Abstract
The heterogeneity of patient phenotype data are an impediment to the research into the origins and progression of neuropsychiatric disorders. This difficulty is compounded in the case of rare disorders such as Phelan‐McDermid Syndrome (PMS) by the paucity of patient clinical data. PMS is a rare syndromic genetic cause of autism and intellectual deficiency. In this paper, we describe the Phelan‐McDermid Syndrome Data Network (PMS_DN), a platform that facilitates research into phenotype–genotype correlation and progression of PMS by: a) integrating knowledge of patient phenotypes extracted from Patient Reported Outcomes (PRO) data and clinical notes—two heterogeneous, underutilized sources of knowledge about patient phenotypes—with curated genetic information from the same patient cohort and b) making this integrated knowledge, along with a suite of statistical tools, available free of charge to authorized investigators on a Web portal https://pmsdn.hms.harvard.edu. PMS_DN is a Patient Centric Outcomes Research Initiative (PCORI) where patients and their families are involved in all aspects of the management of patient data in driving research into PMS. To foster collaborative research, PMS_DN also makes patient aggregates from this knowledge available to authorized investigators using distributed research networks such as the PCORnet PopMedNet. PMS_DN is hosted on a scalable cloud based environment and complies with all patient data privacy regulations. As of October 31, 2016, PMS_DN integrates high‐quality knowledge extracted from the clinical notes of 112 patients and curated genetic reports of 176 patients with preprocessed PRO data from 415 patients.
Keywords: clinical notes, knowledge extraction, knowledge integration, neuropsychiatric disorders, patient reported outcomes, rare
1. INTRODUCTION
Genetic causes of neuropsychiatric disorders are not well understood in general (Kerner, 2015). Research investigations using Genome Wide Association Study (GWAS) (Network and Pathway Analysis Subgroup of the Psychiatric Genomics Consortium, 2015; Wood, 2013), exome‐based sequencing (Girard et al., 2011; Iossifov et al., 2012; O'Roak et al., 2011; Vissers et al., 2010; Xu et al., 2012), and whole genome sequencing (Kong et al., 2012) techniques have revealed several candidate genes that are associated with common neuropsychiatric disorders such as Autism Spectrum Disorder (ASD), intellectual disability, and schizophrenia. However, in the case of rare disorders, understanding the genetic origins and progressions of disorders—one of the key objectives of Precision Medicine research (Collins & Varmus, 2015; Kohane, 2015; Kohane, Churchill, & Murphy, 2012)—is hindered by small patient population size, the consequent paucity of patient data, and the lack of robust phenotyping protocols (Baynam et al., 2015; Delude, 2015; Robinson, Mungall, & Haendel, 2015).
One such rare neuropsychiatric disorder is Phelan‐McDermid Syndrome (PMS) or 22q13 deletion syndrome (OMIM 606232) (Cusmano‐Ozog, Manning, & Eugene Hoyme, 2007; Phelan, 2008; Phelan & McDermid, 2012), with approximately 1,400 cases diagnosed worldwide, mostly in children. PMS is caused by deletion of the terminal end of the long arm of chromosome 22 or by mutation and loss of function of the SHANK3 gene (Macedoni‐Lukšič, Krgović, Zagradišnik, & Kokalj‐Vokač, 2013), which is also implicated in ASD (Gauthier et al., 2008; Uchino & Waga, 2013). Diagnosis is only possible with genetic testing and is often delayed. Early studies have looked at the effect of intranasal insulin therapy (Maxonus, Irnberger, & Rittinger, 2012; Zwanenberg et al., 2016) and the role of Insulin‐like Growth Factor‐1 (IGF‐1) (Kolevzon et al., 2014) in reversing some of the symptoms of PMS, but there is currently no known treatment for the disorder. A wide variety of symptoms have been observed in individuals with PMS, including poor muscle tone, intellectual disability, developmental delays, dysmorphic facial features, vesicoureteral reflux, gastroesophageal reflux, congenital cardiac diseases, and behavioral disorders. Given the scarcity of patient data resulting from the small patient population size, clinical notes and Patient Reported Outcomes (PRO) data—previously underutilized sources of detailed information about patient conditions—assume significant importance in Precision Medicine research into PMS. Comparative analysis of the genetic profiles of the cohort of PMS patients with patient phenotypes reported in the clinical notes and PRO data has the potential to identify correlations between polymorphisms and deletions of specific genes and patient phenotypes, as well as to identify patient subtypes based upon genotypic and phenotypic profiles.
