Abstract
Recent studies of noncrystalline HIV-1 capsid protein (CA) assemblies by our laboratory and by Polenova and coworkers have established the capability of solid state nuclear magnetic resonance (NMR) measurements to provide site-specific structural and dynamical information that is not available from other types of measurements. Nonetheless, the relatively high molecular weight of HIV-1 CA leads to congestion of solid state NMR spectra of fully isotopically labeled assemblies that has been an impediment to further progress. Here we describe an efficient protocol for production of segmentally labeled HIV-1 CA samples in which either the N-terminal domain (NTD) or the C-terminal domain (CTD) is uniformly 15N,13C-labeled. Segmental labeling is achieved by trans-splicing, using the DnaE split intein. Comparisons of two-dimensional solid state NMR spectra of fully labeled and segmentally labeled tubular CA assemblies show substantial improvements in spectral resolution. The molecular structure of HIV-1 assemblies is not significantly perturbed by the single Ser-to-Cys substitution that we introduce between NTD and CTD segments, as required for trans-splicing.
Keywords: solid state nmr, segmental labeling, HIV-1 capsid, intein, protein structure, protein assembly
Introduction
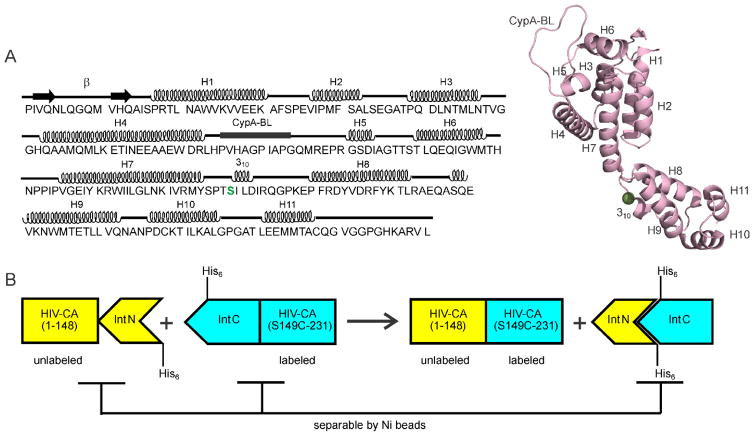
The mature human immunodeficiency virus type 1 (HIV-1) contains a conical capsid shell that encloses the viral genome to ensure its infectivity in a new host cell1–3. The capsid shell is comprised exclusively of approximately 1500 copies of the capsid protein (CA). As shown in Fig. 1A, CA consists of two independently foldable α-helical domains, an N-terminal domain (NTD) that includes helices 1-7 and a C-terminal domain (CTD) that includes helices 8-114–6. These two domains are joined by a short linker that influences the relative orientation of CTD and NTD in CA assemblies7. The capsid shell has been shown to follow a fullerene geometry, comprised largely of CA hexamers6,8–11. Each CA hexamer contains a central ring formed by NTD units and an external ring formed by CTD units. Individual hexamers are stabilized by the intermolecular NTD-NTD and NTD-CTD interactions around six-fold symmetry sites. Inter-hexamer contacts involve CTD-CTD contacts around two-fold and three-fold symmetry sites. Closure of the capsid shell requires twelve CA pentamers at specific locations8,11–13.
Figure 1.
(A) HIV-1 CA sequence, with secondary structure elements indicated above the sequence. The splicing site is at the NTD-CTD linker region, highlighted in green. A cartoon representation of the CA monomer structure (Protein Data Bank file 4XFX) shows the eleven helical segments (H1–H7 of NTD, H8–H11 of CTD), the cyclophilin A binding loop (CypA-BL), and a 310 helix immediately after the linker segment between NTD and CTD. (B) Schematic representation of the plasmid design and the purification strategy for the preparation of segmentally labeled HIV-1 CA. In this case, the CTD segment would be labeled.
Much information about the molecular structure of CA and CA assemblies has been obtained from electron microscopy8,10,13,14, x-ray crystallography4,6,9,12, and multidimensional liquid state nuclear magnetic resonance spectroscopy (NMR)5,7,15–17. Solid state NMR studies by Polenova and coworkers18–22 and by our laboratory23–26 have provided additional information about non-crystalline CA assemblies, including information about site-specific dynamics and conformational variations, the structure of CTD-CTD dimerization interfaces, the molecular basis for variable CA lattice curvature, and structural aspects of the HIV-1 maturation process.
The applicability of solid state NMR methods to HIV-1 CA assemblies has been limited by the relatively high molecular weight of the CA monomer (25.6 kDa) and the resulting spectral congestion. In work from our laboratory, 15N and 13C chemical shift assignments for the majority of immobilized residues in tubular CA assemblies were obtained from three-dimensional (3D) solid state NMR spectra of samples that were uniformly 15N,13C-labeled and samples that were uniformly 15N-labeled and partially 13C-labeled24. Roughly 10% of the solid state NMR signals from immobilized residues did not have definite assignments and various sidechain signals could not be resolved or assigned. Our determination of the CTD-CTD dimerization interface structure in tubular CA assemblies required selective isotopic labeling of certain residue types25. In our study of the immature HIV-1 lattice in virus-like particles formed by a larger HIV-1 Gag protein construct (38.5 kDa), chemical shift assignments were obtained only for a critical segment at the C-terminal end of CA27.
Segmental isotopic labeling is a promising approach for extending the applicability and enhancing the information content of solid state NMR measurements in studies of non-crystalline assemblies that are formed by high-molecular-weight proteins. Although the potential advantages of segmental isotopic labeling in NMR are well known28–35, relatively few successful examples of solid state NMR spectroscopy of segmentally labeled protein assemblies have been reported. These include uniform 15N,13C-labeling of either the N-terminal or the C-terminal portion of the HET-s prion domain in HET-s(218-289) fibrils36, uniform labeling of various N-terminal or C-terminal portions of the low-complexity domain of the fused-in-sarcoma (FUS-LC) protein in FUS-LC fibrils37, uniform labeling of one repeat segment of the CsgA protein in CsgA fibrils38, and uniform labeling of the first 14 residues in the Sup35NM prion protein39. In related work, a 50-residue segment of the influenza M2 protein was prepared by ligation of synthetic transmembrane and extra-membrane segments, with complementary selective labeling of the two segments, and examined in membrane-bound form40; selectively labeled and lipidated peptides were ligated to an N-terminally truncated, unlabeled Ras protein construct, producing full-length, membrane-associated Ras samples that were examined by 13C and 2H NMR41; full-length HIV-1 Vpu was produced for solid state NMR studies of transmembrane Vpu oligomers by ligation of synthetic, selectively labeled transmembrane and extra-membrane segments42; microcrystalline thioredoxin was prepared for solid state NMR studies by co-precipitation of N-terminal and C-terminal fragments that were labeled in a complementary manner43.
