Abstract
BACKGROUND AND PURPOSE:
Prior MR imaging studies, primarily at 1.5T, established hippocampal atrophy as a biomarker for Alzheimer disease. 3T MR imaging offers a higher contrast and signal-to-noise ratio, yet distortions and intensity uniformity are harder to control. We applied our automated hippocampal segmentation technique to 1.5T and 3T MR imaging data, to determine whether hippocampal atrophy detection was enhanced at 3T.
MATERIALS AND METHODS:
We analyzed baseline MR imaging data from 166 subjects from the Alzheimer's Disease Neuroimaging Initiative-1 (37 with Alzheimer disease, 76 with mild cognitive impairment, and 53 healthy controls) scanned at 1.5T and 3T. Using multiple linear regression, we analyzed the effect of clinical diagnosis on hippocampal radial distance, while adjusting for sex. 3D statistical maps were adjusted for multiple comparisons by using permutation-based statistics at a threshold of P < .01.
RESULTS:
Bilaterally significant radial distance differences in the areas corresponding to the cornu ammonis 1, cornu ammonis 2, and subiculum were detected for Alzheimer disease versus healthy controls and mild cognitive impairment versus healthy controls at 1.5T and more profoundly at 3T. Comparison of Alzheimer disease with mild cognitive impairment did not reveal significant differences at either field strength. Subjects who converted from mild cognitive impairment to Alzheimer disease within 3 years of the baseline scan versus nonconverters showed significant differences in the area corresponding to cornu ammonis 1 of the right hippocampus at 3T but not at 1.5T.
CONCLUSIONS:
While hippocampal atrophy patterns in diagnostic comparisons were similar at 1.5T and 3T, 3T showed a superior signal-to-noise ratio and detected atrophy with greater effect size compared with 1.5T.
Alzheimer disease (AD), the most common form of dementia in the elderly, is characterized by progressive cognitive and functional decline. AD is increasingly recognized as one of the most important medical problems facing society today. In the United States alone, approximately 5.2 million people have AD. This number is predicted to rise to 13.8 million by 2050.1 Worldwide, an estimated 35.6 million people lived with dementia in 2010. The worldwide prevalence is expected to reach 65.7 million diagnosed cases in 2030, and 115.4 million, in 2050.2
At the tail end of the cognitive spectrum, AD is often preceded by mild cognitive impairment (MCI), an intermediate stage between normal aging and dementia. Patients with MCI show a milder degree of cognitive impairment and preserved ability for day-to-day activities.3 Patients with MCI are at an increased risk for developing AD. Identifying patients during this prodromal stage allows early interventions. Noninvasive biomarkers for detecting prodromal AD are greatly needed.4
Hippocampal atrophy is the most established structural AD imaging biomarker to date. Hippocampal volume shrinkage is associated with AD-like postmortem hippocampal neuronal loss5,6 and neurofibrillary tangle deposition.7 Atrophy of the cornu ammonis (CA) 1 subfield is characteristic of both AD and MCI.8–11 Greater involvement of CA1 is a risk factor for conversion from MCI to AD.9,10
High-resolution MR imaging has helped us visualize subtle anatomic changes in the brain in the initial stages of disease.12,13 State-of-the-art analytic techniques helped us identify, quantify, and track AD-associated atrophy with great precision. The hippocampus, an area involved early in AD, has been studied via visual rating scales,14–16 regional volumetric analysis,10,11,17–19 and shape-deformation methods.9,20–23 The early studies have used 1.5T, and some of the recent ones have used higher magnetic field strengths. Few studies have compared atrophy detection between 1.5T and 3T field-strength scans.24–27 One tensor-based morphometry whole-brain study reported that analyses of 1.5T and 3T Alzheimer's Disease Neuroimaging Initiative (ADNI) data from the same subjects did not differ statistically in detecting neurodegenerative changes during 12 months.26 Hippocampal volumes extracted from the same study cohort at 1.5T and 3T showed only 3.2% test-retest variability.25 Similar results were reported for manual segmentations in 8 healthy subjects with 1.5T and 3T scans in an epilepsy study.28
Using the ADNI-1 dataset, we sought to directly compare 1.5T and 3T segmentations and to determine how well each magnetic field strength detects anatomic differences between subjects who are cognitively healthy (NC) or have MCI and AD as well as between subjects with MCI who converted from MCI to AD in 3 years (MCI converters) and those who did not (MCI nonconverters). Although distortions and intensity uniformity may be more difficult to control at 3T, 3T MR imaging offers higher contrast and signal-to-noise ratio. We hypothesized that higher field MR imaging will offer greater power to detect hippocampal atrophy in the cognitive spectrum from normal aging to AD.
Materials and Methods
Subjects
ADNI-129–32 is a longitudinal multisite observational study, which started in 2004 and collected cognitive, imaging, and biomarker data from 200 NC subjects, 400 with MCI, and 200 with AD. The goals of ADNI are to determine relationships among clinical, cognitive, imaging, genetic, and biochemical biomarkers as AD evolves from normal aging to MCI to dementia, to discover biomarkers sensitive to early diagnosis, to establish standardized methods for imaging/biomarker collection, and to conduct research on methodologies that could lower the costs of clinical trials.
