Abstract
UDP-glucose pyrophosphorylase (UGPase) is found in all organisms and catalyses the formation of UDP-glucose. In sugarcane, UDP-glucose is a branch-point in the carbon channelling into other carbohydrates, such as sucrose and cellulose, which are the major factors for sugarcane productivity. In most plants, UGPase has been described to be enzymatically active in the monomeric form, while in human and yeast, homo-octamers represent the active form of the protein. Here, we present the crystal structure of UGPase from sugarcane (ScUGPase-1) at resolution of 2.0 Å. The crystals of ScUGPase-1 reveal the presence of two molecules in the asymmetric unit and the multi-angle light scattering analysis shows that ScUGPase-1 forms a mixture of species ranging from monomers to larger oligomers in solution, suggesting similarities with the orthologs from yeast and human.
Introduction
Sugarcane (Saccharum ssp. hybrids) is a highly productive C4 crop used for many centuries to produce sugar and, more recently, other value-added products such as ethanol and bioelectricity, through fermentation and burning of the sugarcane bagasse, respectively. Thereby, the accumulation of sucrose in the culm and the bagasse, cellulosic biomass, are the major yield components [1]. The particular importance of biochemical factors in the regulation of carbon (C)-partitioning to sucrose accumulation in the culm and cellulose synthesis are crucial to improve the sugarcane yield capacity. Among the various enzymes, UDP-glucose pyrophosphorylase (UGPase; EC 2.7.7.9) is important and essential in this carbon regulation, whereas the sugar, UDP-glucose, represents an important branch point in the C channel directing for synthesis of starch, sucrose or cellulose [2–4]. UGPase is an enzyme ubiquitously distributed in all types of organisms and catalyses the formation of UDP-glucose, the key precursor of the sucrose biosynthesis and cell wall metabolism in plants [5,6]. Moreover, UGPase is also involved in the biosynthesis of starch, converting UDP-glucose into ADP-glucose through the reaction coupled to ADP-glucose pyrophosphorylase (AGPase) activity [6]. UGPase also acts in concert with sucrose phosphate synthase (SPS) in the synthesis of sucrose in the source tissues [6], whereas its activity may be linked with the cellulose synthase complex in the formation of cellulose in the sink tissues [7].
To better comprehend the regulation and activity in the reactions involved in the saccharide metabolism, UGPases from different types of organisms have been characterized over the years. It is known that oligomerization plays a role in the regulatory process of these enzymes affecting their function and activity. For instance, UGPase from barley has been shown to exist as a mixture of monomers, dimers and higher oligomeric forms is solution, with the monomer being the most active form [8,9]. Similarly, UGPase from Arabidopsis thaliana also exists as a monomer in solution, although a dimer has been observed in the crystal structure [10]. On the other hand, the yeast and human orthologs have been described to form active octameric complexes [11,12]. Recently, the UGPase from sugarcane (ScUGPase-1) was characterized, showing that the enzymatic activity and regulatory mechanism are similar to those reported for other UGPases [13]. Moreover, small angle X-ray scattering (SAXS) data showed that ScUGPase-1 exists as a combination of monomers, dimers and higher oligomers in solution, with the monomeric envelope very similar to the momoner of the crystal structure of A. thaliana UGPase [13].
In this study, we present the crystal structure of ScUGPase-1 at 2.0 Å resolution. Structural analysis shows high structural similarity with other UGPase orthologs. Multi-angle light scattering (MALS) analysis shows a possible octamer of the recombinant protein in solution, consistent with the crystal structure described for the human and yeast orthologs.
Material and methods
Cloning, expression and purification of ScUGPase-1
The ScUGPase-1 gene (KF278717) was obtained from the Brazilian SUCEST project database (http://www.sucest-fun.org/), with the Sugarcane Assembled Sequence number SCQGLR1062D04.g and cloned into the pENTR-D/TOPO vector, following cloning into the pET160- vector containing a hexa histidine-tag at the N-terminal, as described by [13]. The final construct was transformed into E. coli BL21 (DE3) strain for protein expression.
The expression was performed in 6 L LB medium containing 100 μg/mL ampicillin. Cells were grown at 37°C until OD600 of 0.6 was reached and protein expression was induced by addition of 1 mM isopropyl 1-thio-β-D-galactopyranoside (IPTG) for 4 h at 37°C. The cells were harvested by centrifugation (20 min, 5 000 x g) and resuspended in buffer A (50 mM Tris/HCl pH 8.0; 100 mM NaCl). The cells were lysated by sonication on ice and cell debris were removed by centrifugation (30 min, 20 000 x g). The supernatant was loaded onto a 5 mL HisTrap HP column (GE Healthcare) on an ÄKTATM system (GE Healthcare) for nickel-affinity chromatography. The column was washed with buffer A until the absorbance at 280 nm reached the baseline and the protein was eluted with buffer containing 0.5 M imidazole. The His-tagged ScUGPase-1 was buffer-exchanged into 20 mM Tris/HCl pH 8.0; 20 mM NaCl using a Superdex 200 16/60 gel filtration column (GE Healthcare) and analysed by SDS-PAGE.
Crystallization
The monomeric form of ScUGPase-1 was crystallised by using the hanging-drop vapour-diffusion method. 2 μL of ScUGPase-1 protein solution (8 mg/mL) were mixed with 1 μL or 2 μL of reservoir solution containing 100 mM MES sodium salt pH 6.5, 200 mM (NH4)2SO4 and 23% (w/v) PEG 8000, producing plate-form crystals after two days at 20°C. Crystals were looped-out and soaked in a cryoprotectant solution containing crystallization buffer and ethylene glycol (25% (v/v) before flash-cooling.