In this paper, we report segmental 15N,13C-labeling of NTD and CTD units in full-length HIV-1 CA by a split intein method, formation of morphologically homogeneous tubular assemblies by segmentally-labeled CA on the 10–20 mg scale, and two-dimensional (2D) solid state NMR spectroscopy of the segmentally-labeled CA tubes. Our results show that the single-site mutation required for segmental labeling does not interfere with CA folding and self-assembly and that segmental labeling leads to significant improvements in spectral resolution without otherwise compromising spectral quality. To our knowledge, this work represents the first successful demonstration of segmental labeling of a viral capsid protein. We expect that these results will facilitate future efforts to characterize local structural details and site-specific dynamics in noncrystalline capsid assemblies by solid state NMR.
Materials and methods
Plasmid design
Several different ligation techniques have been used for segmental labeling of proteins, including native chemical ligation44–47, expressed protein ligation32,48–52, protein trans-splicing36,38,53–56, and enzyme based protein ligation57–61. We chose to use the split intein DnaE from Nostoc punctiforme because it tolerates different amino acids at the splicing junction and remains active under a wide range of buffer conditions, including high concentration of denaturants36,62,63.
The splicing site was chosen in the flexible linker region between NTD and CTD of HIV-1 CA to reduce the chances of misfolding and aggregation of the ligated protein. As shown in Fig. 1B, our NTD-intein fusion sequence codes for residues 1-148 of HIV-1 CA along with the N-terminal intein segment of Npu DnaE (total molecular weight = 29.4 kDa for unlabeled material). Our CTD-intein fusion sequence codes for residues 149-231 of HIV-1 CA, with a Ser-to-Cys substitution at residue 149 (see below), along with the C-terminal intein segment of Npu DnaE (total molecular weight = 14.2 kDa for unlabeled material). Our HIV-1 CA sequence corresponds to the HIV-1 group M subtype B, HXB2 isolate. The NTD-intein sequence was cloned into the expression vector pET21 using NdeI and XhoI restriction sites. The CTD-intein sequence was cloned into a pET11a vector using NdeI and BamHI sites. The NTD- and CTD-intein sequences contain His6 tags at their C-terminal and the N-terminal ends, respectively.
Protein expression and purification
Transformed Escherichia coli BL21 (DE3) competent cells were added into 2.0 ml of Luria-Bertani (LB) medium with 100 μg/ml ampicillin and grown at 37° C for 3 h. The culture was then transferred into 100.0 ml of either LB or isotopically labeled minimal medium to prepare the subculture, then grown overnight at 37° C. One liter of the minimal medium contained 33.2 ml of 30X salt solution (1 M Na2HPO4-7H2O, 1 M KH2PO4, and 100 mM Na2SO4), 3.2 g of 13C6-glucose, 1.5 g of 15NH4Cl, 1 ml of 2 M MgCl2, 100 μl of 2 M CaCl2, 50 mg of thiamine, 10 ml of minimum essential medium (MEM) vitamin solution (100X) and 100 μg/ml ampicillin. One liter of LB or minimal medium (for production of either unlabeled or labeled proteins, respectively) was then inoculated with 100 ml of the overnight culture. The cells were grown at 37° C with shaking at 220 rpm. Protein expression was initiated by the addition of 0.2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at the optical density (OD) of 1.2–1.5 for both intein fusion proteins. After induction, NTD-intein-expressing cells were grown at 37° C for 4 h, whereas CTD-intein-expressing cells were grown at 20° C overnight. These growth conditions were selected by screening different IPTG concentrations, temperatures, and expression times, using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis to estimate protein production. The cells were harvested by centrifugation at 9000 rpm (12100 X g) for 30 min. The OD of the culture before harvesting was 4–5. Cell pellets were stored at −80° C or purified immediately.
For purification, cell pellets from each 1 l culture were resuspended in roughly 50 ml of lysis buffer [25 mM Tris, 50 mM NaCl, 1 mM TCEP, 1 tablet of EDTA-free complete protease inhibitor cocktail (Roche), 200 μl of lysozyme from the stock of 20 mg/ml, and 7 M urea at pH 8.0]. The solution was then sonicated (Branson sonifier model 250 with Branson tapered microtip, 3 mm diameter) at a duty cycle of 40% and an amplitude of 4 for 15 min. The cell debris was pelleted by centrifugation at 18000 rpm (26500 X g) for 40 min. The supernatant was then used for affinity-based purification on a Ni-NTA column (Qiagen). His6-tagged fusion proteins were then eluted using elution buffer (25 mM Tris, 50 mM NaCl, 1 mM TCEP and 7 M urea at pH 8.0) with 400 mM imidazole.
Trans-splicing and refolding reactions occurred simultaneously while dialyzing a 1:1 mixture of eluted NTD-intein and CTD-intein fusion proteins at room temperature for 40–42 h. The dialysis buffer contained 25 mM Tris, 50 mM NaCl, 1.0 mM TCEP, and 2.5 M urea at pH 6.8. Protein concentrations, estimated by UV absorption at 280 nm, were kept below 60 μM during dialysis. Large scale preparations were conducted in 30 ml dialysis bags (ThermoFisher Scientific, 3.5 kDa cutoff). Samples for SDS-PAGE and reverse-phase high-performance liquid chromatography (HPLC) were taken at several time points to follow the ligation reaction. The protein mixture was then incubated with Ni-NTA beads, which had been equilibrated with dialysis buffer, and subsequently passed through a gravity column to remove intein proteins and residual fusion proteins, which remained bound to the column. The purified ligated product was then completely refolded by dialyzing overnight at 4° C against Tris buffer (25 mM Tris, 50 mM NaCl, 1.0 mM TCEP) at pH 6.8. Ligation of NTD and CTD was confirmed by mass spectrometry. Segmentally labeled HIV-1 CA was prepared from either labeled NTD-intein and unlabeled CTD-intein or unlabeled NTD-intein and labeled CTD-intein. In both cases, the final yield of ligated HIV-1 CA was approximately 72% (see below).
Fully 15N,13C-labeled, wild-type HIV-1 CA was also prepared using the protocols described previously23,24.
Formation of tubular CA assemblies
Purified, ligated HIV-1 CA was concentrated to 30 mg/ml in Tris buffer (50 mM Tris, 50 mM NaCl, 1 M TCEP, pH 8.0), using centrifugal filter units (Amicon Ultra, 15 ml volume, 3 kDa cutoff or 0.5 ml volume, 10 kDa cutoff). CA self-assembly was triggered by increasing the NaCl concentration to 0.8 M (using a stock solution of 5 M NaCl in 50 mM Tris) while adjusting the final protein concentration to approximately 20 mg/ml. The protein solution became turbid immediately after addition of NaCl. The CA solution was then incubated at 37° C for 15 min to promote nucleation of the structurally ordered CA lattice. Subsequently, the milky white solution was incubated at room temperature for 16–18 h to promote the formation of long, uniform CA tubes64. Formation of wild-type CA tubes was performed as described earlier, at 1.0 M NaCl24. The quality of the tubular CA assemblies was verified by transmission electron microscopy (TEM). To remove CA aggregates that were apparently amorphous, the solution of CA assemblies was then gently centrifuged and washed several times with 0.8 M or 1.0 M NaCl (for ligated or wild-type CA) containing Tris buffer. For solid state NMR, thin-wall 3.2 mm magic-angle spinning (MAS) rotors were packed by centrifugation with approximately 27–35 mg of sample, containing approximately 10–15 mg of CA assemblies.