Our sample consisted of the 187 (22%) of the 842 subjects in ADNI-1 who underwent both 1.5T and 3T MR imaging. 1.5T and 3T brain baseline MR images and their associated clinical data were downloaded from the ADNI public data base (http://adni.loni.usc.edu). The cognitive variables we used to clinically characterize the research cohorts are the global Clinical Dementia Rating (CDR), sum-of-boxes Clinical Dementia Rating,33,34 and the Mini-Mental State Examination (MMSE).35,36 These are the 3 most commonly used global scales to gauge disease severity. The global CDR reflects the overall severity of dementia in which global CDR scores of 0, 0.5, 1, 2, and 3, indicate no cognitive decline, questionable cognitive decline, and very mild, mild, moderate, or severe dementia, respectively. The sum-of-boxes Clinical Dementia Rating ranges from 0 to 18 and reflects clinical deterioration in memory, orientation, judgment and problem solving, community affairs, home and hobbies, and personal care. The MMSE is a 30-point global cognitive screening instrument that evaluates orientation, registration, attention, calculation, recall, and language. All patients with AD met the National Institute of Neurological Disorders and Stroke/Alzheimer's Disease and Related Disorders Association criteria for probable AD.37 Subjects with MCI met the criteria of Petersen et al for MCI.3 Our MCI converter-versus-nonconverter analyses included only subjects with follow-up data for 36 months after baseline (36 were MCI converters, and 31 were MCI nonconverters).
Imaging Data Collection, Image Preprocessing, and Analysis
We acquired 1.5T and 3T scans analyzed in this study from 31 ADNI-1 sites on scanners from 3 MR imaging vendors (GE Healthcare, Milwaukee, Wisconsin; Philips Healthcare, Best, the Netherlands; or Siemens, Erlangen, Germany) with a standardized protocol developed to evaluate 3D T1-weighted sequences for morphometric analyses (http://adni.loni.usc.edu/methods/documents/mri-protocols/).31,38 Participants with structural abnormalities such as hemispheric infarctions, white matter ischemic changes, and focal lesions were excluded from participation.31 Of the 187 subjects who were imaged at both 1.5T and 3T, 111 (59%) subjects were scanned on GE Healthcare, 24 (13%) on a Phillips Healthcare, and 52 (28%) on Siemens 1.5T scanners. At 3T, 29 (15%) subjects were scanned on GE Healthcare, 50 (27%) were scanned on Phillips Healthcare, and 108 (58%), on Siemens scanners.
The 1.5T and 3T scanning protocols used a 3D sagittal volumetric sequence. The typical 1.5T acquisition parameters were TR = 2400 ms, minimum full TE, TI = 1000 ms, flip angle = 8°, FOV = 24 cm, with a 256 × 256 × 170 acquisition matrix in the x-, y-, and z-dimensions, yielding a voxel size of 1.25 × 1.25 × 1.2 mm3. For 3T scans, the typical parameters were a TR = 2300 ms, minimum full TE, TI = 900 ms, flip angle = 8°, FOV = 26 cm, with a 256 × 256 × 170 acquisition matrix in the x-, y-, and z-dimensions, yielding a voxel size of 1.0 × 1.0 × 1.2 mm3. The 3T protocol included an increased receiver bandwidth to compensate for the increase in chemical shift and susceptibility artifacts.31 For a detailed description of the acquisition parameters for the various scanner manufacturers, please see Jack et al (2008).31
The ADNI MR imaging core at the Mayo Clinic preprocessed the data by using Gradwarp (GE Healthcare) for correction of geometric distortion due to gradient nonlinearity,39 “B1-correction” for adjusting image intensity inhomogeneity,31 “N3” bias field correction for reducing residual intensity inhomogeneity,40 and geometric scaling for adjusting scanner- and session-specific calibration errors.31 The raw and preprocessed data are stored at the Laboratory of Neuroimaging at the University of Southern California and are available to the general scientific community for download (http://adni.loni.usc.edu).
MR images were automatically registered by using a 9-parameter transformation to the ICBM53 template.41 The images were resampled in an isotropic space of 220 voxels along x-, y- and z-axes with a final voxel size of 1 mm3.
For each field strength, a minimal deformation target was constructed from 9-parameter linearly aligned scans.42 The minimal deformation target is an unbiased average template image created to represent common features for a group of subjects. This helps to reduce bias by using a template that deviates least from the anatomy of the subjects and to improve statistical power by using a customized template. For this study, 1.5T and 3T baseline scans were nonlinearly aligned to the minimal deformation target developed from 1.5T and 3T images of the NC group.
Two experienced raters manually traced the hippocampal formations of 21 randomly selected subjects (7 NCs, 7 with MCI, and 7 with AD) at 1.5T and 3T by using our in-house hippocampal tracing protocol43 (intrarater reliability, Cronbach α = 0.97; interrater reliability, Cronbach α = 0.9). Traces included the hippocampus proper, dentate gyrus, and subiculum as previously described.44 Tracers were blinded to subject demographics, diagnosis, and the study objective. Anatomic landmarks were followed in all 3 orthogonal viewing planes by using interactive segmentation software. The hippocampi were traced on contiguous coronal sections following a detailed well-established protocol with high intra- and interrater reliability.45 When boundaries were ambiguous, standard neuroanatomic atlases were consulted.46,47 Traces included the whole hippocampal head, body, and tail.
