Abstract
Yersinia polynucleotide phosphorylase (PNPase), a 3’–5’ exoribonuclease, has been shown to affect growth during several stress responses. In E. coli, PNPase is one of the subunits of a multi-protein complex known as the degradosome, but also has degradosome-independent functions. The carboxy–terminus of E. coli ribonuclease E (RNase E) serves as the scaffold upon which PNPase, enolase (a glycolytic enzyme), and RhlB helicase all have been shown to bind. In the yersiniae, only PNPase has thus far been shown to physically interact with RNase E. We show by bacterial two-hybrid and co-immunoprecipitation assays that RhlB and enolase also interact with RNase E. Interestingly, although PNPase is required for normal growth at cold temperatures, assembly of the yersiniae degradosome was not required. However, degradosome assembly was required for growth in the presence of reactive oxygen species. These data suggest that while the Y. pseudotuberculosis PNPase plays a role in the oxidative stress response through a degradosome-dependent mechanism, PNPase’s role during cold stress is degradosome-independent.
Keywords: bacterial stress response, RNase E, cold shock
Introduction
Like other closely related Gram-negative enteric pathogens, Y. pseudotuberculosis employs a type III secretion system (T3SS) to infect host cells, and polynucleotide phosphorylase (PNPase), a phosphorolytic 3’–5’ exo-ribonuclease involved in RNA decay, is required for its optimal functioning (Rosenzweig et al 2005; Rosenzweig et al. 2007). Furthermore, we (and others) have observed that PNPase is required for the cold-shock response and/or acclimation for a number of organisms including: Yersinia pestis and Y. pseudotuberculosis (Rosenzweig et al. 2005; Rosenzweig et al. 2007), Escherichia coli (Jones et al. 1987; Polissi et al. 203; Mathy et al. 2001), and Yersinia enterocolitica (Goverde et al. 1998). Intriguingly, PNPase has been shown to physically interact with an essential endoribonuclease, RNase E, in both E. coli (Carpousis et al. 1994; Vanzo et al. 1998; Khemici and Carpousis 2004) and Y. pseudotuberculosis (Yang et al. 2008) forming a large multi-protein RNA surveillance/quality control complex termed the degradosome. However, the role of the degradosome in various yersiniae stress responses has not been well studied.
RNase E, PNPase, RhlB RNA helicase and enolase have all been identified as components of the E. coli degradosome (Khemici and Carpousis 2004; Rosenzweig et al. 2010). PNPase, enolase, and RhlB each bind to the carboxy terminal domain (CTD) of RNase E that serves as the scaffold. Interestingly, the CTD of RNase E is not well conserved and varies widely in various bacterial species (Erce et al. 2010). Typically a degradosome consists of both an exo- and endoribonuclease (e.g. PNPase and RNase E), and they are thought to work together in concert producing a synergistic effect that optimizes RNA decay of unwanted transcripts (Rosenzweig et al. 2010). However, a degradosome consisting of both RNase R, a cold-inducible exoribonuclease in E. coli (Cairrao et al. 2003; Chen and Deutscher 2010) required for the maturation of SsrA/tmRNA (Cairrao et al. 2003;), and RNase E has also been identified in the psychrotrophic Pseudomonas syringae, possibly suggesting the existence of a specialized cold adapted degradosome (Purusharth et al. 2005). What remains uncertain within the field of RNA biology is the exact contribution the degradosome plays in RNA decay/maturation relative to its individual components. In fact, an inability of E. coli to assemble a degradosome resulted in affecting only some RNA decay outcomes and had only a modest impact on growth kinetics (Kido et al. 1996; Jiang et al. 2000). Perhaps the degradosome specifically degrades subsets of transcripts following periods of induced gene expression (e.g. stress response) while other stress-induced transcripts are degraded by degradosome-independent mechanisms. In support of this, an E. coli PNPase-deficient mutant was found to be more sensitive to oxidative stress in the form of H2O2, and this PNPase requirement for tolerating oxidative stress was independent of degradosome association (Wu et al. 2009). However, a dominant-negative, carboxy-truncated RNase E variant in E. coli (unable to form a degradosome) resulted in poor autoregulation of the rne transcript, suggesting that the degradosome might be required for degradation of specific transcripts in E. coli (Briegel et al. 2006). When a similar carboxy-truncated RNase E variant was expressed in Y. pseudotuberculosis, increased sensitivity to host cell induced stress (HCIS), prompted by macrophage challenge ensued (Yang et al. 2008).
In addition to degradosome constituents’ physical interactions being demonstrated by co-immunoprecipitation (Co-IP) (Coburn et al. 1999; Yang et al. 2008), several bacterial 2 hybrid, B2H, (Karimova et al. 1998) assay studies have supported earlier Co-IP findings. More specifically, the B2H demonstrated an interaction between E. coli-derived PNPase and RhlB helicase (Liou et al. 2002). Additionally, the B2H assay demonstrated interactions between full-length PNPase, enolase, RhlB, and RNase E carboxy- terminal domains (CTD) as well as interactions between micro-domains of RNase E's CTD and the aforementioned full-length binding partners derived from Vibrio angustum S14 (Erce et al. 2009; Erce et al. 2010). Therefore, we sought to characterize the Y. pseudotuberculosis degradosome further since only PNPase has been shown to physically interact with RNase E (Yang et al. 2008) and to determine whether it is required for various abiotic stress responses.