The Phelan‐McDermid Syndrome Data Network (PMS_DN), a Patient Powered Research Network (Avillach et al., 2014; Fleurence, Beal, Sheridan, Johnson, & Selby, 2014; Frank et al., 2015) funded by the Patient Centered Outcomes Research Institute (PCORI, http://www.pcori.org), leverages patient clinical notes and PRO data to achieve its objective of furthering Precision Medicine research into PMS. The PMS_DN project is an example of a patient driven clinical research initiative where patients and their families are the primary stakeholders, managing all aspects of data governance and directing a patient‐centered research agenda in collaboration with academic research institutions. The objective of this paper is to demonstrate how PMS_DN facilitates research into PMS by:
Extracting knowledge from clinical notes by using a combination of Optical Character Recognition (OCR) and Natural Language Processing (NLP) methods.
Ensuring the high‐quality and trustworthiness of the knowledge extracted from clinical notes by allowing experts to crosscheck the knowledge against the de‐identified source raw text.
Integrating the knowledge extracted from clinical notes with PRO data and curated genetic reports from the same cohort of PMS patients, facilitating comparative analyses.
Provisioning free multi‐level access privileges to the integrated knowledge to clinical practitioners and investigators researching into neuropsychiatric disorders over a Web portal and over distributed research networks, while complying with all the stipulations of patient privacy regulations, including the Health Insurance Portability and Accountability Act (HIPAA).
Allaying concerns about long term scalability and viability of the project by adopting a cloud based computation environment.
2. MATERIALS AND METHODS
2.1. Data acquisition
The PMS Foundation (PMSF, http://22q13.org) is a nonprofit foundation founded and run by PMS families that promotes awareness and research into PMS. Through patient outreach activities, PMSF collected patient data from hundreds of families of PMS patients. The collected data included:
Patient Reported Outcomes: Patient Reported Outcomes (PRO) data comprises responses by parents and caregivers of PMS patients to detailed questionnaires about diagnoses, procedures, lab tests, medications, patient behavior, and patient conditions, which were collected and stored in the PMS Information Registry (PMSIR, http://pmsiregistry.patientcrossroads.org).
Clinical notes: The families of PMS patients provided consent to CareSync (http://caresync.com), a third‐party vendor, to request and obtain their health records, including clinical notes, from various healthcare providers on their behalf. CareSync collected the clinical notes and shared the PDF scans with the patients' families and with PMSF. This process greatly simplified the cumbersome and time‐consuming process of patients obtaining access to their health records (Lester, Boateng, Studeny, & Coustasse, 2016).
Curated Genetic Reports: Reports of PMS patients from genetic tests including Comparative Genome Hybridization (CGH) arrays, Single Nucleotide Polymorphism (SNP) arrays, and microarrays were collected, curated by trained genetic counselors, and stored in the PMSIR.
With periodic patient outreach activities, PMSF has progressively improved patient participation in terms of the number of families consenting to share their data with PMSIR and with PMS_DN.
2.2. Data processing
2.2.1. Clinical notes
We used the open source Tesseract OCR tool (Smith, 2007) to extract raw text content from the curated clinical notes. Then, the MITRE MIST tool (Aberdeen et al., 2010) and the Scrubber toolkit (McMurry, Fitch, Savova, Kohane, & Reis, 2013) in the Apache cTAKES NLP engine were used to erase Protected Health Information (PHI) elements from the text. Following de‐identification, the Apache cTAKES NLP engine (Savova et al., 2010) was deployed to extract knowledge by identifying occurrences of concepts defined in the Unified Medical Language System (UMLS) (Bodenreider, 2004) in the text. Apache cTAKES also identifies the context in which the concepts are mentioned in the sentence including negation, patient history, family history, and uncertainty. The identified UMLS concepts were mapped to concept definitions in 20 clinical terminologies (Figure 1) including ICD‐9/10 (http://www.icd9data.com, http://www.icd10data.com), MeSH (Rogers, 1963), SNOMED CT (Schulz & Klein, 2008), and the Human Phenotype Ontology (Robinson et al., 2008).