Data collection and processing
Data collection was carried out on beamline MX2 at the Australian Synchrotron (AS) in Melbourne at a wavelength of 0.9537 Å. The crystal diffracted to 2.0 Å resolution and the collected data were processed (indexing and integration) using XDS [14] and scaled in the Aimless (CCP4) program [15]. The crystals have the symmetry of the P1 space group. There are two molecules in the asymmetric unit. Data collection statistics are listed in Table 1. Diffraction images are available at The SBGrid Data Bank (doi: 10.15785/SBGRID/551).
Table 1. Data-processing and refinement statistics for ScUGPase-1.
| Data Collection | |
| Radiation source | MX2 (AS*) |
| Wavelength (Å) | 0.9537 |
| Space group | P1 |
| Cell dimensions | |
| a, b, c (Å) | 61.35, 66.79, 74.78 |
| Angles (°) | 75.62, 79.00, 77.52 |
| Resolution range (Å) | 42.73–2.00 (2.04–2.00) |
| Rmergea | 0.123 (0.493) |
| Rmeas (within I+/I-) | 0.173 (0.697) |
| Rmeas (all I+ & I-) | 0.138 (0.488) |
| Mean I//σ(I) | 5.6 (2.1) |
| Completeness (%) | 97.5 (95) |
| Multiplicity | 2.0 (1.9) |
| Molecules in ASU | 2 |
| Model used for MR | 1Z90 (Chain A) |
| Refinement | |
| No. reflections (work/test) | 73174 (2000) |
| Rworkb, Rfreec (%) | 19.30/22.90 |
| No. atoms | |
| Protein | 7065 |
| Ligand/ion | 52 |
| Solvent | 353 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 0.838 |
| Ramachandran plot: (% in favoured/outlier regions) | 98.65/0.0 |
| MolProbity clashscore | 2.44 |
| PDB code | 5WEG |
* Australian Synchrotron
a Rmerge = Σhkl(Σi(|Ihkl,i−〈Ihkl〉|))/Σhkl,i〈Ihkl〉, where Ihkl,i is the intensity of an individual measurement of the reflection with Miller indices h, k and l, and 〈Ihkl〉 is the mean intensity of that reflection. Calculated for IN−3 σ(I).
b Rwork = Σhkl(||Fobs,hkl|−|Fcalc,hkl||)/|Fobs,hkl|,where |Fobs,hkl| and |Fcalc,hkl| are the observed and calculated structure factor amplitudes, respectively.
c Rfree is equivalent to Rcryst but calculated with reflections (5%) omitted from the refinement process.
Structure solution and refinement
The structure was solved by molecular replacement using the sequence of UGPase from Arabidopsis thaliana (AtUGPase) (PDB code: 1Z90; chain A), which shares 83% sequence identity with ScUGPase-1, as the search model in Phaser without any modification [16]. The structure was firstly rebuilt through AutoBuild wizard (Phenix) [17] followed by manual building based on Fo-Fc and 2Fo-Fc difference maps using the Coot program [18]. Refinements were carried out using phenix.refine [19], including non-crystallographic symmetry (NCS) and TLS refinement, with chain A divided into three groups and chain B into seven groups. All structural figures were created using PyMOL (Schrödinger).
Size-exclusion chromatography (SEC)-coupled multi-angle light scattering (MALS)
SEC-MALS was performed using a Superdex 200 increase 5/150 column (GE Healthcare) combined with a Dawn Heleos II 11-angle light scattering detector coupled with an Optilab TrEX refractive index detector (Wyatt Technology). The experiments were carried out at room temperature with a protein concentration of 2.0 mg/mL and a flow rate of 0.2 mL/min in 20 mM Tris/HCl pH 8.0; 20 mM NaCl buffer. Molecular mass calculations were performed using the Astra6.1 software (Wyatt Technology). Input of the refractive increment (dn/dc values) was set at 0.186 in the molecular mass calculations, based on the premise that dn/dc is constant for unmodified proteins [20]. The molecular mass was determined across the protein elution peak.
Multiple sequence alignment
Multiple sequence alignment were carried out using the MUSCLE algorithm [21] and optimized in Jalview [22].
Protein Data Bank accession code
Coordinates and structure factors have been deposited in the Protein Data Bank under accession code 5WEG.
Results and discussion
Overall structure of ScUGPase-1
The crystal structure of ScUGPase-1 was determined by molecular replacement using the AtUGPase [10] as the search model. The crystals have the symmetry of the triclinic space group P1 and diffracted to 2.0 Å resolution. The structure, with initial R-work of 29.80% and R-free of 32.7%, was built manually and improved based on Fo-Fc and 2Fo-Fc maps to give final R-work and R-free values of 18.94% and 22.74%, respectively. Data processing and refinement statistics are shown in Table 1.