Results
The RhlB RNA helicase and enolase are subunits of the Yersinia degradosome
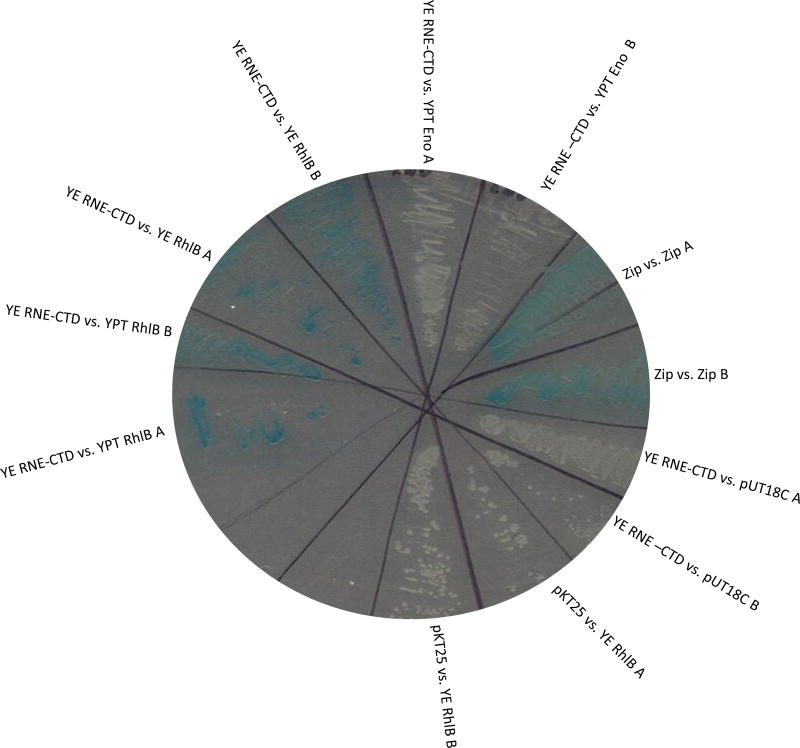
In an attempt to further identify Y. pseudotuberculosis degradosome constituents, we employed the B2H assay (Karimova et al. 1998) to determine whether RhlB and enolase also associate with the RNase E CTD. In this B2H assay, interaction between two proteins results in transcription of the Lac operon, and thus blue color on plates containing X-gal. Our data indicated that the RNase E CTD interacted very strongly with full-length RhlB helicase as evidenced by intensely blue colonies (Fig. 1 panel C). In fact, the intensity of blue mirrored that of the positive control Zip-Zip (compare panel C to B). Blue colonies also appeared when PNPase interacted with RNase E CTD (panel D); however, the overall intensity of blue was less than that of an RhlB-RNase E CTD interaction (compare panel D to B). Little interaction occurred between enolase and the RNase E CTD, as evidenced by weekly blue colonies (panel E). All experimental colonies observed appeared bluer than the empty vector negative control, pKT25RNE-CTD vs. pUT18Cempty vector (compare all panels to panel A).
Figure 1.
Representative B2H experiment testing f or interactions between the Y. pseudotuberculosis RNase E CTD scaffolding region and full length RhlB, enolase, or PNPase. Individual bacterial colony forming units were evaluated, and the positive control was pKT25-Zip vs. pUT18C-Zip. The negative control was pKT25-RNE1–465 vs. pUT18Cempty.
In addition to evaluating degradosome interaction of Y. pseudotuberculosis proteins, we also evaluated degradosome interaction of closely related Y. enterocolitica proteins and tested whether the Y. enterocolitica RNase E CTD interacted with both the Y. pseudotuberculosis and Y. enterocolitica RhlB degradosome-associated proteins. We chose looking at RhlB since it was the strongest binding partner for the Y. pseudotuberculosis RNase E CTD tested earlier (Fig. 1). Interestingly, the Y. enterocolitica RNase E CTD appeared to bind as well to the Y. enterocolitica RhlB protein as it did to the Y. pseudotuberculosis RhlB protein (Fig. 2). As was observed earlier with the Y. pseudotuberculosis RNase E CTD vs. Y. pseudotuberculosis enolase (Fig. 1), the Y. enterocolitica-derived RNase E CTD also interacted poorly with the Y. pseudotuberculosis derived enolase (Fig. 2). The positive control Zip-Zip appeared blue (as expected) while the two empty vector negative controls were white (as expected), pKT25RNE vs. pUT18Cempty and pKT25empty vs. pUT18CRhlB (Fig. 2).
Figure 2.
Representative B2H experiment testing for interactions between the Y. enterocolitica RNase E CTD scaffolding region and full length Y. pseudotuberculosis and Y. enterocolitica RhlB. Single colony bacterial isolates were evaluated, and the positive control was pKT25-Zip vs. pUT18C-Zip. The negative controls were pKT25 vs. pUT18C-Y. enterocolitica RhlB and pKT25- Y. enterocolitica RNE1–465 vs. pUT18C.
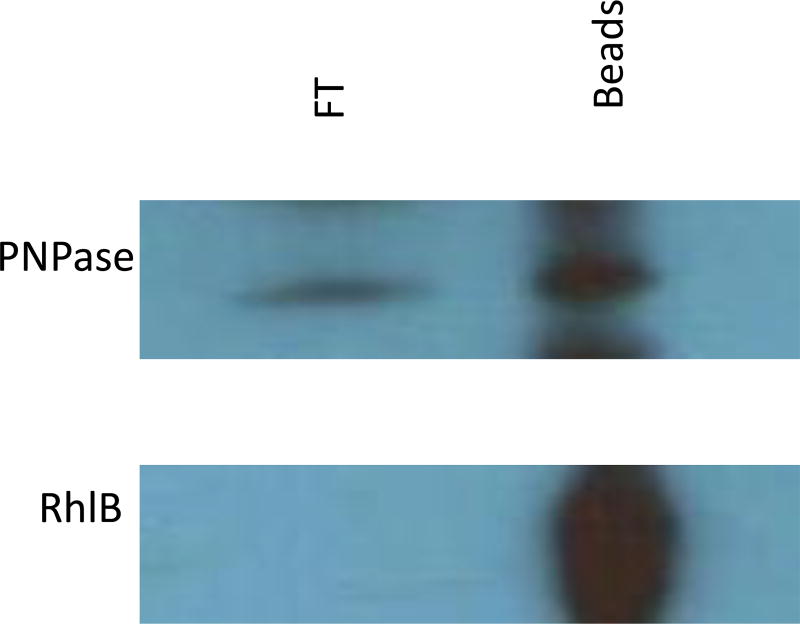
To validate our B2H findings (Fig. 1 and 2), co-immunoprecipitation (Co-IP) assays, utilizing polyclonal anti-RNase E antibodies fused to Protein G agarose beads, were employed. Immunoprecipitated complexes were resolved by SDS-PAGE and probed with polyclonal anti-RhlB or anti-PNPase antibodies. In agreement with our B2H results, RhlB clearly co-immunoprecipitated with RNase E (Fig. 3). PNPase also appeared to co-immunoprecipitate with RNase E (Fig. 3) as was demonstrated in earlier work (Yang et al. 2008). These B2H and co-IP experiments indicate that the RhlB and enolase are conserved subunits of the degradosome in Yersiniae.