Figure 1.
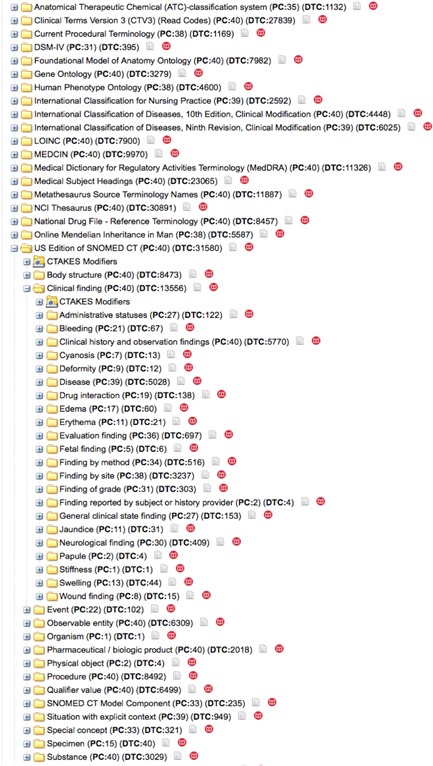
PMS_DN uses the Apache cTAKES NLP engine to extract occurrences of UMLS concepts in the clinical notes of PMS patients. The UMLS concepts are mapped to 20 different terminologies including ICD‐9, ICD‐10, SNOMED, MeSH, and NDFRT. The i2b2/tranSMART user interface allows for easy browsing—starting with broad biomedical concepts and drilling down to find specific patients and data of interest. The i2b2/tranSMART user interface also displays the counts of patients (PC) and distinct terms (DTC) associated with each concept at all levels of the hierarchy [Color figure can be viewed at http://wileyonlinelibrary.com]
2.2.2. Genetic reports
The genetic reports include results from sequencing, CGH arrays, and Fluorescent In‐Situ Hybridization (FISH) probes. Genetic reports are first curated by trained genetic counselors who fill 57 structured fields to represent the genetic abnormalities. Because of the disparity in techniques from which genetic data are obtained, all the curated genetic test result information was manually reviewed to extract the coordinates and genome assembly of the chromosomal abnormalities. Chromosomal coordinates for CGH were extracted from the relevant structured fields (chromosome, gain/loss, start, end), and from the International Society of Cytogenetics Nomenclature (ISCN 2013) standard (Simons, Shaffer, & Hastings, 2013) and comments where necessary. Chromosomal coordinates for FISH results were directly obtained in the GRCh38/hg38 genome assembly (Miga et al., 2014) from the National Center for Biotechnology Information (NCBI) Clone database (Schneider et al., 2013). When multiple assays were available for the same region, the most recent or the most precise—in terms of resolution—assay was used. In order of decreasing resolution of the sequence data, sequencing output was preferred over array CGH, and array CGH was preferred over FISH. Chromosomal coordinates were transformed from each original human genome assembly to the latest one available at the time of this study, GRCh38/hg38, using the University of California—Santa Cruz (UCSC) liftOver tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver). All duplications, deletions, and mutations were retained along with the original fields for the standard nomenclature, karyotype, and parental results; the only exceptions being chromosome alterations with coordinates that did not map to GRCh38/hg38.
2.2.3. Patient reported outcomes (PRO)
The Patient Reported Outcomes (PRO) data stored in the PMSIR comprises 1,300 questions over three distinct questionnaires:
A “clinical” questionnaire with questions regarding diagnosed comorbidities, symptoms, tests, and treatments for the whole range of known pathologies and features associated with PMS,
A “developmental” questionnaire, focusing on physical, motor, behavioral, cognitive, and social development, and
An “adult” questionnaire with specific questions aimed at patients aged 12 or more, regarding the evolution of symptoms after puberty. All the questions from the PRO dataset were manually mapped to UMLS Concept Unique Identifiers (CUIs) by a clinical expert before being preprocessed for statistical analysis.