Hippocampal segmentation used AdaBoost, our automated machine-learning hippocampal segmentation algorithm, based on a statistical method called “Adaptive Boosting.”48 The technique has been previously described in detail in several publications.49,50 Briefly, AdaBoost uses a training set (ie, a small number of representative images) to develop classification rules for hippocampal-versus-nonhippocampal tissue. Our training set for this study consisted of 21 subjects, 7 NCs, 7 with MCI, and 7 with AD, who were scanned at both 1.5T and 3T. AdaBoost analyzes the specific feature information contained in the positive and negative voxels (ie, those belonging and not belonging to the structure of interest) of the training dataset and develops segmentation rules based on the optimal combination of features. AdaBoost uses ∼18,000 local features, such as image gradients, local curvature of image interfaces, tissue classification as gray or white matter, and statistical information on the likely stereotaxic position of the hippocampus. In the training phase, the algorithm applies mathematic approaches from the fields of machine learning and computer vision51 to estimate the optimal weighting of these features in a mathematic formula that computes the probability of any given voxel for being inside the hippocampus. AdaBoost performance has been previously validated and has been found to agree with human raters as well as human raters agree with each other (ie, similar to interrater reliability).49 After statistical rules for hippocampal segmentation are developed, the algorithm is tested in a training set and then applied to the full sample. All segmentations were visually inspected to make sure they appropriately captured the hippocampal anatomy at the given magnetic field strength.
We used an anatomic mesh modeling method called “hippocampal radial distance”44 to match equivalent hippocampal surface points, obtained from the AdaBoost hippocampal segmentations, across subjects. To match the digitized points representing the hippocampus surface traces in each brain volume, we made the AdaBoost contours spatially uniform by modeling them as a 3D parametric surface mesh.44 This procedure allows statistical comparisons in 3D and averaging of hippocampal surface morphology across all individuals belonging to a group. Next, to assess the pattern of regional hippocampal atrophy, a medial core, threading down the center of the hippocampus, was computed for each individual. Radial distance was assessed by measuring the distances from the hippocampal surface points to the medial core of the individual's hippocampal surface model.44 Change in radial distance with time can, therefore, capture localized atrophy. This methodology assures that all hippocampal surfaces are represented by using the same parametric mesh structure. This allows corresponding surface traces and the associated distance measures to be matched across subjects and/or time and averaged across diagnostic groups. Distance fields indexing local expansions or contractions in hippocampal surface morphology can thus be compared statistically between groups.
An ROI was drawn over the lateral hippocampal surface (ie, the area corresponding to CA1) on the 3D hippocampal radial distance model, and the CA1 mean radial distance was computed for each subject at both 1.5T and 3T.
Statistical Methods
We used ANOVA with post hoc Bonferroni correction for multiple comparisons to examine between-group diagnostic differences in age, education, MMSE, CDR, and sum-of-boxes Clinical Dementia Rating. A χ2 test was used to determine differences in sex distribution.
The effect of diagnosis on hippocampal radial distance was studied by using linear regression while correcting for demographic variables that showed significant between-group differences. Our 3D statistical maps were adjusted for multiple comparisons by using permutation-based statistics with a threshold of P < .01. A Student t test was performed for the direct comparison of 1.5T- and 3T-derived radial distance in each diagnostic group.
The observed CA1 regional differences between MCI converters and nonconverters at 3T but not at 1.5T (see “Results”) were further examined by using receiver operating characteristic curves. We studied how well the mean CA1 radial distance (in millimeters) discriminated between MCI converters and nonconverters at 3T and 1.5T. We used the Mann-Whitney Wilcoxon test for independent paired samples to statistically compare the sum of the ranks for the mean CA1 radial distance at 1.5T versus 3T and the DeLong partial area under the curve test to statistically compare the area under the curve of the 2 receiver operating characteristic curves.
Results
Demographics
Twenty-one 3T MR imaging scans failed the automated hippocampal segmentation process and were excluded from further analyses. Thus, our final sample consisted of 166 subjects in ADNI-1 (53 NCs, 76 with MCI, and 37 with AD). Demographic data are shown in the Table. Differences in sex distribution were found (P = .002), with the MCI group having significantly more men than women compared with the NC and AD groups. Sex was included as a covariate in the radial distance multiple regression model. As expected, the NC group had the highest MMSE score (29.3 ± 0.9) compared with MCI (26.8 ± 1.9) and AD groups (23.3 ± 2.1) (P < .0001). NC had the lowest CDR global score (0 ± 0), followed by MCI (0.50 ± 0) and AD (0.69 ± 0.25) (P < .0001). The sum-of-boxes Clinical Dementia Rating followed a similar trend: NC (1.0 ± 0.2), MCI (2.8 ± 1.0), and AD (5.1 ± 1.5) (P < .0001).
Demographic characteristics for the diagnostic-comparisons study
| Variable | NC (n = 53)a | MCI (n = 76)a | AD (n = 37)a | P Value |
|---|---|---|---|---|
| Age (yr) | 75.4 ± 4.7 (60–87) | 75.2 ± 8.2 (55–88) | 74.1 ± 8.7 (57–91) | .671 |
| Education (yr) | 16.1 ± 2.8 (7–20) | 15.9 ± 3.2 (6–20) | 14.6 ± 3.2 (7–20) | .055 |
| Sex, M/F | 18:35 | 47:29 | 13:24 | .002b |
| MMSE | 29.3 ± 0.9 (26–30) | 26.8 ± 1.9 (23–30) | 23.3 ± 2.1 (20–27) | <.0001b |
| CDR global | 0 ± 0 (0) | 0.50 ± 0 (1) | 0.69 ± 0.25 (1) | <.0001b |
| CDR sum of boxes | 1.0 ± 0.2 (1–2) | 2.8 ± 1.0 (2–6) | 5.1 ± 1.5 (3–9) | <.0001b |
| 1.5T Left hippocampus (mm3) | 3859 ± 436 (2707–4730) | 3335 ± 588 (1730–4827) | 3242 ± 615 (1573–4302) | <.0001b |
| 1.5T Right hippocampus (mm3) | 3734 ± 591 (2114–5441) | 3254 ± 641 (850–4796) | 3110 ± 668 (613–4814) | <.0001b |
| 3T Left hippocampus (mm3) | 3897 ± 758 (408–5087) | 3259 ± 601 (1627–4697) | 3123 ± 598 (2013–4437) | <.0001b |
| 3T Right hippocampus (mm3) | 3981 ± 612 (1468–5158) | 3389 ± 638 (1800–5053) | 3237 ± 672 (1873–4577) | <.0001b |
Data are means and ranges.