Figure 3.
Co-immunoprecipitations using anti-RNase E antibody fused Protein G sepharose beads. Depicted is an immunoblot probing the co-precipitated protein complex (bead) and probing the flow through (FT) using either rabbit polyclonal anti-PNPase or -RhlB antibodies.
Degradosome involvement in Y. pseudotuberculosis stress responses
The degradosome and PNPase have previously been implicated in various stress responses, including macrophage induced stress, and cold stress (see discussion). To more completely understand their role during stress, we exposed a Δpnp mutant and a strain over-expressing an rne truncation to a variety of stresses. This rne truncation removed the CTD responsible for interaction with the other degradosome subunits, and its overexpression has previously been shown to interfere with degradosome assembly (Briegel et al. 2006; Yang et al. 2008).
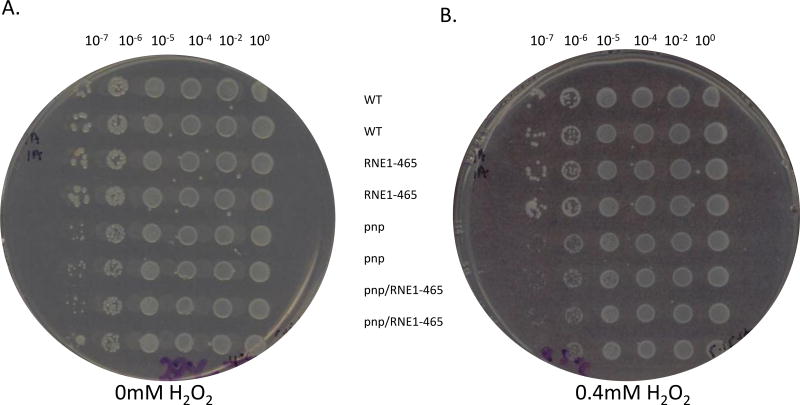
Since the ability of Y. pseudotuberculosis to respond to HCIS was previously shown to be dependent upon PNPase (Rosenzweig et al. 2005; Rosenzweig et al. 2007) as well as upon degradosome assembly (Yang et al. 2008), we were curious as to whether degradosome assembly was required for growth under oxidative stress which would be experienced during macrophage encounters. To test this directly, H2O2 liquid- and plate-based experiments were carried out. For plate-based assays, 0, 0.1, 0.2, 0.3, 0.4, 0.5, 1, 2, 4, 5, 10, 20, 50, and 100mM H2O2 plate concentrations were all evaluated. The Δpnp mutant formed smaller colonies on plates, which was exacerbated by 0.1–0.4mM H2O2 (Fig 4). In a manner similar to how E. coli did not require degradosome assembly during oxidative stress (Wu et al. 2009), interfering with degradosome assembly did not affect growth on H2O2-containing plates (Fig. 4B).
Figure 4.
Representative H2O2 plate experiment. Y. pseudotuberculosis + empty vector pBAD24 (WT), Y. pseudotuberculosis Δpnp + empty vector pBAD24 (pnp), Y. pseudotuberculosis Δpnp + pBAD-RNE1–465 (pnp/RNE), and Y. pseudotuberculosis + pBAD-RNE1–465 (RNE) strains were grown on 0mM H2O2 (A) or 0.4mM H2O2 (B) at 30°C for 16 hours at which point the plates were scanned. All strains were spotted in duplicate for internal dilution and spotting controls.
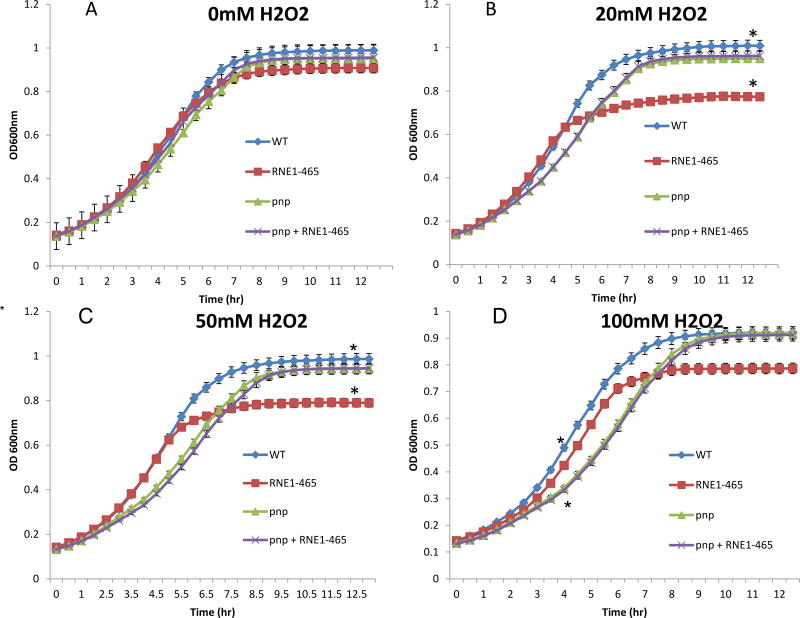
To better quantify H2O2 sensitivity, a liquid-based H2O2 stress test was carried out in which subcultures were optically monitored for growth following the addition of either 0mM, 20mM, 50mM, or 100mM H2O2 (Fig. 5). Remarkably, the more sensitive liquid-based assay revealed two significant effects. First, as indicated by the change in the slope of the graphs in figure 6, the Δpnp mutant had a longer doubling time in H2O2 containing media, but not in control media. In addition, interfering with degradosome assembly caused a reduced culture density as cultures entered stationary phase. Both of these differences were statistically significant. For reasons not well understood, interfering with degradosome assembly in the Δpnp mutant mirrored the phenotype of the Δpnp mutant strain and suppressed the early stationary phenotype when only degradosome assembly was disrupted (Fig. 5).