The knowledge extracted from clinical notes was loaded by dedicated Extract Transform Load (ETL) pipelines into the PMS_DN data repository along with the PRO data and the processed curated genetic reports of the PMS patients.
2.3. Data integration on PMS_DN: Leveraging the i2b2/tranSMART platform
PMS_DN leverages the capabilities of the i2b2/tranSMART knowledge management platform (Patel et al., 2016; Perakslis, van Dam, & Szalma, 2010; Scheufele et al., 2014; Szalma, Koka, Khasanova, & Perakslis, 2010) to integrate heterogeneous datasets—including phenome, exposome, and genome data—and to facilitate browsing and comparative analysis of these datasets. The i2b2/tranSMART platform is layered upon the Informatics for Integrating Biology with Bedside (i2b2) clinical and biomedical data integration platform (Kohane, Churchill, & Murphy, 2012; Murphy et al., 2010). The i2b2 platform uses a simple and intuitive “observation centric” star schema data model that accommodates a variety of longitudinal patient level datasets including clinical data, prescriptions, and laboratory values. Multiple hierarchical ontologies describe the types of data contained within i2b2, allowing users to start with broad biomedical concepts and drill down to find specific patients and data of interest (Figure 1). New data types can be added to i2b2 by modifying the ontology but without changing the underlying database schema or the software. The ease of use of i2b2 has led to its adoption by over 150 University Hospital research centers worldwide.
2.4. Authorized user access to PMS_DN
The primary target audience for PMS_DN are clinical practitioners and researchers working in the areas of autism and other neuropsychiatric disorders. Qualified applicants affiliated with research institutions with an active interest in the research into neuropsychiatric disorders can request access to PMS_DN by filling out a registration form and agreeing to the terms of use. The registration request is reviewed by a Data Network Specialist at PMSF before approval.
Access to PMS_DN is granted at one of two levels: a basic level (Level 1) or an advanced level (Level 2). Level 1 access allows users to browse through and interrogate the patient aggregates of the integrated datasets on PMS_DN's Web portal. Figure 2 demonstrates the use of the i2b2/tranSMART interface to test a hypothesis about the relationship between patient age and hypotonia, a commonly reported symptom in PMS patients. Users with Level 2 access privileges, obtained from PMSF after mandatory Institutional Research Board (IRB) clearances from their institutions of affiliation, can see the raw, de‐identified patient level data (Figure 3) and download it as well. In addition, investigators with Level 2 access privileges can access a novel validation tool, which allows them to verify the accuracy of the knowledge extracted from clinical notes by cross‐checking the identified concepts against the anonymized sentences from which they were extracted (Figure 4). While eliminating residual errors of cTAKES caused by ambiguous context of the raw text, the validation tool improves the trustworthiness of the knowledge by allowing authorized investigators to see the raw text source of the knowledge. The input of the investigators is used to immediately update the knowledge in the PMS_DN repository (Figure 5). In a future release of PMS_DN, we will display the credentials of the experts performing the validation to other users, so the credibility of the validation input can be independently assessed. It must be noted that the knowledge validation step is not an exhaustive review of the entirety of the NLP engine's output. Instead, it is an open‐ended process where experts choose to crosscheck specific concepts of interest identified by the NLP pipeline against the raw anonymized sentences in which they occur.
Figure 2.