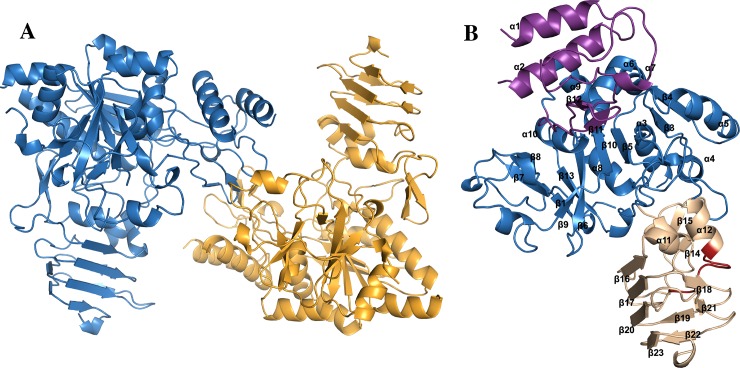
The crystals of ScUGPase-1 contain two molecules per asymmetric unit, labelled monomers A and B (Fig 1A), eight sulfate ions from the crystallization buffer, three molecules of ethylene glycol from the cryoprotectant solution and 353 water molecules. ScUGPase-1 was crystallized fused to an N-terminal His6-tag and TEV protease cleavage site, which added 32 residues to the chain (MHHHHHHGAGGCCPGCCGGGENLYFQGIITSL). Several surfaces loops could not be built due to poor electron density; these include residues A1-A12, A46-A49, A73-A75, A262-A263, B1-12, B47-B50, B72-B75 and B186-B189.
Fig 1. Crystal structure of ScUGPase-1.
(A) Cartoon representation of the two molecules of ScUGPase-1 present in the asymmetric unit. Monomer A is coloured in blue and monomer B in orange. (B) The ScUGPase-1 monomer and its domains. The N-terminal domain is shown in purple, catalytic domain in blue and the C-terminal domain in wheat colour. The RFKS419IPSI motif is shown in red.
Similar to other UGPases [10–12,23,24], each monomer of ScUGPase-1 contains three domains: an N-terminal domain, a catalytic domain and a C-terminal domain (Fig 1B). The N-terminal domain consists of α1, α2, β11, β12, and three loops (Gln169-Gly190; Glu318-Pro328; Leu337-Ala342). The catalytic domain consists of a mixed nine-stranded β-sheet (β1-β6, β9, β10 and β13) as a core, surrounded by six α-helices (α3-α9), and resembles a Rossmann fold (Fig 1B). The C-terminal domain consists of 10 β-strands (β14-β23) and two α-helices (α11-α12), with the motif RFKS419IPSI, essential for the phosphorylation and binding with 14-3-3 protein [13,25], between α12 and β17.
Structural comparison with AtUGPase
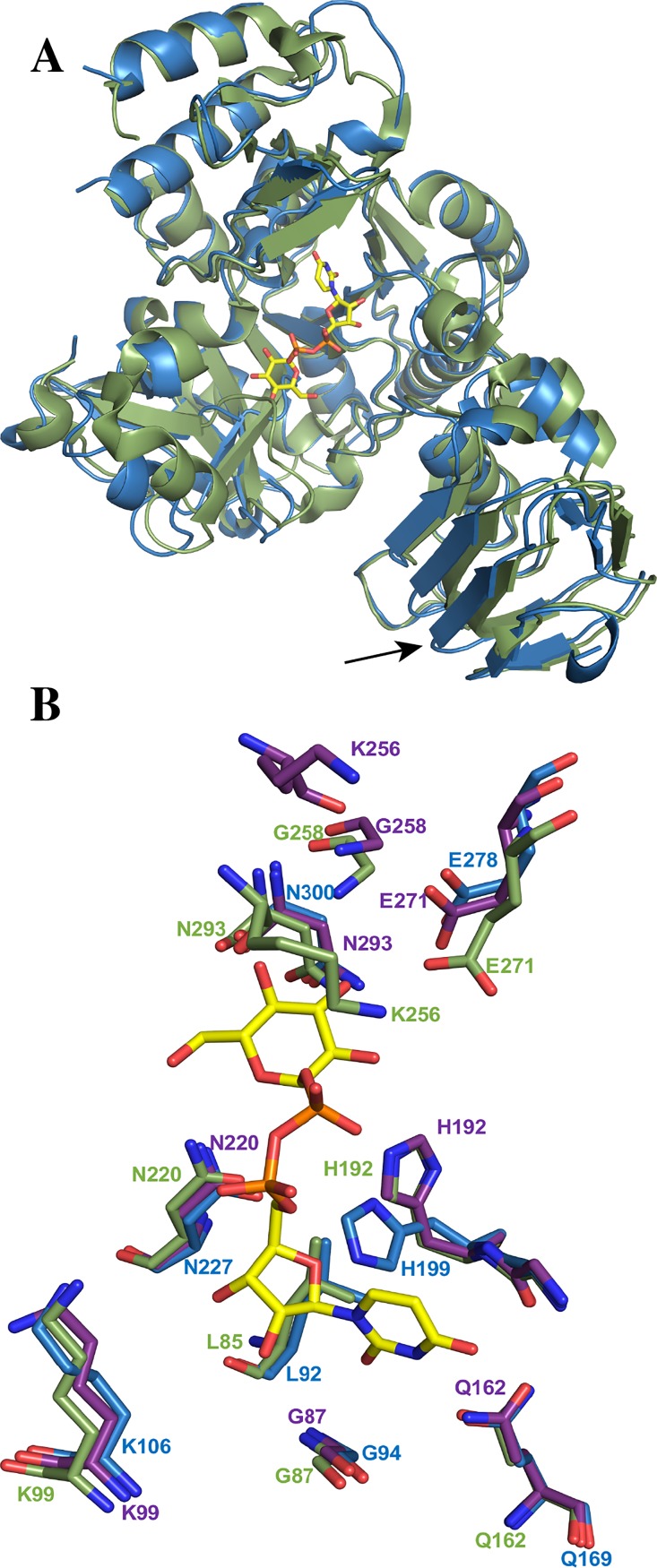
Among all the UGPase structures solved [10–12,23,24], ScUGPase-1 shares the highest sequence identity with UGPase from Arabidopsis thaliana (AtUGPase; 83%), and 54% and 56% with the human and yeast orthologs, respectively. Superposition of monomer A of ScUGPase-1 with the monomer of AtUGPase bound to UDP-glucose [10] (PDB code: 2ICY) shows a very low RMSD (root mean square deviation) value of 0.552 Å for 369 Cα atoms (Fig 2A). The ScUGPase-1 and AtUGPase structures differ in the C-terminal domain, whereas the β-strands β19 and β20 of ScUGPase-1 are replaced by a unique and longer β-strand in the AtUGPase structure (Fig 2A).