Figure 5.
Representative H2O2 liquid growth experiment. Subcultures of Y. pseudotuberculosis + empty vector pBAD24 (WT), Y. pseudotuberculosis Δpnp + empty vector pBAD24 (pnp), Y. pseudotuberculosis Δpnp + pBAD-RNE1–465 (pnp/RNE), and Y. pseudotuberculosis + pBAD-RNE1–465 (RNE) were grown (in triplicate) in 100µl volumes using a 96 well plate. Following 1 hour of static growth, 0mM (A), 20mM (B), 50mM (C), or 100mM (D) H2O2 was a added to the appropriate wells, and plates were constantly agitated while grown at 30°C for 12 hours. Samples were read at optical density 600nm every 30 minutes. Asterisk between the WT and pnp samples in panel D at 4 hours and between WT and RNE in panel B and C at 12 hours denote statistical significance (p< 0.5). All statistical tests employed the Student’s T-test.
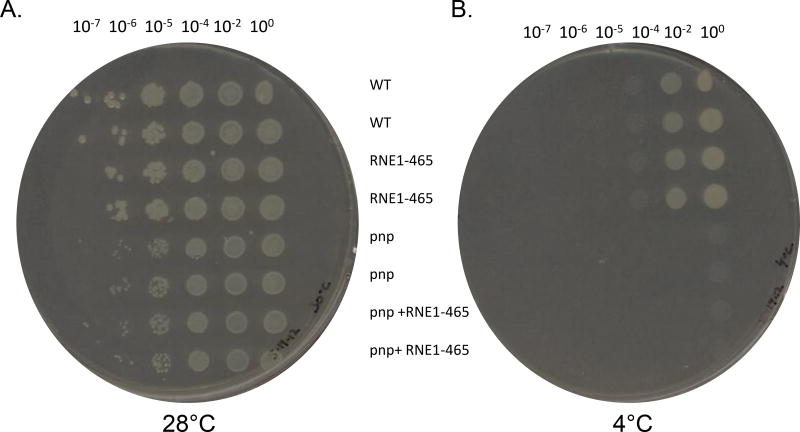
Figure 6.
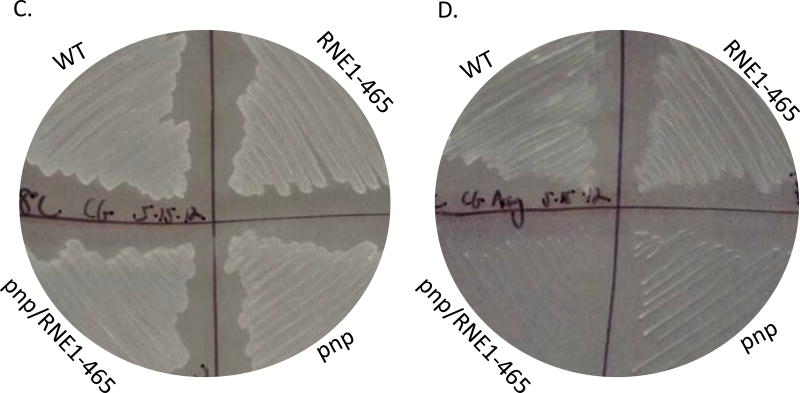
Representative cold-growth experiments. A. Various dilutions of saturated Y. pseudotuberculosis + empty vector pBAD24 (WT), Y. pseudotuberculosis Δpnp + empty vector pBAD24 (pnp), Y. pseudotuberculosis Δpnp + pBAD-RNE1–465 (pnp/RNE), and Y. pseudotuberculosis + pBAD-RNE1–465 (RNE) cultures were spotted (using a pronger) on LB agar plates and grown at 30°C for 16 hours (A) or at 4°C for 11 days (B) at which point the plates were scanned. The same aforementioned strains were streaked on plates and images were acquired following 16 hours of growth at 30°C (C) or following 11 days of growth at 4°C (D).
We also tested growth of these same strains at 4°C (Fig 6). Not surprisingly, and in agreement with previously published data (Rosenzweig et al. 2005; Rosenzweig et al. 2007), the Δpnp mutant was unable to grow at 4°C (Fig. 6B) despite relatively normal growth at 28°C (Fig.6A). When RNE1–465 was expressed, there was no effect on the cold-sensitive phenotype (Fig. 6). These data strongly suggest that the psychrotropic yersiniae’s ability to grow in the cold depends on PNPase in a degradosome-independent manner. To further evaluate the role that degradosome assembly might be playing in yersiniae stress responses, we challenged the strains with several antibiotics that target protein translation, membrane integrity, and cell wall integrity and found that neither the presence of PNPase nor the ability of the yersiniae degradosome to assemble altered antibiotic susceptibility profiles (data not shown).