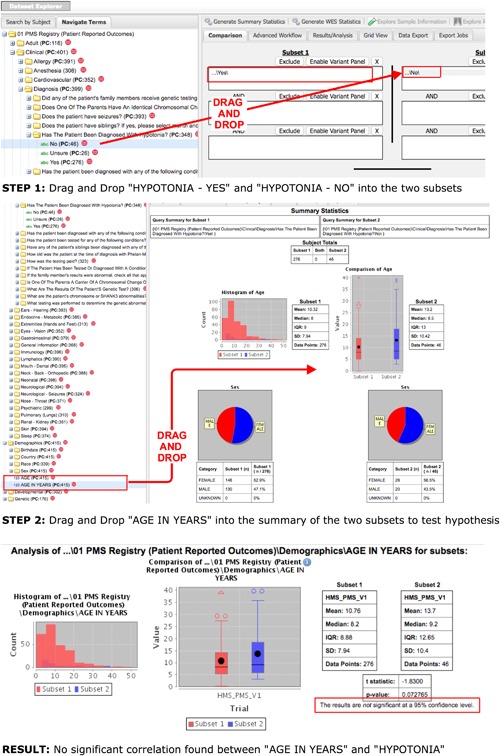
Hypothesis testing on the i2b2/tranSMART interface of PMS_DN. In STEP 1, the user drags and drops the “hypotonia” concept and the “Yes” and “No” values for this concept into the two different subset boxes. Then the user clicks the “Generate Summary Statistics” button. In STEP 2, the user drags and drops the “AGE IN YEARS” concept into the Summary section to test the hypothesis that Hypotonia is correlated with age of the patient. The RESULT shows that no significant correlation can be found [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 3.
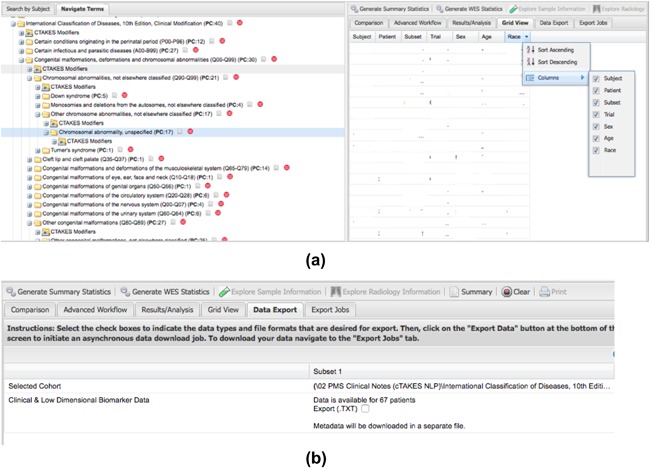
PMS_DN users with advanced access privileges obtained from the PMS Foundation (following appropriate IRB clearances) can view the raw data and perform basic sorting operations on the raw PMS patient data on PMS_DN (a) and also export it (b) [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 4.
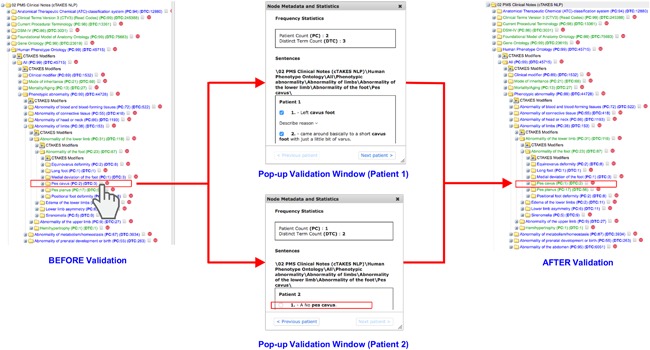
The pop‐up validation window allows clinical experts to cross check the extracted instance of the “Pes Cavus” concept from the Human Phenotype Ontology (“BEFORE Validation” screenshot) against the raw text from which it was extracted (“Pop‐up Validation Window” screenshots). Clicking on the grey icon next to the “Pes Cavus” concept brings up the Pop‐up Validation window where the user can see the raw sentences from which the concept was extracted. Verification by the expert (by deselecting the checkbox against the raw sentence for patient 2) results in the “Pes cavus”concept being displayed in a green colored font (“AFTER Validation” screenshot) indicating to future users that it has been verified by clinical experts. Note the change in the Patient Count value: from 2 in the “BEFORE Validation” screenshot to 1 in the “AFTER Validation” screenshot. This indicates the immediate update of the knowledge base with the expert's input on the validation window [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 5.