Mass spectrometry
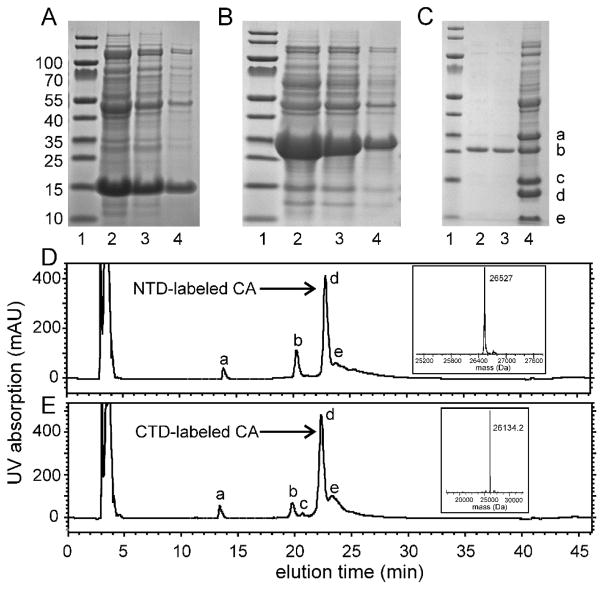
Mass data for CTD-labeled HIV-1 CA were obtained with a Waters LCT Premier time-of-flight mass spectrometer, operating in positive ion mode and using electrospray ionization. Sample introduction was via a Thermo Proswift RP-4H monolithic column (1 mm inner diameter, 50 mm length), using a 100 μl/min flow rate and a solvent gradient from 100% H2O to 80% methanol/20% acetonitrile (with 0.1% trifluoroacetic acid and 0.2% formic acid) in 9 min. The multi-charged m/z spectrum was deconvolved using the MaxEnt I program. Mass data for NTD-labeled HIV-1 CA were obtained with a Thermo MSQ Plus single quadrupole mass spectrometer, also operating in positive ion mode and using electrospray ionization. Sample introduction was via a Thermo Acclaim Pepmap 300 C18 column (1.0 mm inner diameter and 150 mm in length), using a 0.2 ml/min flow rate and a solvent gradient from 88% water/12% acetonitrile to 34% water/66% acetonitrile (with 0.01% trifluoroacetic acid) in 22 min. The multi-charged m/z spectrum was deconvolved using the MagTran program. Expected and measured masses for NTD-labeled CA were 26695.8 Da and 26527 Da respectively. Expected and measured masses for CTD-labeled CA were 26282.85 Da and 26134 Da respectively. From these data, we infer isotopic enrichment levels of about 99.4% for both segmentally labeled products.
Transmission electron microscopy
TEM images were recorded with an FEI Morgagni microscope, operating at 80 keV. Negatively stained samples were prepared on glow-discharged carbon films, supported by lacey carbon on 300-mesh copper grids. After diluting the CA assembly solution by a factor of ten (in 0.8 M NaCl containing Tris buffer), a 10 μl aliquot was adsorbed to the carbon film for 30 s, then blotted with filter paper, rinsed with Tris buffer containing 0.8 M or 1.0 M NaCl, blotted, rinsed with deionized water, blotted, stained with 2% uranyl acetate for 20–30 s, blotted, and dried in air.
Solid state NMR
Solid state NMR measurements were performed with a Varian Infinity spectrometer, operating at a 1H NMR frequency of 746.05 MHz with a Magnex 17.5 T magnet. The three-channel 3.2 mm MAS probe was produced by Peter Gor’kov (Black Fox, Inc., Tallahasse FL), and contains a low electric field design described previously65. Sample temperatures were maintained at 10° C with cooled nitrogen gas. All spectra were recorded with a MAS frequency of 12.0 kHz and with 1H decoupling fields of 85 kHz, using two-pulse phase modulation66. 1H-13C cross-polarization was performed with a 62 kHz 1H field and an amplitude-ramped 13C field centered at 50 kHz, with 1.2 ms contact time. 1H-15N cross-polarization was performed with a 48 kHz 1H field and an amplitude-ramped 15N field centered at 36 kHz, with 1.2 ms contact time. 2D 13C-13C spectra were recorded with 50 ms mixing periods, using radio-frequency-assisted spin diffusion (RAD)67,68. 2D NCACX and NCOCX experiments were recorded with 4.0 ms of band-selective 15N-13C cross polarization, using a 30 kHz 13C field and a 18 kHz 15N field for NCACX spectra (13C carrier frequency centered in the 13Cα region) and a 40 kHz 13C field and a 28 kHz 15N field for NCOCX spectra (13C rf carrier centered in the 13CO region), followed by 50 ms 13C-13C RAD mixing periods. 2D 13C-13C spectra were obtained with 312 complex t1 points and 1024 complex t2 point, with 44.6 kHz and 66.7 kHz spectral widths in the t1 and t2 dimensions. 2D NCACX and NCOCX experiments were recorded with 89 complex t1 points and 768 complex t2 points, with 8.9 kHz and 66.7 kHz spectral widths in the t1 and t2 dimensions. Recycle delays were 2.0 s in all experiments. Each 2D spectrum was acquired as a series of nominally identical 2D blocks, with measurement times of 3.9 h for 2D 13C-13C and 2D NCACX blocks, and 5.5 h for 2D NCOCX blocks. Total measurement times for 2D 13C-13C, NCACX, and NCOCX spectra of NTD-labeled CA assemblies were approximately 16 h, 22 h, and 22 h respectively. Total measurement times for 2D 13C-13C, NCACX, and NCOCX spectra of CTD-labeled CA assemblies were approximately 16 h, 187 h, and 187 h, respectively. (Longer measurement times for the CTD-labeled sample were required in part because of a smaller quantity that was packed in the MAS rotor. In addition, 15N-13C cross-polarization efficiencies were somewhat lower for the CTD-labeled sample, possibly due to differences in molecular motions in the NTD and CTD subunits.)
2D spectra were processed with NMRPipe69. As described below, spectra of NTD-labeled and CTD-labeled samples were compared by subtracting spectra of segmentally labeled samples from spectra of a uniformly labeled sample. This subtraction was done with the addNMR command in NMRpipe, including scale factors that were adjusted to account for differences in sample quantities and total measurement times.
3D NCACX and 3D NCOCX spectra of segmentally-labeled CA assemblies were also obtained at 1H NMR frequencies of 746.05 MHz and 599.09 MHz. Data at the lower frequency were acquired with a Varian InfinityPlus spectrometer, an Oxford Instruments 14.1 T magnet, and a Varian 3.2 mm BioMAS probe. Experimental details are given in Table S1.