Significant.
Hippocampal Regional Volume Differences
Predictably, NC had the largest hippocampal volumes, and the AD group had the smallest (Table). Mean hippocampal volumes at 1.5T were significantly different among NC, MCI, and AD both on the left and the right (ANOVA, P < .0001). Between-group differences after Bonferroni correction for multiple comparisons were significant for NC versus MCI (left and right, P < .0001) and NC versus AD (left and right, P < .0001), but they were not significant when comparing MCI with AD (left, P = l; right, P = .77). 3T-derived left hippocampal volumes showed significant differences among NC, MCI, and AD (ANOVA, P < .0001). 3T-derived hippocampal volumes were significantly different between NC and MCI (left and right, P < .0001) and NC and AD (left and right, P < .0001) but not between MCI versus AD (left, P = .90; right, P = .71).
We used a paired-samples t test to directly compare 1.5T versus 3T hippocampal volumes. In the pooled sample, we observed significantly larger right (P = .03) but not left (P = .50) hippocampal volumes at 3T compared with 1.5T. The intraclass correlation coefficient between 1.5T and 3T volumes was 0.89, and the absolute mean volumetric difference was 1.8%. Within diagnostic groups, significant volumetric differences were seen in the NC group on the right (right, P = .04; left, P = .43) but not in the MCI (right, P = .20; left, P = .43) or AD groups (right, P = .42; left, P = .40).
Surface-Mapping Atrophy Patterns
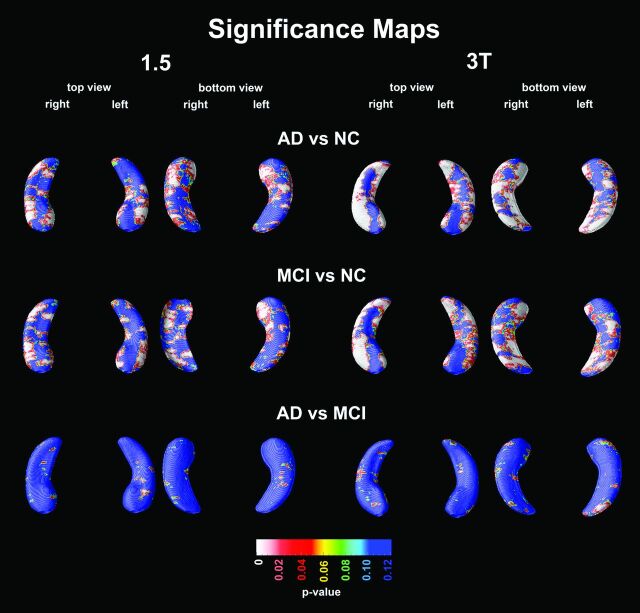
Bilaterally significant atrophy in the CA1, CA2, and subiculum regions was detected for AD versus NC at both field strengths but was more extensive at 3T (1.5T: left Pcorrected = .0015, right Pcorrected = .0015; 3T: left Pcorrected = .0001, right Pcorrected = .0001; Fig 1). The pattern of atrophy seen in MCI compared with NC was similar and was similarly more extensive at 3T (1.5T: left Pcorrected = .0019, right Pcorrected = .0020; 3T: left Pcorrected = .0008, right Pcorrected = .0001). No significant differences in radial distance between AD and MCI were found (1.5T: left Pcorrected = .49, right Pcorrected = 1.0; 3T: left Pcorrected = .31, right Pcorrected = .65).
Fig 1.
3D statistical maps of diagnostic comparisons by using 1.5T and 3T. Red and white areas in the significance maps correspond to P < .05.
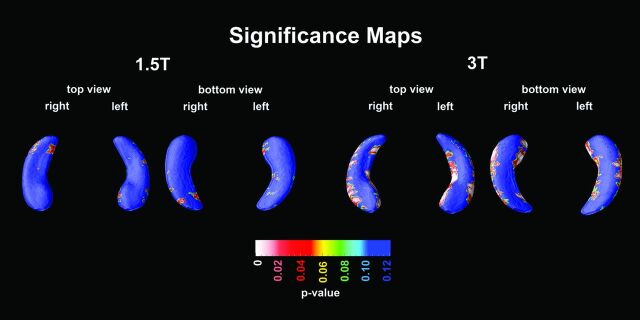
The hippocampal radial distance of MCI converters versus MCI nonconverters was not significantly different at 1.5T (left Pcorrected = .80, right Pcorrected = .74), yet at 3T, we observed differences in the CA1 radial distance that were significant on the right (right Pcorrected = .01) and trend-significant on the left (left Pcorrected = .06, Fig 2). Mann-Whitney Wilcoxon comparison of mean CA1 radial distance between MCI converters and nonconverters showed significant differences at 3T (P = .003) and trend-significance at 1.5T (P = .098).