Fig 2. Structural comparison of ScUGPase-1 and AtUGPase.
(A) Superposition of monomers of ScUGPase-1 (blue) and AtUGPase (green) (PDB code: 2ICY; RMSD value of 0.552 Å for 369 Cα atoms). UDP-glucose (yellow sticks) is shown in the active site. The arrow indicates β19 and β20 of ScUGPase-1; they are replaced by a unique β-strand in the AtUGPase structure. (B) Comparison of the active site of apo-ScUGPase-1 and AtUGPase containing UDP-glucose. Residues of AtUGPase involved in the stabilization of UDP-glucose are shown as green sticks and residues of ScUGPase-1 likely important for ligand-binding are shown in blue. The active site of the apo-AtUGPase (PDB code: 1Z90) is also included (purple sticks), showing the same arrangement as for AtUGPase containing UDP-glucose.
Analysis of the active site of AtUGPase shows that the UDP-glucose molecule is bound by several residues. The uridinyl group is coordinated by residues Gln162, Gly191 and Gly87 whereas the glucose portion of the molecule is coordinated by Asn220, Gly258, Glu271, Asn293 and Leu85. The β-phosphate is stabilized by His192 and Lys256 and the α-phosphate by Lys99 (Fig 2B).
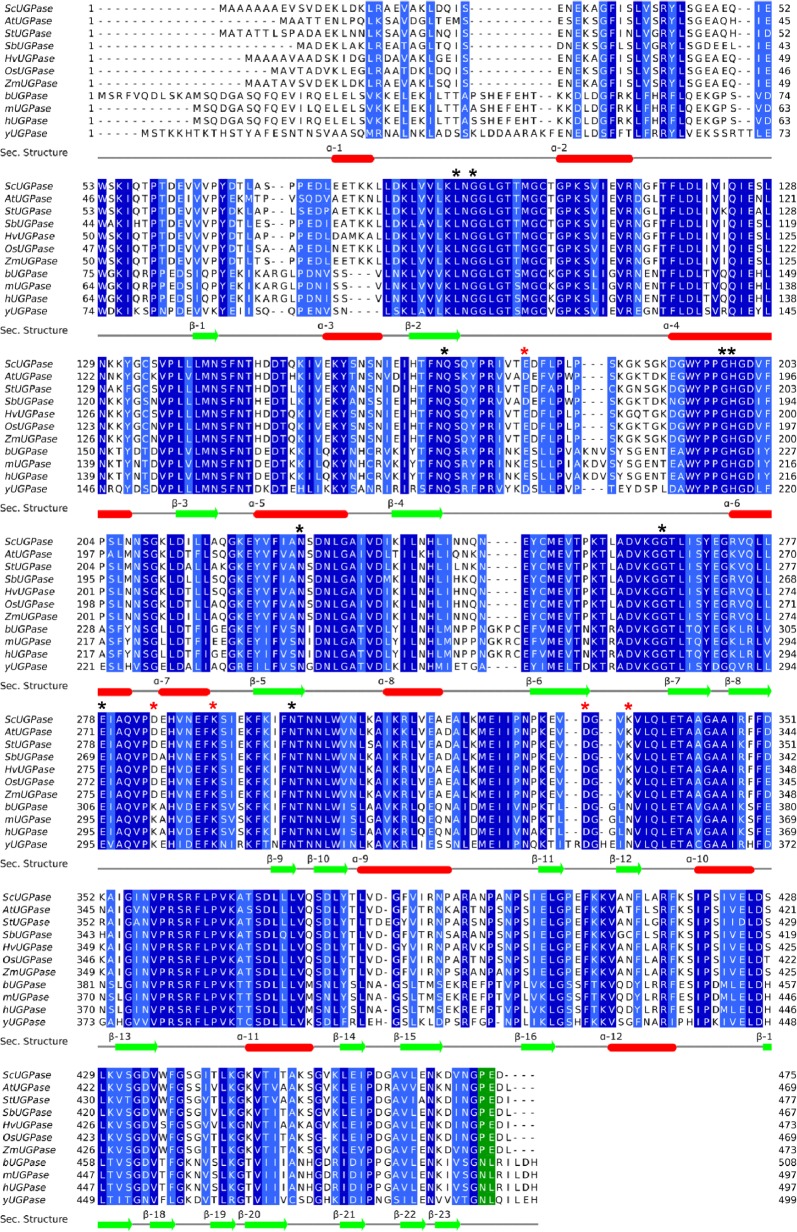
Multiple sequence alignments show that all these residues are highly conserved in all UGPase orthologs analysed (Fig 3). Structural comparison of AtUGPase and ScUGPase-1 suggests that UDP-glucose likely binds to ScUGPase-1 active site in a similar way. Hence, the uridinyl group is likely stabilized by Gly94, Gln169 and Gly198, whereas the glucose portion would be coordinated by Asn227, Gly265, Glu278, Asn300 and Leu92, though no electron density was observed for Gly265 in the structure of ScUGPase-1.