Results
In vitro trans-splicing for preparation of segmentally labeled HIV-1 CA assemblies
The objective of this work was to prepare tubular HIV-1 CA assemblies (which are known to contain the same triangular lattice of CA hexagons as conical capsids within mature HIV-113,14) with segmental labeling of either their NTD or their CTD segments, and without introducing significant chemical or structural differences from wild-type CA assemblies. Intein-based trans-splicing techniques have been used previously for this purpose36,38,70. Cysteine is required as the first residue of the C-terminal extein because of its efficient involvement in the protein trans-thioesterification and acyl transfer reactions during trans-splicing71,72. HIV-1 CA contains only two native cysteines (C198 and C218), which are involved in intramolecular disulfide bond formation in the CTD. The location and importance in domain structure of these cysteines make them inappropriate for use as splicing sites. Accordingly, a serine-to-cysteine substitution was planned to create the splicing site, with serine chosen because it differs from cysteine by only one atom. The linker segment of HIV-1 CA, between NTD and CTD segments, was chosen as a desirable segment for the splicing site to minimize the chance of adverse effects on protein structure and function, even considering the importance of linker region in viral core assembly and stability73. The linker segment contains two serine residues, S146 and S149. S149C has been reported to be a viable CA mutant74, suggesting that cysteine substitution at residue 149 does not interfere with CA lattice formation. S146 was disfavored as a substitution site on the grounds that P147 would become the +2 residue of the C-terminal extein and that proline at this position may not be favorable for the splicing reaction.
Consequently, HIV-1 CA was divided into two parts, namely (i) NTD plus linker residues up to T148 and (ii) C149 to the C-terminus of CTD. As shown in Fig. 1B, the NTD-containing part of CA was fused with the first 105 residues of Npu DnaE intein (NTD-intein fusion), while the C-terminal 35 amino acids of Npu DnaE were fused with the CTD-containing part of CA (CTD-intein fusion). The fusion proteins were purified from inclusion bodies with high yield. Approximately 290 mg of the NTD-intein fusion protein was obtained per liter of LB medium, and approximately 100 mg per liter of minimal medium. Approximately 120 mg of the CTD-intein fusion protein was obtained per liter of LB medium, and approximately 105 mg per liter of minimal medium. (For comparison, 46 mg of full-length HIV-1 CA was obtained per liter of minimal medium, indicating that higher expression in inclusion bodies was obtained with the intein fusions.) Figs. 2A and 2B show SDS-PAGE analyses of the over-expression profiles for the two fusion proteins. The trans-splicing activity of the intein was restored by dialyzing the ligation mixture against buffer with a lower denaturant concentration36. Progress of the trans-splicing reaction was regularly monitored by SDS-PAGE and reverse-phase HPLC. After 40–42 h of incubation at room temperature, the precursor fusion proteins and ligated product remained soluble in the ligation mixture, with some precipitation of the N-terminal and C-terminal intein fragments (Fig. 2C, lane 4). The ligated product was readily separable from other components of the ligation mixture by Ni-NTA bead-based purification (Fig. 2C, lanes 2 and 3). Approximately 28 mg of the ligated product (1.1 μmol) was obtained from a total of 64 mg of the two fusion proteins in an equimolar mixture (1.5 μmol each), corresponding to a yield of approximately 72%.
Figure 2.
(A) SDS-PAGE analysis of the cell lysate from overexpresssion of the CTD-intein fusion protein. Lane 1 contains molecular weight markers. Lanes 2–4 contain decreasing quantities of the lysate. The band near 15 kDa is the CTD-intein fusion protein. Gels were stained using Imperial protein stain from ThermoFisher Scientific. (B) Same as panel A, but for the NTD-intein fusion protein, which is the band near 30 kDa. (C) SDS-PAGE analysis of the ligation mixture after 40 h of incubation (lane 4) and the purified ligation product (lanes 2 and 3). Bands marked a–e are the NTD-intein fusion protein, ligated HIV-1 CA, the CTD-intein fusion protein, the N-terminal intein segment of Npu DnaE, and the C-terminal intein segment, respectively. (D) Reverse phase HPLC chromatogram of the ligation mixture after 40 h of incubation to produce full-length CA with a uniformly 15N,13C-labeled NTD segment. UV absorption peaks (280 nm) marked a–e are the C-terminal intein segment, the CTD-intein fusion protein, the N-terminal intein segment, the ligated CA product, and the NTD-intein fusion protein, respectively. The inset shows the mass spectrum of peak d, after deconvolution of multi-charged mass data from electrospray ionization. (E) Same as panel D, but for production of full-length CA with a uniformly 15N,13C-labeled CTD segment.
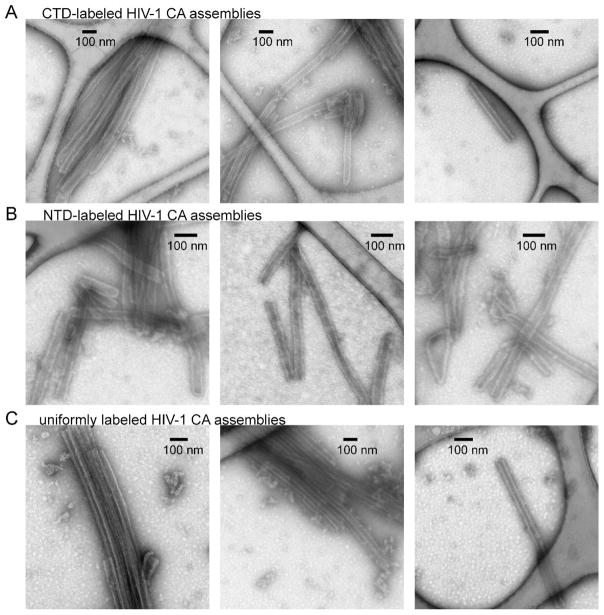
Segmentally labeled HIV-1 CA samples were prepared by expressing either the NTD-intein or the CTD-intein fusion protein in labeled minimal media. Figs. 2D and 2E show reverse-phase HPLC analyses of the segmentally labeled ligation mixtures after incubation for 40 hr. The HPLC profiles indicate the high conversion rate from precursors to product protein. Mass spectra confirm the successful preparation of segmentally labeled HIV-1 CA (insets to Figs. 2D and 2E). Segmentally labeled HIV-1 CA assemblies with tubular morphology were then prepared by spontaneous self-assembly of the CA monomers upon addition of 0.8 M NaCl to the protein solution at pH 8.0. TEM images of the CA tubes with negative staining are shown in Fig. 3. The overall appearance and dimensions of segmentally labeled and wild-type CA tubes were similar. A minor component of CA assemblies with cone-like morphologies was also observed, especially in the NTD-labeled CA sample (e.g., Fig. 3B, right-hand panel).
Figure 3.
Characterization of the HIV-1 CA assemblies by TEM. Rows A–C are images of negatively stained tubular assemblies formed by CTD-labeled CA, NTD-labeled CA, and uniformly labeled CA, respectively.