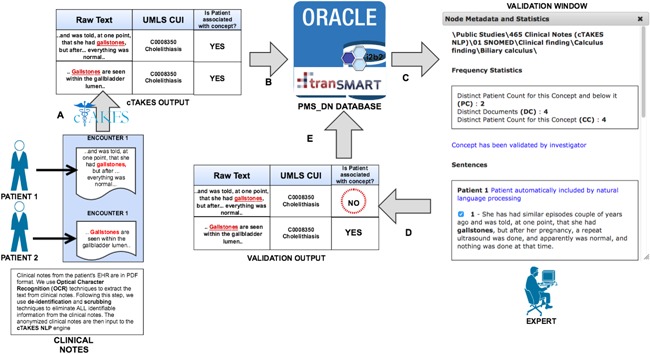
The novel validation tool that can be used by clinical experts to crosscheck the identified concepts against the sentences from which they were extracted. a) Apache cTAKES extracts instances of UMLS concepts from the raw text of clinical notes. b) The output of cTAKES is loaded into the PMS_DN database. c) An expert uses the validation tool to verify the extracted UMLS concept against the raw text source. d) The expert verifies that the extraction of the UMLS concept is valid (or otherwise). e) The input of the experts is used to update the knowledge in the PMS_DN database immediately [Color figure can be viewed at http://wileyonlinelibrary.com]
PMS_DN uses the single sign on feature of the OAuth2 authorization protocol (http://oauth.net/2/) to leverage the login credentials from: a) Harvard Medical School, Boston Children's Hospital, or the University of Pittsburgh, or b) NIH eRA Commons or c) Google Mail or d) GitHub (http://github.com) to login to the i2b2/tranSMART Web portal. The OAuth2 based single sign on feature obviates a potential security loophole associated with the storage of user login credentials on PMS_DN.
To foster collaborative research with similar Patient Powered Research Networks, snapshots of PMS_DN data in the form of patient counts for queried parameters are available to authorized investigators using distributed research networks such as SHRINE (Weber et al., 2009) and the PCORnet PopMedNet (http://www.popmednet.org).
2.5. Cloud hosting
To ensure long‐term scalability and to eliminate concerns about data archival and hardware maintenance and procurement, we have ported the PMS_DN application to a HIPAA compliant cloud based environment hosted by Amazon Web Services (AWS, http://aws.amazon.com). The PMS_DN data repository is hosted on a Relational Data Service (RDS) instance of AWS. The ETL pipelines are hosted on dedicated Elastic Compute Cloud (EC2) instances of AWS. The raw clinical notes are stored in a secure Simple Storage Service (S3) instance of AWS prior to processing. Figure 6 displays the entire cloud‐based architecture and data flows of PMS_DN.
Figure 6.
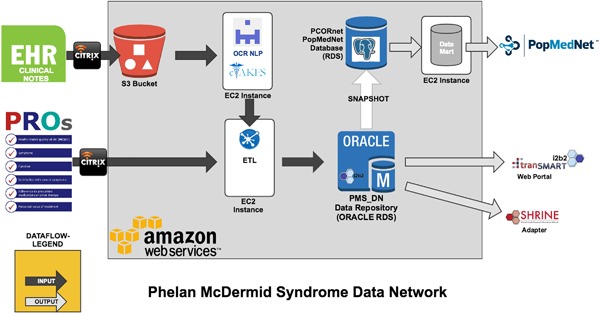
Cloud‐hosted architecture and data flow of PMS_DN [Color figure can be viewed at http://wileyonlinelibrary.com]
3. RESULTS
As of October 31, 2016, 623 families (334 in the USA) provided consent to PMSF to share their data with PMS_DN. PMS_DN integrates: a) the knowledge extracted by Apache cTAKES from the clinical notes of 112 patients comprising 40,320 pages in 2202 files, b) preprocessed PRO data from 415 patients, and c) curated genetic information from 176 patients. Following integration, 70 patients were linked across the three datasets, that is, PMS_DN has the full complement of clinical notes, genetic reports, and PRO data for 70 patients, enabling comparative analyses across the datasets (Figure 7). This number is expected to increase as more patient data becomes available. Authorized users can access and interrogate the integrated PMS patient data on the i2b2/tranSMART Web user interface of PMS_DN at https://pmsdn.hms.harvard.edu. Level 2 users with advanced access privileges and the appropriate IRB clearances can: i) obtain advanced, raw data download privileges on PMS_DN from PMSF and also ii) verify the accuracy of the knowledge extracted from clinical notes by cross‐checking the identified concepts with anonymized sentences from which they were extracted using the validation tool.