Fig 3. Multiple sequence alignment of UGPase orthologs.
Proteins were aligned by MUSCLE [21] and the alignment optimized in Jalview [22]. The aligned sequences from top to bottom with their accession numbers are: ScUGPase-1 from Saccharum spp—sugarcane (A0A075E2Q1); AtUGPase from Arabidopsis thaliana (Q9M9P3); StUGPase from Solanum tuberosum–potato (P19595); SbUGPase from Sorghum bicolor (C5XSC5); HvUGPase from Hordeum vulgare–barley (Q43772); OsUGPase from Oryza sativa–rice (Q93X08); ZmUGPase from Zea mays–maize (B6T4R3); bUGPase from bovine–Bos taurus (Q07130); mUGPase from mouse–Mus musculus (Q91ZJ5-2); hUGPase from human–Homo sapiens (Q16851-2) and yUGPase from yeast–Saccharomyces cerevisiae (P32861). Region coloured in green corresponds to the residues in the C-terminal region involved in the end-to-end interactions in the yeast and human orthologs. Black asterisks (*) indicate the residues important for ligand binding, and red asterisks (*) indicate residues involved in the dimer interface in the crystal of ScUGPase-1. Elements of secondary structure are shown based on the crystal structure of ScUGPase-1. β-sheets and α-helices are shown in green and red, respectively.
A slight difference observed in the active site of ScUGPase-1 is the orientation of the side chain of His199. In the AtUGPase structure, the side chain is orientated closer to the β-phosphate, whereas in the ScUGPase-1 structure it is orientated closer to the α-phosphate. To evaluate whether this orientation could be caused by conformational changes induced by ligand, the structure of apo-AtUGPase (PDB code: 1Z90) was also analysed (S1A Fig). Structural comparison (Fig 2B) shows that the His192 assumes the same conformation in the apo-structure as in the UDP-glucose bound-structure. The side-chain likely has to rotate to allow substrate binding. Interestingly, in contrast to plant UGPases, which do not undergo large conformational changes (S1A Fig), Leishmania major (LmUGPase) and human UGPase (hUGPase) have been reported to undergo conformational changes induced by ligands [23,26,27]. Upon substrate binding, active monomers of LmUGPase suffers two processes that lead to large conformational modification. The first one is a significant relocation of the NB loop (nucleotide binding loop) towards the ligand, whereas the second one involves a movement of the SB loop (substrate binding loop) over the sugar moiety. These two conformational changes lead to a handle-like extension formed by β9, β10 and the connecting loop that adopt very different conformations in the apo- and ligand bond UGPase. They also demonstrated that the residues at the beginning and at the end of the handle, as well as the adjacent residues perform a 12° turn toward the sugar moiety in the catalytic domain (S1 Fig). Comparison of this region in the structure of ScUGPase shows that β7 and β8 (corresponding to β9 and β10 in LmUGPase) and the connecting loop are too short to undergo similar conformational change [23,26]. On the other hand, hUGPase is known to form active octamers that limit the conformational flexibility of subunits. To overcome this limitation, hUGPase stabilizes the ligand via an intermolecular mechanism named interlock, which involves R287 of one subunit and D456 of the neighbouring subunit [26]. These differences show that UGPases have diverse mechanisms to stabilize the substrate, depending of the organism.
Oligomeric state of ScUGPase-1
The oligomeric state of UGPase has been extensively studied, since oligomerization plays a regulatory role in the activity of this protein [9]. Studies have shown that other UGPases from plants exists as a mixture of monomers, dimers and higher oligomeric forms, with the monomer as the most active form [8,9]. In vitro analyses have also demonstrated that the oligomerization of UGPase is affected by buffer composition, with phosphate and Tris buffers promoting the appearance of several oligomers of different sizes, while MOPS and HEPES lead to UGPase de-oligomerization [9].
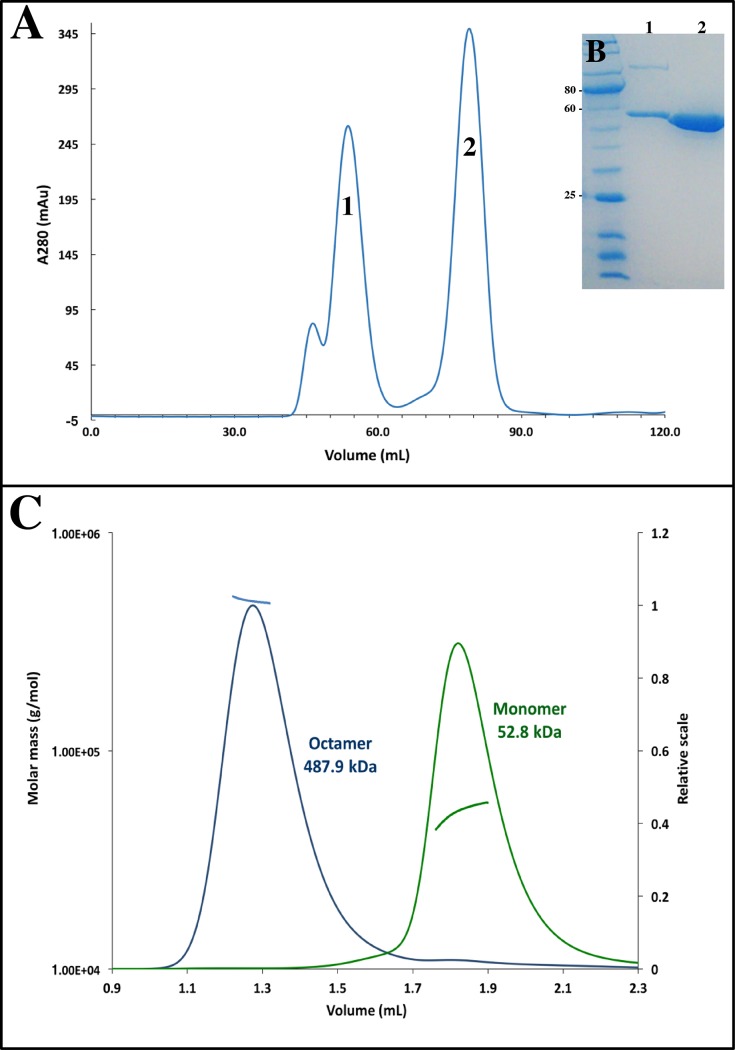
Size-exclusion chromatography conducted during the purification of ScUGPase-1, in the presence of Tris buffer, showed the presence of two main peaks (Fig 4A), indicating the existence of ScUGPase-1 as a mixture of monomers and higher oligomeric forms in solution. The crystal structure revealed the presence of two molecules per asymmetric unit, suggesting a possible dimer (Fig 2A). A putative dimer formation was also observed in the structure of AtUGPase [10], but with a different arrangement (S2 Fig).