Fig 2.
3D statistical maps for MCI converters versus nonconverters at 1.5T and 3T.
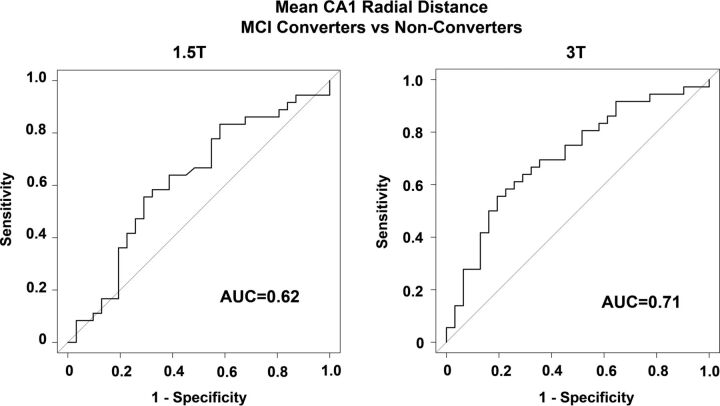
The receiver operating characteristic analyses examining the ability of mean CA1 radial distance to discriminate MCI converters and nonconverters resulted in an area under the curve of 0.62 at 1.5T and 0.71 at 3T (Fig 3). While the absolute difference in the area under the curve between the 2 field strengths (Δ area under the curve = 0.09) was relatively large, it was not statistically significant (DeLong test, Z = 1.5361, P = .125).
Fig 3.
Receiver operator characteristic curves of the ability of mean CA1 radial distance to differentiate between MCI converters versus nonconverters at 1.5T and 3T. AUC indicates area under the curve.
Discussion
The bilateral atrophy in the CA1, CA2, and subiculum regions for NC versus AD and NC versus MCI in this present study agrees with findings in prior studies.9–11,20,52–54 At 3T, MCI converters versus nonconverters presented significant hippocampal atrophy along the lateral and medial edges of the hippocampus corresponding to the CA1 and subicular regions—in concurrence with prior reports by our group based on independent samples.10
To date, several groups have compared the performance of 1.5T with 3T for detecting brain atrophy. In a strictly signal-to-noise comparison study, Fushimi et al24 reported no significant differences in signal-to-noise ratios between 1.5T and 3T in multisection images with a 0-mm gap. Applying tensor brain morphometry and Structural Image Evaluation Using Normalization of Atrophy algorithms to ADNI-1 1.5T and 3T data, Ho et al26 reported no significant advantage of 3T over 1.5T for detecting 1-year whole-brain atrophy rates. Perhaps most importantly, 2 groups compared the performance of their automated multiatlas hippocampal segmentation techniques at 1.5T and 3T in small samples derived from ADNI-1. Lötjönen et al25 used a sample consisting of 10 NCs, 10 subjects with MCI, and 10 subjects with AD and reported an intraclass correlation coefficient of 0.98 and an absolute volumetric difference between 1.5T and 3T of 3.2%. Macdonald et al,27 by using 18 subjects with AD and 18 NCs from ADNI-1, reported an intraclass correlation coefficient of 0.97 between 1.5T and 3T hippocampi segmented with their hippocampal multiatlas propagation and segmentation algorithm. Here by using 166 subjects with ADNI-1 imaged at 1.5T and 3T, we report an intraclass correlation coefficient of 0.89 and a mean volumetric difference between 1.5T and 3T measurements across the sample of 1.8%. In addition, here we reveal that 3T seems to show an advantage in detecting subtle region-specific morphometric changes over 1.5T.
The strengths of this study include the large sample size from many ADNI sites across the country, standardized MR imaging protocol, systematic postacquisition analysis, and quality control of phantom-based monitoring of all scanners. Some limitations resulted from the susceptibility-induced geometric distortion and signal losses, which could increase noise, especially in the temporal lobe regions at 3T. Additionally, higher field-strength imaging is more susceptible to chemical shift artifacts.55,56 Despite these limitations, 3T proved superior to 1.5T for shape analysis and between-group comparisons.
For this study, we had access to only 1.5T and 3T data. It is possible that even higher magnetic field57,58 strengths such as 7T or 11T may reveal finer scale features and provide superior power for detecting between-group differences. The future directions of this study include conducting longitudinal analyses to show the progressive atrophy that occurs with the disease with time.
Conclusions
Our 1.5T and 3T hippocampal volumes obtained after automated segmentation with the AdaBoost algorithm were not significantly different between MCI and AD but were significant in NC versus MCI and NC versus AD, with the 3T segmentations resulting in larger mean hippocampal volumes in the NC group. This difference could potentially be explained by the larger CSF pool around the hippocampal formation in subjects with atrophy, which provides good contrast for hippocampal versus extrahippocampal tissue discriminability even at 1.5T—this provides a comparable segmentation in MCI and AD but not in NCs who lack large CSF pools surrounding the hippocampus in the temporal horn. Our data suggest that 3T images, with their higher contrast and higher signal-to-noise ratio, may enhance the topographic localization of atrophy.
Acknowledgments
Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative data base (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
The ADNI was launched in 2003 by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration, private pharmaceutical companies, and nonprofit organizations, as a $60 million, 5-year public-private partnership. The primary goal of the ADNI has been to test whether serial MR imaging, positron-emission tomography, other biologic markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment and early Alzheimer disease. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians in developing new treatments and to monitor their effectiveness, as well as lessen the time and cost of clinical trials.