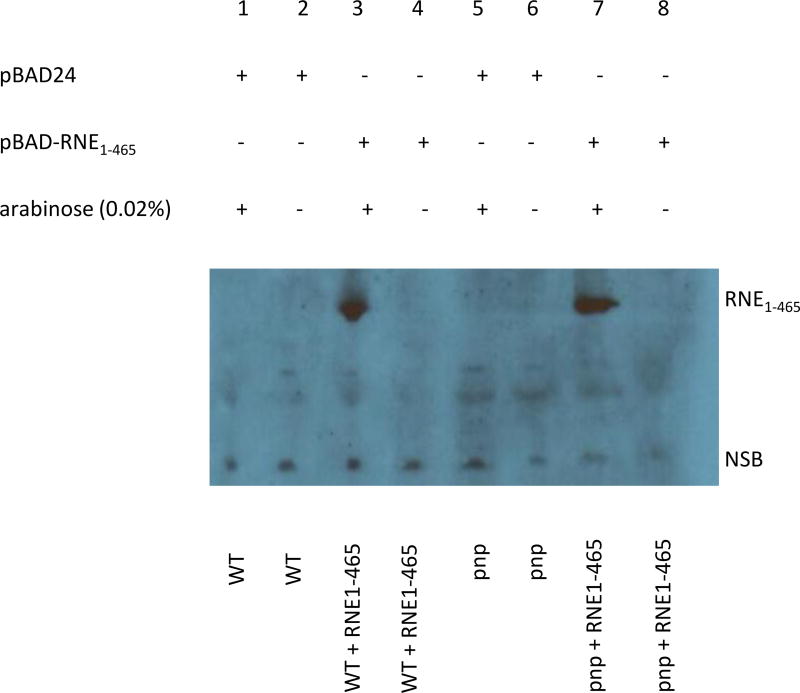
Since we observed that over-expression of RNE1–465 led to a significant reduction in biomass during oxidative stress, but that there was no similar reduction in biomass when expressed in the Δpnp background (Fig. 7), we hypothesized that perhaps PNPase affected expression of the plasmid-encoded RNE1–465. Following a 1.5 hour induction of RNE1–465 in both strains and Western blot analysis, we concluded that the truncated RNE1–465 was expressed similarly in both strains, and that PNPase was not modulating RNE1–465 expression levels. More specifically, the Y. pseudotuberculosis + empty vector pBAD24 (WT) and Y. pseudotuberculosis Δpnp + empty vector pBAD24 (pnp) controls did not express RNE1–465 when either induced with 0.02% arabinose or not (lanes 1, 2, 5, and 6). However, the Y. pseudotuberculosis + pBAD-RNE1–465 (RNE) and the Y. pseudotuberculosis Δpnp + pBAD-RNE1–465 (pnp/RNE) both expressed the ~ 52KDa RNE1–465 when induced with 0.02% arabinose (lanes 3 and 7).
Figure 7.
Arabinose induction of the truncated RNE1–465. Y. pseudotuberculosis + empty vector pBAD24 (WT), Y. pseudotuberculosis Δpnp + empty vector pBAD24 (pnp), Y. pseudotuberculosis Δpnp + pBAD-RNE1–465 (pnp + RNE), and Y. pseudotuberculosis + pBAD-RNE1–465 (WT + RNE) cultures were grown and induced for 1.5 hours with 0.02% arabinose. An immunoblot was performed employing polyclonal anti-RNase E antibody. CTD-deficient RNE1–465 was expressed in strains that contained the appropriate plasmid and that were induced with 0.02% arabinose. NSB= non-specific band.
Discussion
Y. pseudotuberculosis is a very close relative of the etiological agent of plague, Y. pestis, which diverged from Y. pseudotuberculosis between 15,000–20,000 years ago (Achtman et al. 1999). In fact, their RNase E, PNPase, RhlB and enolase proteins are 97–100% identical. Unlike Y. pestis (which has caused three major human pandemics), Y. pseudotuberculosis (like the more distantly related Y. enterocolitica) causes a relatively benign self-limiting gastro-intestinal disease in humans (Galindo et al. 2011). Being psychrotropic and a human pathogen, a better understanding of Y. pseudotuberculosis stress responses could result in the discovery of novel targets for chemotherapeutic design. Both temperature (i.e. cold) and oxidative stress responses have been characterized in this manuscript, the former potentially experienced by Y. pseudotuberculosis or Y. enterocolitica during food processing and shipping and the latter experienced when attacked by host innate immune cells during an infection. Knowing that the exoribonuclease, PNPase, is required for cold growth of several organisms (Jones et al. 1987; Goverde et al. 1998) including Y. pseudotuberculosis (Rosenzweig et al. 2005), we strove to evaluate whether the PNPase requirement for cold growth of Y. pseudotuberculosis was degradosome-dependent. Similarly, we chose to characterize the Y. pseudotuberculosis oxidative stress response since PNPase had already been implicated in the E. coli H2O2 stress response in a degradosome-independent manner (Wu et al. 2009). In fact, PNPase has already been shown to promote yersiniae virulence and is required for optimal T3SS function (Rosenzweig et al. 2005 & 2007), so identifying the exact constituents of the Y. pseudotuberculosis degradosome improves our understanding of how RNA metabolism impacts bacterial virulence as well.
