulation of Epithelial-Mesenchymal Transition Induced by Growth Factors. Journal of Cellular Physiology. 2016;231(1):120–129. doi: 10.1002/jcp.25060. [DOI] [PubMed] [Google Scholar]
- 72.Lin X. J., Chong Y., Guo Z.-W., et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: A multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. The Lancet Oncology. 2015;16(7):804–815. doi: 10.1016/S1470-2045(15)00048-0. [DOI] [PubMed] [Google Scholar]
- 73.Chen X., Ruan A., Wang X., et al. miR-129-3p, as a diagnostic and prognostic biomarker for renal cell carcinoma, attenuates cell migration and invasion via downregulating multiple metastasis-related genes. Journal of Cancer Research and Clinical Oncology. 2014;140(8):1295–1304. doi: 10.1007/s00432-014-1690-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kang M., Li Y., Liu W., et al. miR-129-2 suppresses proliferation and migration of esophageal carcinoma cells through downregulation of SOX4 expression. International Journal of Molecular Medicine. 2013;32(1):51–58. doi: 10.3892/ijmm.2013.1384. [DOI] [PubMed] [Google Scholar]
- 75.Pronina I. V., Loginov V. I., Burdennyy A. M., et al. Expression and DNA methylation alterations of seven cancer-associated 3p genes and their predicted regulator miRNAs (miR-129-2, miR-9-1) in breast and ovarian cancers. Gene. 2016;576(1):483–491. doi: 10.1016/j.gene.2015.10.059. [DOI] [PubMed] [Google Scholar]
- 76.Tang X., Tang J., Liu X., et al. Downregulation of MIR-129-2 by promoter hypermethylation regulates breast cancer cell proliferation and apoptosis. Oncology Reports. 2016;35(5):2963–2969. doi: 10.3892/or.2016.4647. [DOI] [PubMed] [Google Scholar]
- 77.Xiao Y., Li X., Wang H., Wen R., He J., Tang J. Epigenetic regulation of miR-129-2 and its effects on the proliferation and invasion in lung cancer cells. Journal of Cellular and Molecular Medicine. 2015;19(9):2172–2180. doi: 10.1111/jcmm.12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang Y. W., Liu J. C., Deatherage D. E., et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Research. 2009;69(23):9038–9046. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shen R., Pan S., Qi S., Lin X., Cheng S. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 in gastric cancer. Biochemical and Biophysical Research Communications. 2010;394(4):1047–1052. doi: 10.1016/j.bbrc.2010.03.121. [DOI] [PubMed] [Google Scholar]
- 80.Long X. H., Zhou Y. F., Peng A. F., et al. Demethylation-mediated miR-129-5p up-regulation inhibits malignant phenotype of osteogenic osteosarcoma by targeting Homo sapiens valosin-containing protein (VCP) Tumor Biology. 2015;36(5):3799–3806. doi: 10.1007/s13277-014-3021-7. [DOI] [PubMed] [Google Scholar]
- 81.Yang Y., Huang J. Q., Zhang X., Shen L. F. MiR-129-2 functions as a tumor suppressor in glioma cells by targeting HMGB1 and is down-regulated by DNA methylation. Molecular and Cellular Biochemistry. 2015;404(1-2):229–239. doi: 10.1007/s11010-015-2382-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu C. Y., Lin K. Y., Tien M. T., Wu C. T., Uen Y. H., Tseng T. L. Frequent DNA methylation of MiR-129-2 and its potential clinical implication in hepatocellular carcinoma. Genes, Chromosomes and Cancer. 2013;52(7):636–643. doi: 10.1002/gcc.22059. [DOI] [PubMed] [Google Scholar]
- 83.Chen X., Zhang L., Zhang T., et al. Methylation-mediated repression of microRNA 129-2 enhances oncogenic SOX4 expression in HCC. Liver International. 2013;33(3):476–486. doi: 10.1111/liv.12097. [DOI] [PubMed] [Google Scholar]
- 84.Wong T. S., Liu X. B., Wong B. Y. H., Ng R. W. M., Yuen A. P. W., Wei W. I. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clinical Cancer Research. 2008;14(9):2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 85.Yao S., Zhao T., Jin H. Expression of MicroRNA-325-3p and its potential functions by targeting HMGB1 in non-small cell lung cancer. Biomedicine & Pharmacotherapy. 2015;70(C):72–79. doi: 10.1016/j.biopha.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 86.Chan J. A., Krichevsky A. M., Kosik K. S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Research. 2005;65(14):6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 87.Si M. L., Zhu S., Wu H., Lu Z., Wu F., Mo Y.