The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California, San Francisco. The ADNI is the result of effort of many coinvestigators from a broad range of academic institutions and private corporations, and subjects have been recruited from >50 sites across the United States and Canada. The initial goal of ADNI was to recruit 800 subjects, but ADNI has been followed by ADNI–Grand Opportunity and ADNI-2. To date, these 3 protocols have recruited >1500 adults, 55–90 years of age, to participate in the research, consisting of cognitively healthy older individuals, those with early or late MCI, and those with early AD. The follow-up duration of each group is specified in the protocols for ADNI-1, ADNI-2, and ADNI–Grand Opportunity. Subjects originally recruited for ADNI-1 and ADNI–Grand Opportunity had the option of being followed in ADNI-2. For up-to-date information, see www.adni-info.org.
Data collection and sharing for this project were funded by the Alzheimer's Disease Neuroimaging Initiative (National Institutes of Health Grant U01 AG024904) and the Department of Defense ADNI (Department of Defense award number W81XWH-12–2–0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer's Association; Alzheimer's Drug Discovery Foundation; BioClinica; Biogen Idec; Bristol-Myers Squibb; Eisai; Elan Pharmaceuticals; Eli Lilly; F. Hoffmann-La Roche and its affiliated company Genentech; GE Healthcare; Innogenetics; IXICO; Janssen Alzheimer Immunotherapy Research & Development; Johnson & Johnson Pharmaceutical Research & Development; Medpace; Merck & Co; Meso Scale Diagnostics; NeuroRx Research; Novartis Pharmaceuticals; Pfizer; Piramal Imaging; Servier; Synarc; and Takeda Pharmaceutical. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
ABBREVIATIONS:
- AD
Alzheimer disease
- ADNI
Alzheimer's Disease Neuroimaging Initiative
- CA
cornu ammonis
- CDR
Clinical Dementia Rating
- MCI
mild cognitive impairment
- MMSE
Mini-Mental State Examination
- NC
healthy control
Footnotes
Disclosures: Paul M. Thompson—RELATED: Grant: We have an ADNI grant to cover costs of MRI processing.* David A. Elashoff—RELATED: Grant: National Institutes of Health. Clifford R. Jack—RELATED: Grant: National Institutes of Health*; UNRELATED: Consultancy: Janssen.* Michael Weiner—Scientific Advisory Boards: 2013–2014, Pfizer, Eli Lilly; Consulting: 2013, Alzheimer's Drug Discovery Foundation, Avid Radiopharmaceuticals, Eli Lilly, ClearView Healthcare Partners, Perceptive Informatics, Smartfish AS, Decision Resources, Araclon, Synarc, Merck, Genentech, Defined Health; Funding for Travel: 2013, Kenes International, Alzheimer's Disease Research Center University of California, San Diego, University of California, Los Angeles, University of California, San Diego, Sanofi-Aventis Groupe, University Center Hospital Toulouse, Araclon, AC Immune, Eli Lilly, New York Academy of Sciences; 2014, National Brain Research Center, India (for Johns Hopkins Medicine, Baltimore); Editorial Advisory Boards: Alzheimer's and Dementia, MRI; Research Support from Government Entities: Department of Defense, Veterans Administration; Organizations Contributing to the Foundation for National Institutes of Health and Thus to the National Imaging Associates–Funded Alzheimer's Disease Neuroimaging Initiative: Abbott, Alzheimer's Association, Alzheimer's Drug Discovery Foundation, Anonymous Foundation, AstraZeneca, Bayer Healthcare, BioClinica (ADNI 2), Bristol-Myers Squibb, Cure Alzheimer's Fund, Eisai, Elan, Gene Network Sciences, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson & Johnson, Eli Lilly, Medpace, Merck, Novartis, Pfizer, Roche, Schering Plough, Synarc, Wyeth. Liana G. Apostolova—RELATED: Grant: National Institutes of Health R01 AG040770,* National Imaging Associates K23 AG026803,* National Imaging Associates P50 AG16570,* National Imaging Associates U01AG024904*; UNRELATED: Consultancy: Eli Lilly; Payment for Lectures (including service on Speakers Bureaus): Eli Lilly; Payment for Development of Educational Presentations: Eli Lilly. *Money paid to the institution.
Research was conducted at Department of Neurology, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, California.
This work was generously supported by the Easton Consortium for Alzheimer Drug Discovery and Biomarkers, National Institutes of Health R01 AG040770, National Imaging Associates K23 AG026803, National Imaging Associates P50 AG16570, National Institute of Biomedical Imaging and Bioengineering EB008561, National Institute of Biomedical Imaging and Bioengineering EB01651, National Library of Medicine LM05639, National Center for Research Resources RR019771 and National Institutes of Health RR021813, and the ADNI National Imaging Associates U01AG024904.