Solid state NMR of segmentally labeled HIV-1 CA assemblies
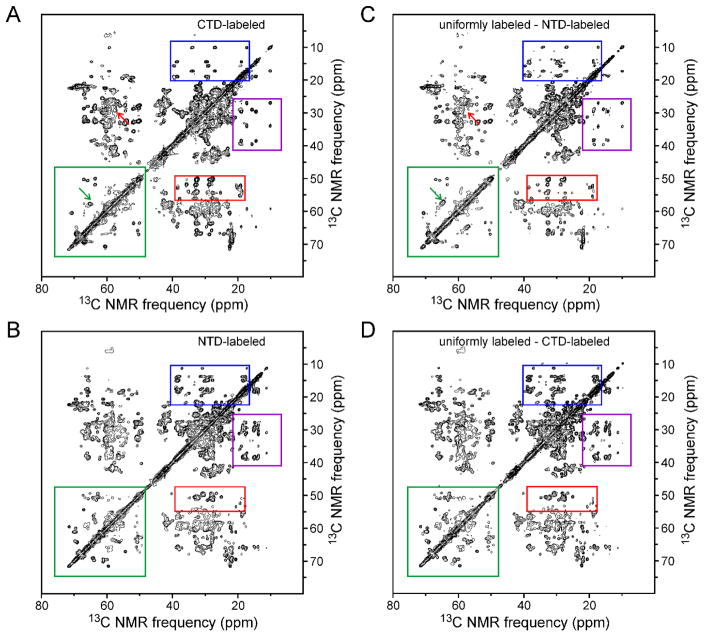
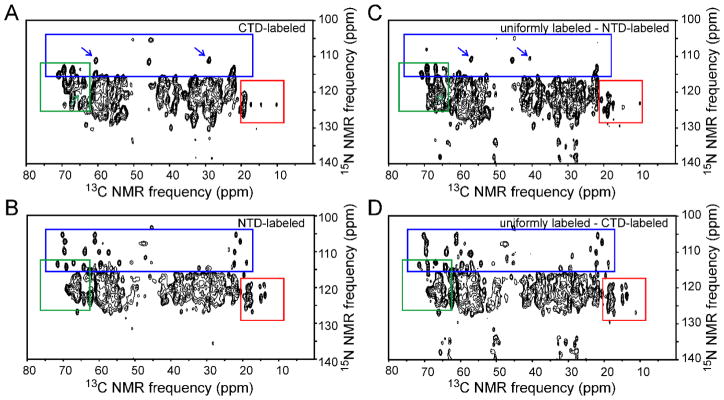
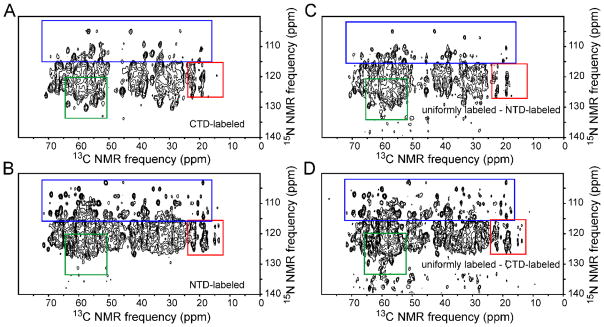
Tubular CA assemblies prepared from both uniformly labeled and segmentally labeled samples were washed 3–4 times with Tris buffer (pH 8.0, 1.0 or 0.8 M NaCl). CA tubes were then pelleted directly into 3.2 mm MAS NMR rotors. 2D solid state NMR spectra were recorded to indicate the sample quality (i.e., labeling efficiency, structural homogeneity) and to compare the spectral complexity of uniformly and segmentally labeled CA assemblies. Figs. 4, 5, and 6 show the 2D 13C-13C, NCACX, and NCOCX spectra of CTD-labeled and NTD-labeled assemblies (panels A and B, respectively), as well as 2D difference spectra in which spectra of segmentally labeled assemblies were subtracted from spectra of uniformly labeled assemblies (panels C and D). In general, 2D spectra of the CTD-labeled assemblies are different from 2D spectra of NTD-labeled assemblies, as expected. Significantly, 2D spectra of the CTD-labeled assemblies closely resemble 2D difference spectra obtained by subtracting spectra of NTD-labeled assemblies from spectra of uniformly labeled assemblies. This observation shows that segmentally labeled CA tubes have the same molecular structure and level of structural order as uniformly labeled CA tubes, despite the S149C substitution required for intein trans-splicing and minor differences in optimal assembly conditions.
Figure 4.
(A,B) Aliphatic regions of 2D solid state 13C-13C NMR spectra of CTD-labeled CA assemblies and NTD-labeled CA assemblies, respectively. Color-coded rectangles indicate regions where clear differences in crosspeak patterns are observed. (C) 2D spectrum obtained by subtracting the spectrum of NTD-labeled CA assemblies from that of fully-labeled CA assemblies. (D) 2D spectrum obtained by subtracting the spectrum of CTD-labeled CA assemblies from that of fully-labeled CA assemblies. Green arrows in panels A and C indicate a crosspeak from S149 that is present in panel C but absent from panel A, due to the S149C substitution in the segmentally labeled samples. Red arrows indicate a crosspeak from C149 or C198 that is present in panel A but absent from panel C. Contour levels increase by successive factors of 1.7.
Figure 5.
Same as Fig. 4, but for 2D NCACX spectra of segmentally and fully labeled HIV-1 CA assemblies. Arrows in panels A and C indicate crosspeaks from C218 that are shifted due to the difference in C198-C218 disulfide formation in the segmentally labeled and fully labeled samples, resulting from reducing conditions maintained during CA self-assembly. Contour levels increase by successive factors of 1.5.
Figure 6.
Same as Fig. 4, but for 2D NCOCX spectra of segmentally and fully labeled HIV-1 CA assemblies. Contour levels increase by successive factors of 1.5.
For comparison, 2D spectra of uniformly labeled CA assemblies (which were used to generate the difference spectra in Figs. 4–6) are shown in Figs. S1–S3 of Supporting Information. As expected, 2D spectra of segmentally labeled assemblies are less congested than 2D spectra of uniformly labeled assemblies, especially for CTD-labeled assemblies, which are labeled at approximately one third of the roughly 186 residues that contribute to solid state NMR signals from uniformly labeled assemblies24. Figs. S1–S3 also show one-dimensional (1D) slices from all 2D spectra and 2D difference spectra.
The S149C substitution does produce certain differences in the spectra, most prominently the absence of S149 signals and presence of signals from C149 or C198 (ambiguous assignment) in the 2D 13C-13C spectrum of CTD-labeled assemblies (green and red arrows in Figs. 4A and 4C, blue arrows in Figs. 5A and 5C). The fact that segmentally labeled assemblies were prepared under reducing conditions, whereas uniformly labeled assemblies were prepared under oxidizing conditions, leads to a shift in the well-resolved NCACX signals from C218 (blue arrows in Figs. 5A and 5C). Minor chemical shift perturbations at other sites arising from the S149C substitution and the use of reducing conditions probably contribute to differences in the 1D slices in Figs. S1–S3.
From a preliminary analysis of 3D NCACX and NCOCX spectra of the NTD-labeled and CTD-labeled assemblies (Table S2 and Figs. S4–S6), it was possible to obtain chemical shift assignments for nine residues that lacked unambiguous assignments in earlier work by Bayro et al.24 Newly assigned signals are relatively weak, for the most part, arising from residues in inter-helical loops (G106, E175) or near the ends of helical segments (I141, V142, S146, R173, A174, E212, E213), which may be partially disordered in these noncrystalline assemblies.