Fig 4. Size exclusion chromatography and MALS analysis.
(A) Size exclusion chromatogram of ScUGPase-1. Peaks between 40 and 60 mL correspond presumably to higher oligomeric forms of ScUGPase-1, whereas the peak at 75 mL corresponds to the monomeric form. (B) Coomassie-stained gel under reducing conditions showing the purity of ScUGPase-1. (C) SEC-MALS analysis of ScUGPase-1. Blue and green lines indicate the trace from the refractive index detector during SEC for the octamer and monomer, respectively. Black lines on each peak correspond to the averaged molecular weight (Mw; y axis) distribution across the peak.
Analysis of the possible dimer of ScUGPase-1 found in the crystal shows that the putative dimer interface is small and stabilized by hydrogen bonds between Glu178 (monomer A) and Lys291 (monomer B); Lys335 (monomer A) and Asp284 (monomer B); and between Asp332 (monomer A) and Lys335 (monomer B). These residues are conserved in all analysed UGPase orthologs from plants (Fig 3, shown with red asterisk), however, except for Asp284, none of these residues are relevant for the putative dimer interaction observed for AtUGPase. The “Protein interface and assemblies” (PISA) server [28] has been used to access potential interfaces relevant for protein-protein interactions. It has been reported that a minimum contact area of ~ 600 Å2 is required for protein-protein complexes. Analysis of ScUGPase-1 revealed a buried surface area of 480 Å2 between monomers A and B, and a complexation significance score (CSS) of 0, indicating the corresponding dimer is unlikely to be stable. A small buried surface area of 600 Å2 was also observed for the putative dimer of AtUGPase [10].
In order to get further insights into the oligomerization properties of ScUGPase-1, we performed multi-angle light scattering (MALS) using the samples corresponding to the monomer and the larger oligomeric form peaks (Fig 4A), at concentration of 2.0 mg/mL. Analysis of the monomer peak showed a molecular mass of 52.8 kDa (± 0.9%) (Fig 4B), close to the expected mass of 56.1 kDa. On the other hand, analysis of the peak corresponding to the higher oligomeric form showed a molecular mass of 487.9 kDa (± 0.7%) (Fig 4B), which would be consistent with an octamer (theoretical mass 448.8 kDa). Octamers have been found in solution and in the crystal structure of the yeast (yUGPase) and human (hUGPase) orthologs [11,12]. The crystal structure of both proteins revealed that the C-terminal domain is essential for the dimer formation through an end-to-end arrangement, and four dimers assemble into an octamer, the fully active form of the protein (S3A and S3B Fig) [11,12]. The RMSD values between the monomer structures of ScUGPase-1 and hUGPase and yUGPase are 1.14 Å (376 Cα atoms) and 0.804 Å (399 Cα atoms), respectively, showing a high structural similarity (S4 Fig).
We could not identify in the crystal structure ScUGPase-1 any interaction among the symmetry-related molecules that are equivalent to the interactions between human and yeast molecules in the octamers. Interestingly, using small angle x-ray scattering (SAXS) analysis, Soares et al., 2014 have shown that at low concentrations (0.64 mg/mL) oxidized ScUGPase-1 exists as dimer in solution and that the dimer envelope observed suggests a dimer interface via C-terminal domain in an arrangement very similar to yUGPase and hUGPase. Yu & Zheng 2012 have reported that in humans and possibly in other higher eukaryotes, Asn491 and Leu492 are essential for dimer formation. Multiple sequence alignment shows that indeed these two residues are conserved in human, yeast, bovine and mouse proteins, but not in plant proteins, which have a proline and glutamic acid instead (Fig 3, green). Thereby, the residues and the mechanism involved in the putative end-to-end interactions in ScUGPase-1 are still unknown, but unlikely to be the same as observed in the structures from human and yeast. The role of the C-terminal domain in the oligomerization of UGPase has been investigated in barley, which shares a sequence identity of 93% with ScUGPase-1. In studies using truncated proteins, it has been demonstrated that deletion of the last eight residues of the C-terminal region not only increases the UGPase activity, but also keeps the protein mostly as a monomer, suggesting that this region may play a role in the oligomerization of UGPase protein [29].