REFERENCES
- 1. Alzheimer's Association. 2013 Alzheimer's disease facts and figures. Alzheimers Dement 2013;9:208–45 [DOI] [PubMed] [Google Scholar]
- 2. Wortmann M. Dementia: a global health priority—highlights from an ADI and World Health Organization report. Alzheimers Res Ther 2012;4:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Petersen RC, Doody R, Kurz A, et al. Current concepts in mild cognitive impairment. Arch Neurol 2001;58:1985–92 [DOI] [PubMed] [Google Scholar]
- 4. Appel J, Potter E, Shen Q, et al. A comparative analysis of structural brain MRI in the diagnosis of Alzheimer's disease. Behav Neurol 2009;21:13–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bobinski M, de Leon MJ, Wegiel J, et al. The histological validation of post mortem magnetic resonance imaging-determined hippocampal volume in Alzheimer's disease. Neuroscience 2000;95:721–25 [DOI] [PubMed] [Google Scholar]
- 6. Zarow C, Vinters HV, Ellis WG, et al. Correlates of hippocampal neuron number in Alzheimer's disease and ischemic vascular dementia. Ann Neurol 2005;57:896–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Csernansky JG, Hamstra J, Wang L, et al. Correlations between antemortem hippocampal volume and postmortem neuropathology in AD subjects. Alzheimer Dis Assoc Disord 2004;18:190–95 [PubMed] [Google Scholar]
- 8. Frisoni GB, Ganzola R, Canu E, et al. Mapping local hippocampal changes in Alzheimer's disease and normal ageing with MRI at 3 Tesla. Brain 2008;131:3266–76 [DOI] [PubMed] [Google Scholar]
- 9. Apostolova LG, Thompson PM, Green AE, et al. 3D comparison of low, intermediate, and advanced hippocampal atrophy in MCI. Hum Brain Mapp 2010;31:786–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Apostolova LG, Dutton RA, Dinov ID, et al. Conversion of mild cognitive impairment to Alzheimer disease predicted by hippocampal atrophy maps. Arch Neurol 2006;63:693–99 [DOI] [PubMed] [Google Scholar]
- 11. Apostolova LG, Dinov ID, Dutton RA, et al. 3D comparison of hippocampal atrophy in amnestic mild cognitive impairment and Alzheimer's disease. Brain 2006;129:2867–73 [DOI] [PubMed] [Google Scholar]
- 12. Mueller SG, Laxer KD, Barakos J, et al. Subfield atrophy pattern in temporal lobe epilepsy with and without mesial sclerosis detected by high-resolution MRI at 4 Tesla: preliminary results. Epilepsia 2009;50:1474–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mueller SG, Chao LL, Berman B, et al. Evidence for functional specialization of hippocampal subfields detected by MR subfield volumetry on high resolution images at 4 T. Neuroimage 2011;56:851–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Scheltens P, Launer LJ, Barkhof F, et al. Visual assessment of medial temporal lobe atrophy on magnetic resonance imaging: interobserver reliability. J Neurol 1995;242:557–60 [DOI] [PubMed] [Google Scholar]
- 15. Visser PJ, Verhey FR, Hofman PA, et al. Medial temporal lobe atrophy predicts Alzheimer's disease in patients with minor cognitive impairment. J Neurol Neurosurg Psychiatry 2002;72:491–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Korf ES, Wahlund LO, Visser PJ, et al. Medial temporal lobe atrophy on MRI predicts dementia in patients with mild cognitive impairment. Neurology 2004;63:94–100 [DOI] [PubMed] [Google Scholar]
- 17. Jack CR Jr, Petersen RC, Xu YC, et al. Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology 1999;52:1397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Devanand DP, Pradhaban G, Liu X, et al. Hippocampal and entorhinal atrophy in mild cognitive impairment: prediction of Alzheimer disease. Neurology 2007;68:828–36 [DOI] [PubMed] [Google Scholar]
- 19. Mueller SG, Schuff N, Yaffe K, et al. Hippocampal atrophy patterns in mild cognitive impairment and Alzheimer's disease. Hum Brain Mapp 2010;31:1339–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morra JH, Tu Z, Apostolova LG, et al. Automated 3D mapping of hippocampal atrophy and its clinical correlates in 400 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. Hum Brain Mapp 2009;30:2766–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Apostolova LG, Morra JH, Green AE, et al. Automated 3D mapping of baseline and 12-month associations between three verbal memory measures and hippocampal atrophy in 490 ADNI subjects. Neuroimage 2010;51:488–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Apostolova LG, Hwang KS, Andrawis JP, et al. 3D PIB and CSF biomarker associations with hippocampal atrophy in ADNI subjects. Neurobiol Aging 2010;31:1284–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Colliot O, Chetelat G, Chupin M, et al. Discrimination between Alzheimer disease, mild cognitive impairment, and normal aging by using automated segmentation of the hippocampus. Radiology 2008;248:194–201 [DOI] [PubMed] [Google Scholar]
- 24. Fushimi Y, Miki Y, Urayama S, et al. Gray matter-white matter contrast on spin-echo T1-weighted images at 3 T and 1.5 T: a quantitative comparison study. Eur Radiol 2007;17:2921–25 [DOI] [PubMed] [Google Scholar]
- 25. Lötjönen J, Wolz R, Koikkalainen J, et al. Fast and robust extraction of hippocampus from MR images for diagnostics of Alzheimer's disease. Neuroimage 2011;56:185–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ho AJ, Hua X, Lee S, et al. Comparing 3 T and 1.5 T MRI for tracking Alzheimer's disease progression with tensor-based morphometry. Hum Brain Mapp 2010;31:499–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Macdonald KE, Leung KK, Bartlett JW, et al. Automated template-based hippocampal segmentations from MRI: the effects of 1.5T or 3T field strength on accuracy. Neuroinformatics 2014;12:405–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Briellmann RS, Syngeniotis A, Jackson GD. Comparison of hippocampal volumetry at 1.5 Tesla and at 3 Tesla. Epilepsia 2001;42:1021–24 [DOI] [PubMed] [Google Scholar]