-Y. miR-21-mediated tumor growth. Oncogene. 2007;26(19):2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 88.Zhu S., Wu H., Wu F., Nie D., Sheng S., Mo Y.-Y. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Research. 2008;18(3):350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 89.Löffler D., Brocke-Heidrich K., Pfeifer G., et al. Interleukin-6-dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110(4):1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 90.Wang P., Zou F., Xiaodong Z., et al. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Research. 2009;69(20):8157–8165. doi: 10.1158/0008-5472.CAN-09-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Volinia S., Calin G. A., Liu C. G., et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Acadamy of Sciences of the United States of America. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Medina P. P., Nolde M., Slack F. J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467(7311):86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 93.Hatley M. E., Patrick D. M., Garcia M. R., et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18(3):282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pfeffer S. R., Yang C. H., Pfeffer L. M. The Role of MIR-21 in Cancer. Drug Development Research. 2015;76(6):270–277. doi: 10.1002/ddr.21257. [DOI] [PubMed] [Google Scholar]
- 95.Wang X., Zhang J., Zhou L., et al. Significance of serum microRNA-21 in diagnosis of hepatocellular carcinoma (HCC): clinical analyses of patients and an HCC rat model. Int J Clin Exp Pathol. 2015;8(2):1466–78. [PMC free article] [PubMed] [Google Scholar]
- 96.Huang C. S., Yu W., Cui H., et al. Increased expression of miR-21 predicts poor prognosis in patients with hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8(6):p. 7234. [PMC free article] [PubMed] [Google Scholar]
- 97.Guo X., Lv X., Lv X., Ma Y., Chen L., Chen Y. Circulating miR-21 serves as a serum biomarker for hepatocellular carcinoma and correlated with distant metastasis. Oncotarget . 2017;8(27):44050–44058. doi: 10.18632/oncotarget.17211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hu L., Ye H., Huang G., et al. Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via miR-21. Tumor Biology. 2016;37(2):2691–2702. doi: 10.1007/s13277-015-4111-x. [DOI] [PubMed] [Google Scholar]
- 99.Najafi Z., Sharifi M., Javadi G. Degradation of miR-21 induces apoptosis and inhibits cell proliferation in human hepatocellular carcinoma. Cancer Gene Therapy. 2015;22(11):530–535. doi: 10.1038/cgt.2015.51. [DOI] [PubMed] [Google Scholar]
- 100.Jiang J., Yang P., Guo Z., et al. Overexpression of microRNA-21 strengthens stem cell-like characteristics in a hepatocellular carcinoma cell line. World Journal of Surgical Oncology. 2016;14(1, article no. 278) doi: 10.1186/s12957-016-1028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li Z. B., Li Z. Z., Li L., Chu H. T., Jia M. MiR-21 and miR-183 can simultaneously target SOCS6 and modulate growth and invasion of hepatocellular carcinoma (HCC) cells. European Review of Medical and Pharmacological Sciences. 2015;19(17):3208–3217. [PubMed] [Google Scholar]
- 102.Qiu X., Dong S., Qiao F., et al. HBx-mediated miR-21 upregulation represses tumor-suppressor function of PDCD4 in hepatocellular carcinoma. Oncogene. 2013;32(27):3296–3305. doi: 10.1038/onc.2013.150. [DOI] [PubMed] [Google Scholar]
- 103.Li C. H., Xu F., Chow S., et al. Hepatitis B virus X protein promotes hepatocellular carcinoma transformation through interleukin-6 activation of microRNA-21 expression. European Journal of Cancer. 2014;50(15):2560–2569. doi: 10.1016/j.ejca.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 104.Liu Y., Yan W., Tohme S., et al. Hypoxia induced HMGB1 and mitochondrial DNA interactions mediate tumor growth in hepatocellular carcinoma through Toll-like receptor 9. Journal of Hepatology. 2015;63(1, article no. 5550):114–121. doi: 10.1016/j.jhep.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tsung A., Tohme S., Billiar T. R. High-mobility group box-1 in sterile inflammation. Journal of Internal Medicine. 2014;276(5):425–443. doi: 10.1111/joim.12276. [DOI] [PubMed] [Google Scholar]