Our data have identified RhlB, PNPase, and RNase E as components of the Y. pseudotuberculosis degradosome, which previously had been shown to only inlcude PNPase and RNase E (Yang et al. 2008). Furthermore, using the B2H assay, we demonstrated how the carboxy-terminus of a Y. enterocolitica derived RNase E protein can also interact with Y. pseudotuberculosis RhlB helicase strongly supporting the notion that all pathogenic yersiniae can assemble a degradosome. We further characterized the role the Y. pseudotuberculosis degradosome plays in various stress responses and surprisingly found that the Y. pseudotuberculosis degradosome is not implicated in all stress responses that require PNPase involvement. More specifically, we determined that the Y. pseudotuberculosis cold-growth requirement for PNPase (Rosenzweig et al. 2005; Rosenzweig et al. 2007) is degradosome independent. However, Y. pseudotuberculosis degradosome assembly was required for the oxidative stress response. Degradosome involvement with oxidative stress is in agreement with a previously published report of its requirement for macrophage-induced stress (Yang et al. 2008) and in contrast to its dispensability in the E. coli oxidative stress response (Wu et al. 2009). This is a shining example of how even closely related Gram-negative, enteric bacteria, e.g. E. coli and Y. pseudotuberculosis, might behave differently to various stressors and employ different coping mechanisms to overcome the stress itself.
Unexpectedly, PNPase and the degradosome affect growth during H2O2 stress in different phases of growth. PNPase appeared important during log-phase growth of Y. pseudotuberculosis, while degradosome assembly affected biomass accumulation resulting in an early stationary phase. Even more unexpected was that the absence of PNPase suppressed the H2O2 sensitive phenotype of RNE1–465. Furthermore, the deletion of the PNPase-encoding gene did not diminish expression levels of RNE1–465, so the observation remains both intriguing and unexplained. In one scenario, PNPase responds to oxidative stress in Y. pseudotuberculosis independently during early growth; however, during later growth PNPase associates with the degradosome to overcome the stress and enter into an acclimation phase. Of course, such a scenario fails to explain the surprising and unexplained phenomenon in which the absence of PNPase suppressed the H2O2-sensitive phenotype of RNE1–465. Perhaps a global evaluation of transcript abundance in each strain during oxidative stress is warranted and could reveal clues to help explain why PNPase and the degradosome affect growth during H2O2 stress differently despite PNPase not diminishing expression levels of RNE1–465.
Taken together, these data have expanded our understanding of the Y. pseudotuberculosis degradosome by clearly identifying RhlB helicase as a member of the multi-protein complex. Additionally, these data have delineated the role of the Y. pseudotuberculosis degradosome in various stress responses. Whereas PNPase seemingly affects growth at 4°C in a degradosome-independent manner, the Y. pseudotuberculosis oxidative stress response clearly requires degradosome assembly to achieve optimal biomass during late log-phase growth. Realizing the unique contributions made by the degradosome during various stress responses could enable us to uncover novel chemotherapeutic targets more specifically aimed at disarming pathogens and making them more vulnerable/susceptible to those agents.
Materials and Methods
Strains and plasmids
DHM1 is a cya-deleted E. coli, slow growing, temperature sensitive mutant strain that is used for the B2H screening.
For the immunoprecipitations, Yersinia pseudotuberculosis ATCC® 6902™ was used.
YPT YPIII pIB102 (Bölin and Wolf-Watz 1984) (WT) and YPIII pIB100Δpnp (Rosenzweig et al. 2005) were used for the cold growth and H2O2 plate-based assays.
The arabinose-inducible promoter containing pBAD24 (Guzman et al. 1995) plasmid was used as a cloning vector into which a carboxy-truncated RNase E (encoding only the first 465 amino acid residues in the amino terminus) was cloned (Yang et al. 2008). For all inductions 0.02% arbinose was used unless otherwise noted. Ampicillin working concentrations were 100µg/mL.
Y. pseudotuberculosis Primers for B2H cloning
RNaseE CTD:
Forward: tcaggattcctccagcattggctacc
Reverse: tcagaattcttactcaacagattgc
PNPase:
Forward: tcaggatcctttgctgactccgattattcg
Reverse: tcagaattcttactctgctgctgcttc
RhlB helicase:
Forward: tcaggatcctatgagcaaaacacacttg
Reverse: tcagaattctcagcctggtcgcttacgg
Enolase:
Forward: tcaggatcctatgtccaaaattgttaaag
Reverse: tcagaattcttactggcctttaacttc
Y. enterocolitica Primers for B2H cloning
RNE CTD:
Forward: tcaggatcctctggcgacgttctctctg
Reverse: tcagaattcctattcaaccgattgtg
RhlB helicase:
Forward: tcaggatcctttgaccgaacagaag
Reverse: tcagaattctcagctcggtcgcttac
Cloning of Proteins for B2H
RNase E CTD was cloned into the plasmid pKT25 while full-length enolase, PNPase, and RhlB were cloned into plasmid pUT18C. PCR products were generated using a 2× PCR master mix (New England Biolabs), and all cloning was done using BamH1 and EcoR1 high fidelity enzymes (New England Biolabs). All constructs were sequenced to confirm that they were correct.
DHM1 E. coli transformations and B2H assay
After DHM1 were harvested at OD600 of 0.5–0.6, pellets were washed twice in 1.0mL of ice cold water, washed once in ice cold 10% polyethalene glycol (PEG), and resuspended in ~750µl 10% PEG. To introduce plasmids, the cells were electroporated at 1,700 V (Biorad Inc.). Following a 1 hour recovery at 30°C with agitation, transformations were plated on LB agar plates containing 40µg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranosid), IPTG (0.5mM), 100µg/ml of ampicillin, and 50µg/ml of kanamycin. Plates were placed at 30°C, and colonies were observed between 48–72 hours later (Euromedex Inc.).