Discussion
Information obtained in previous solid state NMR studies of HIV-1 CA assemblies using fully labeled CA has been limited by spectral congestion and associated ambiguities in signal assignments. Results presented above demonstrate an efficient method for preparation of segmentally labeled CA. The high expression level of the intein fusion proteins, high yield of ligated HIV-1 CA, and minimal modification in the native sequence make this strategy suitable for solid state NMR studies. NMR linewidths in spectra of segmentally labeled samples are indistinguishable from linewidths in spectra of uniformly labeled samples (Figs. 4–5 and S1–S3). There is a significant reduction in spectral congestion, especially in case of CTD-labeled CA assemblies. Further reductions in line widths and spectral congestion can be obtained by combining this ligation technique with specific amino acid labeling or sparse labeling schemes. A similar ligation approach can in principle be used to prepare segmentally labeled samples of the HIV-1 Gag polyprotein for studies of the immature HIV-1 protein lattice27,75,76.
The high yield of segmentally labeled CA obtained in this work may be attributed to the following factors: (i) Expression levels of the NTD-intein and CTD-intein fusion proteins from the inclusion bodies were high (approximately 290 mg/l and 124 mg/l, respectively), so that adequate material was available for optimization of the trans-splicing reaction and for subsequent production of samples for solid state NMR; (ii) The stability of the fusion proteins and ligated HIV-1 CA during the long reaction time (40–42 h) at room temperature allowed the trans-splicing reaction to proceed to near completion (approximately 72% of the theoretical yield). Recent developments in intein engineering may accelerate the trans-splicing reaction and improve the final yield of ligated product77; (iii) Implementation of His6 tags on the C-terminus of the NTD-intein fusion protein and the N-terminus of the CTD-intein fusion protein segment allowed a single-step separation of the ligated CA product from the unligated precursor proteins and the cleaved intein segments, thereby minimizing product loss in the purification process; (iv) The HIV-1 CA protein refolds readily from urea-denatured components, leading to only minor losses from misfolding or amorphous aggregation during the trans-splicing, purification, refolding, and self-assembly steps.
Together with other recent demonstrations of segmental labeling36–39, results presented above indicate that intein-based methods have the potential to expand the applicability of solid state NMR to high-molecular-weight protein assemblies, by allowing the NMR measurements to focus on protein segments that are of particular interest or by allowing different segments within a protein to be labeled in complementary ways that facilitate signal assignments or the detection of intramolecular or intermolecular contacts. Segmental labeling approaches may also be of particular interest in low-temperature solid state NMR studies, including studies that employ dynamic nuclear polarization, where inhomogeneous broadening of solid state NMR lines often interferes with the interpretation of multidimensional spectra of fully labeled proteins.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases and the Intramural AIDS Targeted Antiviral Program of the National Institutes of Health. We thank Dr. Marvin J. Bayro for generous assistance and helpful advice regarding HIV-1 CA assembly conditions, electron microscopy, and solid state NMR spectroscopy. We thank John R. Lloyd for performing mass spectrometry of our ligated products.
References
- 1.Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr Opin Struct Biol. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briggs JAG, Kräusslich HG. The molecular architecture of HIV. J Mol Biol. 2011;410:491–500. doi: 10.1016/j.jmb.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 3.Engelman A, Cherepanov P. The structural biology of HIV-1: Mechanistic and therapeutic insights. Nature Rev Microbiol. 2012;10:279–290. doi: 10.1038/nrmicro2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gamble TR, Yoo SH, Vajdos FF, vonSchwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 5.Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 6.Gres AT, Kirby KA, KewalRamani VN, Tanner JJ, Pornillos O, Sarafianos SG. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Science. 2015;349:99–103. doi: 10.1126/science.aaa5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deshmukh L, Schwieters CD, Grishaev A, Ghirlando R, Baber JL, Clore GM. Structure and dynamics of full-length HIV-1 capsid protein in solution. J Am Chem Soc. 2013;135:16133–16147. doi: 10.1021/ja406246z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 9.Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganser-Pornillos BK, Cheng A, Yeager M. Structure of full-length HIV-1 CA: A model for the mature capsid lattice. Cell. 2007;131:70–79. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 11.Mattei S, Glass B, Hagen WJH, Kräusslich HG, Briggs JAG. The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science. 2016;354:1434–1437. doi: 10.1126/science.aah4972. [DOI] [PubMed] [Google Scholar]
- 12.Pornillos O, Ganser-Pornillos BK, Yeager M. Atomic-level modelling of the HIV capsid. Nature. 2011;469:424–427. doi: 10.1038/nature09640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao G, Perilla JR, Yufenyuy EL, Meng X, Chen B, Ning J, Ahn J, Gronenborn AM, Schulten K, Aiken C, Zhang P. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature. 2013;497:643–646. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 15.Wong HC, Shin R, Krishna NR. Solution structure of a double mutant of the carboxy-terminal dimerization domain of the HIV-1 capsid protein. Biochemistry. 2008;47:2289–2297. doi: 10.1021/bi7022128. [DOI] [PubMed] [Google Scholar]
- 16.Shin R, Tzou YM, Krishna NR. Structure of a monomeric mutant of the HIV-1 capsid protein. Biochemistry. 2011;50:9457–9467. doi: 10.1021/bi2011493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byeon IJL, Meng X, Jung J, Zhao G, Yang R, Ahn J, Shi J, Concel J, Aiken C, Zhang P, Gronenborn AM. Structural convergence between cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell. 2009;139:780–790. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han Y, Ahn J, Concel J, Byeon IJL, Gronenborn AM, Yang J, Polenova T. Solid state NMR studies of HIV-1 capsid protein assemblies. J Am Chem Soc. 2010;132:1976–1987. doi: 10.1021/ja908687k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byeon IJL, Hou G, Han Y, Suiter CL, Ahn J, Jung J, Byeon CH, Gronenborn AM, Polenova T. Motions on the millisecond time scale and multiple conformations of HIV-1 capsid protein: Implications for structural polymorphism of CA assemblies. J Am Chem Soc. 2012;134:6455–6466. doi: 10.1021/ja300937v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han Y, Hou G, Suiter CL, Ahn J, Byeon IJL, Lipton AS, Burton S, Hung I, Gorkov PL, Gan Z, Brey W, Rice D, Gronenborn AM, Polenova T. Magic angle spinning NMR reveals sequence-dependent structural plasticity, dynamics, and the spacer peptide 1 conformation in HIV-1 capsid protein assemblies. J Am Chem Soc. 2013;135:17793–17803. doi: 10.1021/ja406907h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu M, Hou G, Zhang H, Suiter CL, Ahn J, Byeon IJL, Perilla JR, Langmead CJ, Hung I, Gor’kov PL, Gan Z, Brey W, Aiken C, Zhang P, Schulten K, Gronenborn AM, Polenova T. Dynamic allostery governs cyclophilin A-HIV capsid interplay. Proc Natl Acad Sci USA. 2015;112:14617–14622. doi: 10.1073/pnas.1516920112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Hou G, Lu M, Ahn J, Byeon IJL, Langmead CJ, Perilla JR, Hung I, Gor’Kov PL, Gan Z, Brey WW, Case DA, Schulten K, Gronenborn AM, Polenova T. HIV-1 capsid function is regulated by dynamics: Quantitative atomic-resolution insights by integrating magic-angle-spinning NMR, QM/MM, and MD. J Am Chem Soc. 2016;138:14066–14075. doi: 10.1021/jacs.6b08744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen B, Tycko R. Structural and dynamical characterization of tubular HIV-1 capsid protein assemblies by solid state nuclear magnetic resonance and electron microscopy. Protein Sci. 2010;19:716–730. doi: 10.1002/pro.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayro MJ, Chen B, Yau WM, Tycko R. Site-specific structural variations accompanying tubular assembly of the HIV-1 capsid protein. J Mol Biol. 2014;426:1109–1127. doi: 10.1016/j.jmb.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bayro MJ, Tycko R. Structure of the dimerization interface in the mature HIV-1 capsid protein lattice from solid state NMR of tubular assemblies. J Am Chem Soc. 2016;138:8538–8546. doi: 10.1021/jacs.6b03983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu JX, Bayro MJ, Tycko R. Major variations in HIV-1 capsid assembly morphologies involve minor variations in molecular structures of structurally ordered protein segments. J Biol Chem. 2016;291:13098–13112. doi: 10.1074/jbc.M116.720557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bayro MJ, Ganser-Pornillos BK, Zadrozny KK, Yeager M, Tycko R. Helical conformation in the CA-SP1 junction of the immature HIV-1 lattice determined from solid state NMR of virus-like particles. J Am Chem Soc. 2016;138:12029–12032. doi: 10.1021/jacs.6b07259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yagi H, Tsujimoto T, Yamazaki T, Yoshida M, Akutsu H. Conformational change of H+-ATPase β monomer revealed on segmental isotope labeling NMR spectroscopy. J Am Chem Soc. 2004;126:16632–16638. doi: 10.1021/ja045279o. [DOI] [PubMed] [Google Scholar]
- 29.Williams FP, Milbradt AG, Embrey KJ, Bobby R. Segmental isotope labelling of an individual bromodomain of a tandem domain BRD4 using sortase A. PLoS ONE. 2016;11:e0154607. doi: 10.1371/journal.pone.0154607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minato Y, Ueda T, MacHiyama A, Shimada I, Iwaï H. Segmental isotopic labeling of a 140 kDa dimeric multi-domain protein chea from Escherichia coli by expressed protein ligation and protein trans-splicing. J Biomol NMR. 2012;53:191–207. doi: 10.1007/s10858-012-9628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michel E, Skrisovska L, Wüthrich K, Allain FHT. Amino acid-selective segmental isotope labeling of multidomain proteins for structural biology. Chem Bio Chem. 2013;14:457–466. doi: 10.1002/cbic.201200732. [DOI] [PubMed] [Google Scholar]
- 32.Camarero JA, Shekhtman A, Campbell EA, Chlenov M, Gruber TM, Bryant DA, Darst SA, Cowburn D, Muir TW. Autoregulation of a bacterial σ factor explored by using segmental isotopic labeling and NMR. Proc Natl Acad Sci USA. 2002;99:8536–8541. doi: 10.1073/pnas.132033899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu R, Ayers B, Cowburn D, Muir TW. Chemical ligation of folded recombinant proteins: Segmental isotopic labeling of domains for NMR studies. Proc Natl Acad Sci USA. 1999;96:388–393. doi: 10.1073/pnas.96.2.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu DS, Cowburn D. Segmental isotopic labeling of proteins for NMR study using intein technology. In: Mootz HD, editor. Split inteins: Methods and protocols. Vol. 1495. 2017. pp. 131–145. [DOI] [PubMed] [Google Scholar]
- 35.Liu DS, Yuan Y, Xu R, Cowburn D. Domain interactions of C-terminal Src kinase determined through NMR spectroscopy with segmental isotope labeling. Protein Cell. 2017;8:67–71. doi: 10.1007/s13238-016-0333-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schubeis T, Lührs T, Ritter C. Unambiguous assignment of short- and long-range structural restraints by solid state NMR spectroscopy with segmental isotope labeling. Chem Bio Chem. 2015;16:51–54. doi: 10.1002/cbic.201402446. [DOI] [PubMed] [Google Scholar]
- 37.Murray DT, Kato M, Lin Y, Thurber KR, Hung I, McKnight SL, Tycko R. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell. 2017 doi: 10.1016/j.cell.2017.08.048. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schubeis T, Yuan P, Ahmed M, Nagaraj M, Van Rossum BJ, Ritter C. Untangling a repetitive amyloid sequence: Correlating biofilm-derived and segmentally labeled curli fimbriae by solid state NMR spectroscopy. Angew Chem Int Ed. 2015;54:14669–14672. doi: 10.1002/anie.201506772. [DOI] [PubMed] [Google Scholar]
- 39.Frederick KK, Michaelis VK, Caporini MA, Andreas LB, Debelouchina GT, Griffin RG, Lindquist S. Combining DNP NMR with segmental and specific labeling to study a yeast prion protein strain that is not parallel in-register. Proc Natl Acad Sci USA. 2017;114:3642–3647. doi: 10.1073/pnas.1619051114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwon B, Tietze D, White PB, Liao SY, Hong M. Chemical ligation of the influenza M2 protein for solid state NMR characterization of the cytoplasmic domain. Protein Sci. 2015;24:1087–1099. doi: 10.1002/pro.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reuther G, Tan KT, Vogel A, Nowak C, Arnold K, Kuhlmann J, Waldmann H, Huster D. The lipidated membrane anchor of full length N-Ras protein shows an extensive dynamics as revealed by solid state NMR spectroscopy. J Am Chem Soc. 2006;128:13840–13846. doi: 10.1021/ja063635s. [DOI] [PubMed] [Google Scholar]
- 42.Kochendoerfer GG, Jones DH, Lee S, Oblatt-Montal M, Opella SJ, Montal M. Functional characterization and NMR spectroscopy on full-length Vpu from HIV-1 prepared by total chemical synthesis. J Am Chem Soc. 2004;126:2439–2446. doi: 10.1021/ja038985i. [DOI] [PubMed] [Google Scholar]
- 43.Marulanda D, Tasayco ML, McDermott A, Cataldi M, Arriaran V, Polenova T. Magic angle spinning solid state NMR spectroscopy for structural studies of protein interfaces. Resonance assignments of differentially enriched Escherichia coli thioredoxin reassembled by fragment complementation. J Am Chem Soc. 2004;126:16608–16620. doi: 10.1021/ja0464589. [DOI] [PubMed] [Google Scholar]
- 44.Dawson PE, Muir TW, Clark-Lewis I, Kent SBH. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 45.Dawson PE, Kent SBH. Synthesis of native proteins by chemical ligation. Annu Rev Biochem. 2000;69:923–960. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