Altogether, our results suggest the existence of ScUGPase-1 as a mixture of monomers, dimers and octamers in solution, which might adopt a similar structural arrangement as described for human and yeast orthologs. Further studies are necessary in order to get more insights about the oligomeric structure of ScUGPase-1.
Conclusions
In this study, we present the first crystal structure of UGPase-1 from sugarcane at a resolution of 2.0 Å. Structural comparisons to the UGPase from Arabidopsis thaliana (AtUGPase) reveals a high structural similarity, providing insights into the active site of ScUGPase-1. Multi-angle light scattering (MALS) results indicate the presence of a possible octamer in solution, which might be formed by four dimers through end-to-end arrangements of two monomers similar to yeast and human proteins.
Supporting information
(A) Surface representation of apo-AtUGPase (green) (PDB code: 1Z90) and AtUGPase bound to UDP-glucose (purple) (PDB code: 2ICX). RMSD value of 0.576 Å for 373 Cα atoms indicates small conformational changes induced by ligand. (B) Surface representation of LmUGPase comparing the open (brown) (PDB code: 2OEF) and closed conformation (light green) (PDB code: 2OEG). RMSD of 1.826 Å for 373 Cα atoms.
(TIF)
Superposition of the ScUGPase-1 putative dimer (blue and orange) with the dimer of AtUGPase (green and grey) (PDB code: 2ICX). RMSD value of 20.88 Å for 875 Cα atoms.
(TIF)
(A) Diagram showing the dimer formation through an end-to-end arrangement in the C-terminal. (B) Top and side view of the hUGPase octamer. The four dimers are shown in red, cyan, green and yellow. (PDB code: 3R2W).
(TIF)
(A) Superposition of ScUGPase-1 (blue) and hUGPase (red) (PDB code 3RW2) shows RMSD value of 1.14 Å over 376 Cα atoms. (B) Superposition of ScUGPase-1 (blue) with yUGPase (pink) (PDB code: 2I5K). RMSD value of 0.804 Å over 399 Cα atoms.
(TIF)
Acknowledgments
We thank Prof. Dr. Milton T. Stubbs and Dr. Christoph Parthier (Martin Luther University Halle-Wittenberg, Germany) and the University of Queensland Remote Operation Crystallization and X-ray (UQ-ROCX) for the crystallization facility used for the initial screening. We acknowledge Australian Synchrotron, the beamline staff and Dr. Thomas Ve for the support during data collection. We also thank Dr. Simon Williams for the help with the MALS analysis.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Grant 2013/15576-5 - Fundação de Amparo à Pesquisa do Estado de São Paulo - http://www.fapesp.br/ to MM; grant 2008/06767-3 - Fundação de Amparo à Pesquisa do Estado de São Paulo - http://www.fapesp.br/ to JSMS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Waclawovsky AJ, Sato PM, Lembke CG, Moore PH, Souza GM. Sugarcane for bioenergy production: an assessment of yield and regulation of sucrose content. Plant Biotechnol J. 2010;8: 263–276. doi: 10.1111/j.1467-7652.2009.00491.x [DOI] [PubMed] [Google Scholar]
- 2.Payyavula RS, Tschaplinski TJ, Jawdy SS, Sykes RW, Tuskan GA, Kalluri UC. Metabolic profiling reveals altered sugar and secondary metabolism in response to UGPase overexpression in Populus. BMC Plant Biol. 2014;14: 265 doi: 10.1186/s12870-014-0265-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibeaut DM. Nucleotide sugars and glycosyltransferases for synthesis of cell wall matrix polysaccharides. Plant Physiol Biochem. 2000;38: 69–80. doi: 10.1016/S0981-9428(00)00167-4 [Google Scholar]
- 4.Bar-Peled M, O’Neill MA. Plant Nucleotide Sugar Formation, Interconversion, and Salvage by Sugar Recycling*. Annu Rev Plant Biol. 2011;62: 127–155. doi: 10.1146/annurev-arplant-042110-103918 [DOI] [PubMed] [Google Scholar]
- 5.Amor Y, Haigler CH, Johnson S, Wainscott M, Delmer DP. A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proc Natl Acad Sci U S A. National Academy of Sciences; 1995;92: 9353–7. Available: http://www.ncbi.nlm.nih.gov/pubmed/7568131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kleczkowski LA. Glucose activation and metabolism through UDP-glucose pyrophosphorylase in plants. Phytochemistry. 1994;37: 1507–1515. doi: 10.1016/S0031-9422(00)89568-0 [Google Scholar]
- 7.Winter H, Huber SC. Regulation of Sucrose Metabolism in Higher Plants: Localization and Regulation of Activity of Key Enzymes. Crit Rev Biochem Mol Biol. 2000;35: 253–289. doi: 10.1080/10409230008984165 [DOI] [PubMed] [Google Scholar]
- 8.Martz FO, Wilczynska M, Kleczkowski LA. Oligomerization status, with the monomer as active species, defines catalytic efficiency of UDP-glucose pyrophosphorylase. Biochem J. 2002;367: 295–300. doi: 10.1042/BJ20020772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleczkowski LA, Martz F, Wilczynska M. Factors affecting oligomerization status of UDP-glucose pyrophosphorylase. Phytochemistry. 2005;66: 2815–2821. doi: 10.1016/j.phytochem.2005.09.034 [DOI] [PubMed] [Google Scholar]