Co-immunoprecipitation
YPT was grown in 100mL of LB medium to OD 620nm of 0.7. Cells were harvested by centrifugation at 5000g for 15 minutes at 4°C. Pellets were then resuspended in 5mL 1× IP Buffer as part of a commercially available Protein G immunoprecipiation Kit (Sigam Aldrich IP50). Complete EDTA-free protease inhibitor cocktail (Roche) (one tablet per 5mL of solution) was added to the resuspended cells. Cells were lysed via sonication and 600µl of sonicate/lysate was used for downstream IP reaction (Sigma IP Kit protcol).
Immunoblotting
Polyacrylamide gel electrophoresis (PAGE) was carried out using pre-cast, 10 well, 4–12% gradient BIS-TRIS gels (Invitrogen Life Technologies), and proteins were transferred unto nitrocellulose membranes (Biorad) and blocked with non-fat milk. Polyclonal rabbit anti-PNPase, anti-RNase E, and/or anti-RhlB helicase antibodies were used to probe RNase E complexes or whole cell extracts (at a dilution of 1:3000) for 1 hour at room temperature.
Cold growth assay
A previously published protocol (Rosenzweig et al. 2005) was used with several modifications. In short, 10-fold serial dilutions of saturated bacterial cultures were spotted in duplicate in ~ 2µl volumes (using a pronger) on 2 LB agar (Difco) plates containing 100µg/mL ampicillin (Sigma) and 0.02% arabinose (Sigma). One plate was placed at 30°C while the other was placed at 4°C and monitored for an 11 day period. Alternatively, cultures were streaked out on the aforementioned plates and monitored for their growth over an 11 day period.
H2O2 plate and liquid-based assays
Previously published protocols (Wu et al. 2009) were employed. In short, saturated cultures were diluted, and subcultures of OD 600nm ~0.2 were established in triplicate 100µl volumes of LB medium (Difco) in 96 well plates. Following static growth @ 30°C for 1.0 hours (with the appropriate antibiotic added and arabinose @ 0.02%), a stock 0.88 M H2O2 was added to the various cultures yielding H2O2 concentrations of either 0mM, 20mM, 50mM, or 100mM, respectively. Growth in the liquid cultures was monitored every 30 minutes over a 12 hour period with continuous agitation. Growth curves were plotted, and the Student’s T-test was used to determine statistical significance with p values < 0.05 considered significant.
For plate-based H2O2 assays, 10-fold serial dilutions of saturated bacterial cultures were spotted in duplicate (using a pronger) in ~2µl volumes on 2 LB agar (Difco) plates containing 100µg/mL Ampicillin (Sigma) and 0.02% Arabinose (Sigma). Plate H2O2 concentrations were 0mM, 0.4mM, 1mM, 2mM, 4mM, and 100mM.
Acknowledgments
We gratefully acknowledge the generosity of William Margolin for B2H strains and plasmids, Kevin Morano for use of a 96-well plate reader for the growth curves, Kurt Schesser for the YPT strains and pBAD-RNE1–465 and AJ Carpousis for anti-RNase E, -PNPase, and -RhlB polyclonal antibodies used for IPs and immunoblotting. We would also like to thank Melissa Erce for her helpful suggestions and Ashok K. Chopra for stimulating discussion. We would also like to acknowledge our funding from NASA Cooperative Agreement NNXO8B4A47A (JAR) and NSF Research Opportunity Award MCB-1020739 001 (AVH).
References
- Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. Yersina pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proceedings of the National Academy of Science of the United States of America. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel KJ, Baker A, Jain C. Identification and analysis of Escherichia coli ribonuclease E dominant-negative mutants. Genetics. 2006;172(1):7–15. doi: 10.1534/genetics.105.048553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölin I, Wolf-Watz H. Molecular cloning of the temperature-inducible outer membrane proteins of yersiniae. Infect Immun. 1984;43:72–78. doi: 10.1128/iai.43.1.72-78.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairrão F, Cruz A, Mori H, Arraiano CM. Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol Microbiol. 2003;50(4):1349–60. doi: 10.1046/j.1365-2958.2003.03766.x. [DOI] [PubMed] [Google Scholar]
- Carpousis AJ. The Escherichia coli RNA degradosome: structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem Soc Trans. 2002;30(2):150–5. [PubMed] [Google Scholar]
- Carpousis AJ, Van Houwe G, Ehretsmann C, Krisch HM. Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994;11;76(5):889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- Chen C, Deutscher MP. RNase R is a highly unstable protein regulated by growth phase and stress. RNA. 2010;16:667–72. doi: 10.1261/rna.1981010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn GA, et al. Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3’ exonuclease and a DEAD-box RNA helicase. Genes & Development. 1999;13:2594–2603. doi: 10.1101/gad.13.19.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erce MA, Low JK, March PE, Wilkins MR, Takayama KM. Identification and functional analysis of RNase E of Vibrio angustum S14 and two-hybrid analysis of its interaction partners. Biochim Biophys Acta. 2009 Aug;1794(8):1107–14. doi: 10.1016/j.bbapap.2009.03.016. 2009. Epub 2009 Apr 2. [DOI] [PubMed] [Google Scholar]
- Erce MA, Low JK, Wilkins MR. Analysis of the RNA degradosome complex in Vibrio angustum S14. FEBS J. 2010 Dec;277(24):5161–73. doi: 10.1111/j.1742-4658.2010.07934.x.5173. 2010. [DOI] [PubMed] [Google Scholar]