- 46.Balambika R, Inui T, Sargsyan H, Arshava B, Cohen LS, Ding FX, Becker JM, Naider F. Synthesis of a double transmembrane domain fragment of Ste2p by native chemical ligation. Int J Pept Res Ther. 2007;13:251–263. [Google Scholar]
- 47.Rajagopal S, Kent SBH. Total chemical synthesis and biophysical characterization of the minimal isoform of the KChIP2 potassium channel regulatory subunit. Protein Sci. 2007;16:2056–2064. doi: 10.1110/ps.072876107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.David R, Richter MPO, Beck-Sickinger AG. Expressed protein ligation: Method and applications. Eur J Biochem. 2004;271:663–677. doi: 10.1111/j.1432-1033.2004.03978.x. [DOI] [PubMed] [Google Scholar]
- 49.Severinov K, Muir TW. Expressed protein ligation, a novel method for studying protein-protein interactions in transcription. J Biol Chem. 1998;273:16205–16209. doi: 10.1074/jbc.273.26.16205. [DOI] [PubMed] [Google Scholar]
- 50.Vitali F, Henning A, Oberstrass FC, Hargous Y, Auweter SD, Erat M, Allain FHT. Structure of the two most C-terminal RNA recognition motifs of PTB using segmental isotope labeling. EMBO J. 2006;25:150–162. doi: 10.1038/sj.emboj.7600911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Vasudevan S, Sojitrawala R, Zhao W, Cui C, Xu C, Fan D, Newhouse Y, Balestra R, Jerome WG, Weisgraber K, Li Q, Wang J. A monomeric, biologically active, full-length human apolipoprotein E. Biochemistry. 2007;46:10722–10732. doi: 10.1021/bi700672v. [DOI] [PubMed] [Google Scholar]
- 52.Skrisovska L, Allain FHT. Improved segmental isotope labeling methods for the NMR study of multidomain or large proteins: Application to the RRMs of NpI3p and hnRNP L. J Mol Biol. 2008;375:151–164. doi: 10.1016/j.jmb.2007.09.030. [DOI] [PubMed] [Google Scholar]
- 53.Muralidharan V, Muir TW. Protein ligation: An enabling technology for the biophysical analysis of proteins. Nat Methods. 2006;3:429–438. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- 54.Muona M, Aranko AS, Iwai H. Segmental isotopic labelling of a multidomain protein by protein ligation by protein trans-splicing. Chem Bio Chem. 2008;9:2958–2961. doi: 10.1002/cbic.200800604. [DOI] [PubMed] [Google Scholar]
- 55.Li Y. Split-inteins and their bioapplications. Biotechnol Lett. 2015;37:2121–2137. doi: 10.1007/s10529-015-1905-2. [DOI] [PubMed] [Google Scholar]
- 56.Volkmann G, Iwaï H. Protein trans-splicing and its use in structural biology: Opportunities and limitations. Mol Bio Sys. 2010;6:2110–2121. doi: 10.1039/c0mb00034e. [DOI] [PubMed] [Google Scholar]
- 57.Jackson DY, Burnier J, Quan C, Stanley M, Tom J, Wells JA. A designed peptide ligase for total synthesis of ribonuclease A with unnatural catalytic residues. Science. 1994;266:243–247. doi: 10.1126/science.7939659. [DOI] [PubMed] [Google Scholar]
- 58.Kobashigawa Y, Kumeta H, Ogura K, Inagaki F. Attachment of an NMR-invisible solubility enhancement tag using a sortase-mediated protein ligation method. J Biomol NMR. 2009;43:145–150. doi: 10.1007/s10858-008-9296-5. [DOI] [PubMed] [Google Scholar]
- 59.Nguyen GKT, Cao Y, Wang W, Liu CF, Tam JP. Site-specific N-terminal labeling of peptides and proteins using butelase 1 and thiodepsipeptide. Angew Chem Int Ed. 2015;54:15694–15698. doi: 10.1002/anie.201506810. [DOI] [PubMed] [Google Scholar]
- 60.Freiburger L, Sonntag M, Hennig J, Li J, Zou P, Sattler M. Efficient segmental isotope labeling of multi-domain proteins using sortase A. J Biomol NMR. 2015;63:1–8. doi: 10.1007/s10858-015-9981-0. [DOI] [PubMed] [Google Scholar]
- 61.Antos JM, Truttmann MC, Ploegh HL. Recent advances in sortase-catalyzed ligation methodology. Curr Opin Struct Biol. 2016;38:111–118. doi: 10.1016/j.sbi.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iwai H, Züger S, Jin J, Tam PH. Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett. 2006;580:1853–1858. doi: 10.1016/j.febslet.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 63.Zettler J, Schütz V, Mootz HD. The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett. 2009;583:909–914. doi: 10.1016/j.febslet.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 64.Barklis E, Alfadhli A, McQuaw C, Yalamuri S, Still A, Barklis RL, Kukull B, Lopez CS. Characterization of the in vitro HIV-1 capsid assembly pathway. J Mol Biol. 2009;387:376–89. doi: 10.1016/j.jmb.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McNeill SA, Gor’kov PL, Shetty K, Brey WW, Long JR. A low-E magic angle spinning probe for biological solid state NMR at 750 MHz. J Magn Reson. 2009;197:135–144. doi: 10.1016/j.jmr.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- 67.Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344:631–637. [Google Scholar]
- 68.Morcombe CR, Gaponenko V, Byrd RA, Zilm KW. Diluting abundant spins by isotope edited radio frequency field assisted diffusion. J Am Chem Soc. 2004;126:7196–7197. doi: 10.1021/ja047919t. [DOI] [PubMed] [Google Scholar]
- 69.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRpipe: A multidimensional spectral processing system based on Unix pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 70.Mehler M, Eckert CE, Busche A, Kulhei J, Michaelis J, Becker-Baldus J, Wachtveitl J, Dötsch V, Glaubitz C. Assembling a correctly folded and functional heptahelical membrane protein by protein trans-splicing. J Biol Chem. 2015;290:27712–27722. doi: 10.1074/jbc.M115.681205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lockless SW, Muir TW. Traceless protein splicing utilizing evolved split inteins. Proc Natl Acad Sci USA. 2009;106:10999–11004. doi: 10.1073/pnas.0902964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shah NH, Muir TW. Inteins: Nature’s gift to protein chemists. Chem Sci. 2014;5:446–461. doi: 10.1039/C3SC52951G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang J, Ablan SD, Derebail S, Hercík K, Soheilian F, Thomas JA, Tang S, Hewlett I, Nagashima K, Gorelick RJ, Freed EO, Levin JG. The interdomain linker region of HIV-1 capsid protein is a critical determinant of proper core assembly and stability. Virology. 2011;421:253–265. doi: 10.1016/j.virol.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rihn SJ, Wilson SJ, Loman NJ, Alim M, Bakker SE, Bhella D, Gifford RJ, Rixon FJ, Bieniasz PD. Extreme genetic fragility of the HIV-1 capsid. PLoS Path. 2013;9:e1003461. doi: 10.1371/journal.ppat.1003461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schur FKM, Obr M, Hagen WJH, Wan W, Jakobi AJ, Kirkpatrick JM, Sachse C, Krausslich HG, Briggs JAG. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science. 2016;353:506–508. doi: 10.1126/science.aaf9620. [DOI] [PubMed] [Google Scholar]
- 76.Wagner JM, Zadrozny KK, Chrustowicz J, Purdy MD, Yeager M, Ganser-Pornillos BK, Pornillos O. Crystal structure of an HIV assembly and maturation switch. eLife. 2016;5:e17063. doi: 10.7554/eLife.17063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stevens AJ, Brown ZZ, Shah NH, Sekar G, Cowburn D, Muir TW. Design of a split intein with exceptional protein splicing activity. J Am Chem Soc. 2016;138:2162–2165. doi: 10.1021/jacs.5b13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.