- 29. Mueller SG, Weiner MW, Thal LJ, et al. The Alzheimer's Disease Neuroimaging Initiative. Neuroimaging Clin N Am 2005;15:869–77, xi–xii [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mueller SG, Weiner MW, Thal LJ, et al. Ways toward an early diagnosis in Alzheimer's disease: the Alzheimer's Disease Neuroimaging Initiative (ADNI). Alzheimers Dement 2005;1:55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jack CR Jr, Bernstein MA, Fox NC, et al. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging 2008;27:685–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weiner MW, Veitch DP, Aisen PS, et al. The Alzheimer's Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement 2013;9:e111–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hughes CP, Berg L, Danziger WL, et al. A new clinical scale for the staging of dementia. Br J Psychiatry 1982;140:566–72 [DOI] [PubMed] [Google Scholar]
- 34. Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology 1993;43:2412–14 [DOI] [PubMed] [Google Scholar]
- 35. Cockrell JR, Folstein MF. Mini-Mental State Examination (MMSE). Psychopharmacol Bull 1988;24:689–92 [PubMed] [Google Scholar]
- 36. Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”: a practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 1975;12:189–98 [DOI] [PubMed] [Google Scholar]
- 37. McKhann G, Drachman D, Folstein M, et al. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology 1984;34:939–44 [DOI] [PubMed] [Google Scholar]
- 38. Leow AD, Klunder AD, Jack CR Jr, et al. Longitudinal stability of MRI for mapping brain change using tensor-based morphometry. Neuroimage 2006;31:627–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jovicich J, Czanner S, Greve D, et al. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage 2006;30:436–43 [DOI] [PubMed] [Google Scholar]
- 40. Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 1998;17:87–97 [DOI] [PubMed] [Google Scholar]
- 41. Mazziotta J, Toga A, Evans A, et al. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos Trans R Soc Lond B Biol Sci 2001;356:1293–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hua X, Leow AD, Parikshak N, et al. Tensor-based morphometry as a neuroimaging biomarker for Alzheimer's disease: an MRI study of 676 AD, MCI, and normal subjects. Neuroimage 2008;43:458–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Narr KL, van Erp TG, Cannon TD, et al. A twin study of genetic contributions to hippocampal morphology in schizophrenia. Neurobiol Dis 2002;11:83–95 [DOI] [PubMed] [Google Scholar]
- 44. Thompson PM, Hayashi KM, De Zubicaray GI, et al. Mapping hippocampal and ventricular change in Alzheimer disease. Neuroimage 2004;22:1754–66 [DOI] [PubMed] [Google Scholar]
- 45. Narr KL, Thompson PM, Sharma T, et al. Three-dimensional mapping of temporo-limbic regions and the lateral ventricles in schizophrenia: gender effects. Biol Psychiatry 2001;50:84–97 [DOI] [PubMed] [Google Scholar]
- 46. Mai JK, Paxinos G, Assheuer JK. Atlas of the Human Brain. San Diego: Elsevier Academic Press; 2004 [Google Scholar]
- 47. Duvernoy HM. The Human Hippocampus: An Atlas of Applied Anatomy. Munich: Springer Verlag; 1988 [Google Scholar]
- 48. Freund Y, Shapire R. A decision-theoretic generalization of online learning and an application to boosting. Journal of Computer and System Sciences 1997;55:119–39 [Google Scholar]
- 49. Morra JH, Tu Z, Apostolova LG, et al. Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer's disease mild cognitive impairment, and elderly controls. Neuroimage 2008;43:59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Morra JH, Tu Z, Apostolova LG, et al. Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. Neuroimage 2009;45:S3–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Morra JH, Tu Z, Toga AW, et al. Machine learning for brain image segmentation. In: Gonzalez FA, Romero E, eds. Biomedical Image Analysis and Machine Learning Technologies. Hershey: Medical Information Science References; 2009 [Google Scholar]
- 52. Wang L, Swank JS, Glick IE, et al. Changes in hippocampal volume and shape across time distinguish dementia of the Alzheimer type from healthy aging. Neuroimage 2003;20:667–82 [DOI] [PubMed] [Google Scholar]
- 53. Becker JT, Davis SW, Hayashi KM, et al. Three-dimensional patterns of hippocampal atrophy in mild cognitive impairment. Arch Neurol 2006;63:97–101 [DOI] [PubMed] [Google Scholar]
- 54. Frisoni GB, Sabattoli F, Lee AD, et al. In vivo neuropathology of the hippocampal formation in AD: a radial mapping MR-based study. Neuroimage 2006;32:104–10 [DOI] [PubMed] [Google Scholar]
- 55. Frayne R, Goodyear BG, Dickhoff P, et al. Magnetic resonance imaging at 3.0 Tesla: challenges and advantages in clinical neurological imaging. Invest Radiol 2003;38:385–402 [DOI] [PubMed] [Google Scholar]
- 56. Tanenbaum LN. Clinical 3T MR imaging: mastering the challenges. Magn Reson Imaging Clin N Am 2006;14:1–15 [DOI] [PubMed] [Google Scholar]
- 57. Das SR, Avants BB, Pluta J, et al. Measuring longitudinal change in the hippocampal formation from in vivo high-resolution T2-weighted MRI. Neuroimage 2012;60:1266–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mueller SG, Stables L, Du AT, et al. Measurement of hippocampal subfields and age-related changes with high resolution MRI at 4T. Neurobiol Aging 2007;28:719–26 [DOI] [PMC free article] [PubMed] [Google Scholar]