- Galindo CL, Rosenzweig JA, Kirtley ML, Chopra AK. Pathogenesis of Y. enterocolitica and Y. pseudotuberculosis in Human Yersiniosis. Journal of Pathogens. 2011:16. doi: 10.4061/2011/182051. Article ID 182051, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goverde RL, Huis in't Veld JH, Kusters JG, Mooi FR. The psychrotrophic bacterium Yersinia enterocolitica requires expression of pnp, the gene for polynucleotide phosphorylase, for growth at low temperature (5 degrees C) Mol Microbiol. 1998;28(3):555–69. doi: 10.1046/j.1365-2958.1998.00816.x. [DOI] [PubMed] [Google Scholar]
- Guzman L-M, Belin D, Carson M, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Diwa A, Belasco JG. Regions of RNase E important for 5'-end-dependent RNA cleavage and autoregulated synthesis. J Bacteriol. 2000;182(9):2468–75. doi: 10.1128/jb.182.9.2468-2475.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PG, VanBogelen RA, Neidhardt FC. Induction of proteins in response to low temperature in Escherichia coli. J Bacteriol. 1987;169(5):2092–5. doi: 10.1128/jb.169.5.2092-2095.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khemici V, Carpousis AJ. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol Microbiol. 2004;51(3):777–90. doi: 10.1046/j.1365-2958.2003.03862.x. [DOI] [PubMed] [Google Scholar]
- Kido M, Yamanaka K, Mitani T, Niki H, Ogura T, Hiraga S. RNase E polypeptides lacking a carboxyl-terminal half suppress a mukB mutation in Escherichia coli. J Bacteriol. 1996 Jul;178(13):3917–25. doi: 10.1128/jb.178.13.3917-3925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawal A, Jejelowo O, Chopra AK, Rosenzweig JA. Ribonucleases and Bacterial Virulence. Microbial Biotechnology. 2010a doi: 10.1111/j.1751-7915.2010.00212.x. Article first published online: 14 OCT 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawal A, Jejelowo OA, Rosenzweig JA. The Effects of Low-Shear Mechanical Stress on Yersinia pestis Virulence ASTROBIOLOGY. 2010b;10(9):881–888. doi: 10.1089/ast.2010.0493. 2010. [DOI] [PubMed] [Google Scholar]
- Liou GG, Chang HU, Lin CS, Lin-Chao S. DEAD Box RhlB RNA Helicase Physically Associates with Exoribonuclease PNPase to Degrade Double-stranded RNA Independent of the Degradosome-assembling Region of RNase E. THE JOURNAL OF BIOLOGICAL CHEMISTRY. 2002;277(43):41157–41162. doi: 10.1074/jbc.M206618200. [DOI] [PubMed] [Google Scholar]
- Mathy N, Jarrige AC, Robert-Le Meur M, Portier C. Increased expression of Escherichia coli polynucleotide phosphorylase at low temperatures is linked to a decrease in the efficiency of autocontrol. J Bacteriol. 2001;183:3848–54. doi: 10.1128/JB.183.13.3848-3854.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus K, Anastasov N, Kaberdin V, Francis KP, Miller VL, Scherer S. The AGUAAA motif in cspA1/A2 mRNA is important for adaptation of Yersinia enterocolitica to grow at low temperature. Mol Microbiol. 2003;50(5):1629–45. doi: 10.1046/j.1365-2958.2003.03795.x. [DOI] [PubMed] [Google Scholar]
- Neuhaus K, Rapposch S, Francis KP, Scherer S. Restart of exponential growth of cold-shocked Yersinia enterocolitica occurs after down-regulation of cspA1/A2 mRNA. J Bacteriol. 2000;182(11):3285–8. doi: 10.1128/jb.182.11.3285-3288.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polissi A, De Laurentis W, Zangrossi S, Briani F, Longhi V, Pesole G, Dehò G. Changes in Escherichia coli transcriptome during acclimatization at low temperature. Res Microbiol. 2003;154(8):573–80. doi: 10.1016/S0923-2508(03)00167-0. [DOI] [PubMed] [Google Scholar]
- Purusharth RI, Klein F, Sulthana S, Jäger S, Jagannadham MV, Evguenieva-Hackenberg E, Ray MK, Klug G. Exoribonuclease R interacts with endoribonuclease E and an RNA helicase in the psychrotrophic bacterium Pseudomonas syringae Lz4W. J Biol Chem. 2005;280(15):14572–8. doi: 10.1074/jbc.M413507200. [DOI] [PubMed] [Google Scholar]
- Rosenzweig JA, Abogunde O, Thomas K, Lawal A, Nguyen YU, Sodipe A, Jejelowo O. Spaceflight and modeled microgravity effects on microbial growth and virulence. Appl Microbiol Biotechnol. 2010 Jan;85(4):885–91. doi: 10.1007/s00253-009-2237-8. 2010. Epub 2009 Oct 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig JA, Chromy B, Echeverry A, Yang J, Adkins B, Plano GV, McCutchen-Maloney S, Schesser K. Polynucleotide phosphorylase independently controls virulence factor expression levels and export in Yersinia spp. FEMS Microbiol Lett. 2007;270(2):255–64. doi: 10.1111/j.1574-6968.2007.00689.x. 2007. [DOI] [PubMed] [Google Scholar]
- Rosenzweig JA, Weltman G, Plano GV, Schesser K. Modulation of yersinia type three secretion system by the S1 domain of polynucleotide phosphorylase. J Biol Chem. 2005;280(1):156–63. doi: 10.1074/jbc.M405662200. [DOI] [PubMed] [Google Scholar]
- Vanzo NF, Li YS, Py B, Blum E, Higgins CF, Raynal LC, Krisch HM, Carpousis AJ. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 1998;12(17):2770–81. doi: 10.1101/gad.12.17.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Jiang Z, Liu M, Gong X, Wu S, Burns CM, Li Z. Polynucleotide phosphorylase protects Escherichia coli against oxidative stress. Biochemistry. 2009;48(9):2012–20. doi: 10.1021/bi801752p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Inouye M. Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli. J Bacteriol. 2001;183(9):2808–16. doi: 10.1128/JB.183.9.2808-2816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Jain C, Schesser K. RNase E regulates the Yersinia type 3 secretion system. J Bacteriol. 2008;190(10):3774–8. doi: 10.1128/JB.00147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]