- 10.McCoy JG, Bitto E, Bingman CA, Wesenberg GE, Bannen RM, Kondrashov DA, et al. Structure and Dynamics of UDP–Glucose Pyrophosphorylase from Arabidopsis thaliana with Bound UDP–Glucose and UTP. J Mol Biol. 2007;366: 830–841. doi: 10.1016/j.jmb.2006.11.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roeben A, Plitzko JM, Körner R, Böttcher UMK, Siegers K, Hayer-Hartl M, et al. Structural Basis for Subunit Assembly in UDP-glucose Pyrophosphorylase from Saccharomyces cerevisiae. J Mol Biol. 2006;364: 551–560. doi: 10.1016/j.jmb.2006.08.079 [DOI] [PubMed] [Google Scholar]
- 12.Yu Q, Zheng X. The crystal structure of human UDP-glucose pyrophosphorylase reveals a latch effect that influences enzymatic activity. Biochem J. 2012;442: 283–291. doi: 10.1042/BJ20111598 [DOI] [PubMed] [Google Scholar]
- 13.Soares JSM, Gentile A, Scorsato V, Lima ADC, Kiyota E, Dos Santos ML, et al. Oligomerization, membrane association, and in vivo phosphorylation of sugarcane UDP-glucose pyrophosphorylase. J Biol Chem. American Society for Biochemistry and Molecular Biology Inc.; 2014;289: 33364–33377. doi: 10.1074/jbc.M114.590125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. International Union of Crystallography; 2010;66: 125–32. doi: 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69: 1204–14. doi: 10.1107/S0907444913000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallographica Section D: Biological Crystallography. 2006. pp. 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams PD, Afonine P V, Bunkóczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. International Union of Crystallography; 2010;66: 213–21. doi: 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60: 2126–32. doi: 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 19.Afonine P V., Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr Sect D Biol Crystallogr. 2012;68: 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen J, Arakawa T, Philo JS. Size-Exclusion Chromatography with On-Line Light-Scattering, Absorbance, and Refractive Index Detectors for Studying Proteins and Their Interactions. Anal Biochem. Academic Press; 1996;240: 155–166. doi: 10.1006/abio.1996.0345 [DOI] [PubMed] [Google Scholar]
- 21.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32: 1792–7. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25: 1189–91. doi: 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steiner T, Lamerz A-C, Hess P, Breithaupt C, Krapp S, Bourenkov G, et al. Open and Closed Structures of the UDP-glucose Pyrophosphorylase from Leishmania major. J Biol Chem. 2007;282: 13003–13010. doi: 10.1074/jbc.M609984200 [DOI] [PubMed] [Google Scholar]
- 24.Mariño K, Güther MLS, Wernimont AK, Amani M, Hui R, Ferguson MAJ. Identification, subcellular localization, biochemical properties, and high-resolution crystal structure of Trypanosoma brucei UDP-glucose pyrophosphorylase. Glycobiology. Oxford University Press; 2010;20: 1619–30. doi: 10.1093/glycob/cwq115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toroser D, Athwal GS, Huber SC. Site-specific regulatory interaction between spinach leaf sucrose-phosphate synthase and 14-3-3 proteins. FEBS Lett. 1998;435: 110–4. Available: http://www.ncbi.nlm.nih.gov/pubmed/9755869 [DOI] [PubMed] [Google Scholar]
- 26.Führing JI, Cramer JT, Schneider J, Baruch P, Gerardy-Schahn R, Fedorov R. A Quaternary Mechanism Enables the Complex Biological Functions of Octameric Human UDP-glucose Pyrophosphorylase, a Key Enzyme in Cell Metabolism. Sci Rep. 2015;5: 9618 doi: 10.1038/srep09618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Führing J, Cramer JT, Routier FH, Lamerz A-C, Baruch P, Gerardy-Schahn R, et al. Catalytic Mechanism and Allosteric Regulation of UDP-Glucose Pyrophosphorylase from Leishmania major. ACS Catal. American Chemical Society; 2013;3: 2976–2985. doi: 10.1021/cs4007777 [Google Scholar]
- 28.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372: 774–97. doi: 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 29.Meng M, Fitzek E, Gajowniczek A, Wilczynska M, Kleczkowski LA. Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley UDP-glucose pyrophosphorylase. Biochim Biophys Acta—Proteins Proteomics. 2009;1794: 1734–1742. doi: 10.1016/j.bbapap.2009.08.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Surface representation of apo-AtUGPase (green) (PDB code: 1Z90) and AtUGPase bound to UDP-glucose (purple) (PDB code: 2ICX). RMSD value of 0.576 Å for 373 Cα atoms indicates small conformational changes induced by ligand. (B) Surface representation of LmUGPase comparing the open (brown) (PDB code: 2OEF) and closed conformation (light green) (PDB code: 2OEG). RMSD of 1.826 Å for 373 Cα atoms.
(TIF)
Superposition of the ScUGPase-1 putative dimer (blue and orange) with the dimer of AtUGPase (green and grey) (PDB code: 2ICX). RMSD value of 20.88 Å for 875 Cα atoms.
(TIF)
(A) Diagram showing the dimer formation through an end-to-end arrangement in the C-terminal. (B) Top and side view of the hUGPase octamer. The four dimers are shown in red, cyan, green and yellow. (PDB code: 3R2W).
(TIF)
(A) Superposition of ScUGPase-1 (blue) and hUGPase (red) (PDB code 3RW2) shows RMSD value of 1.14 Å over 376 Cα atoms. (B) Superposition of ScUGPase-1 (blue) with yUGPase (pink) (PDB code: 2I5K). RMSD value of 0.804 Å over 399 Cα atoms.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.