Figure 7.
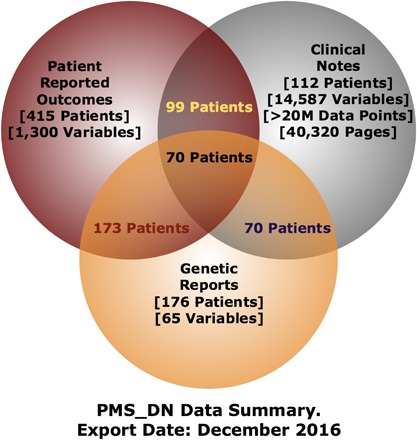
As of Oct 31, 2016, PMS_DN integrates the knowledge extracted from the clinical notes of 112 patients with the curated genetic reports of 176 patients and the Patient Reported Outcomes data obtained from 415 patients. PMS_DN contains all three datasets—PRO, clinical notes, and curated genetic reports—of 70 patients [Color figure can be viewed at http://wileyonlinelibrary.com]
4. DISCUSSION
PMS_DN facilitates research into the origins and treatment of PMS by making high quality, trustworthy knowledge available to clinical practitioners and investigators researching neuropsychiatric disorders, while safeguarding patient privacy through rigorous patient de‐identification methods.
4.1. Patient de‐identification
PMS_DN uses a combination of two independent anonymizers—the MITRE MIST anonymizer (Aberdeen et al., 2010) and the Scrubber toolkit (McMurry et al., 2013) in the Apache cTAKES NLP engine—to remove PHI elements from the clinical notes. In a study (McMurry et al., 2013), the Scrubber toolkit in Apache cTAKES identified and removed approximately 98% of the PHI elements (Recall = 98%) from a test corpus of clinical notes selected from the i2b2 De‐Identification Challenge dataset (Uzuner, Luo, & Szolovits, 2007). However, the same study reported a very low precision score, that is, a number of useful non‐PHI elements were removed from the clinical notes by the Apache cTAKES Scrubber in addition to the PHI elements. Another investigation studied the effectiveness of the MITRE MIST tool in removing PHI elements from clinical notes (Deleger, Molnar, Savova, Xia, Lingren, Li, & Solti, 2013) and reported F‐Scores (the harmonic mean of precision and recall metrics) (Hripcsak & Rothschild, 2005) of 93.48% and 95.2% at sentence‐level and word‐level de‐identification. These performance metrics were comparable with the performance of human experts in identifying PHI elements in the same corpus of clinical notes.
Because a maximally effective de‐identifier with maximal precision and recall performance metrics has yet to be developed, the PMS_DN combines the two independent anonymizers to try and remove PHI elements from the PMS patients' clinical notes to the maximum extent possible. Despite these efforts, the likelihood of the appearance of PHI elements in the clinical notes cannot be ruled out. We have attempted to mitigate this limitation by restricting the visibility of the anonymized raw text of the clinical notes (on the validation window) to only those users with Level 2 advanced access privileges.
Given the early stage of deployment of PMS_DN, the patient de‐identification pipeline has not been observed to adversely impact the comprehensibility of the content of the clinical notes so far. A typical example of an anonymized sentence from the clinical notes can be seen in Figure 4 as displayed in the validation window to a Level 2 user for verification. At present, only the exact sentence from which the concept was extracted is displayed in the validation window. In a future version of PMS_DN, we plan to display, in addition to the source sentence for the concept, the sentences immediately preceding and following this source sentence to try and make the context clearer to the user accessing the validation window.
4.2. Knowledge extraction from clinical notes and expert validation
From the anonymized text in clinical notes, the Apache cTAKES NLP engine identified the mentions of concepts defined in the UMLS in addition to the appropriate context—including negation, uncertainty, patient history, and family history—in which the extracted concept is mentioned. The validation tool allows experts to cross‐check whether the context has been correctly identified by the NLP engine and make corrections where necessary. This is intended to be an open‐ended process and not an exhaustive review of the functional efficiency of the NLP engine. The credentials of the experts—including research background and interests, institutions of affiliation, and the relevance of their proposed research work with PMS patient data to the objective of PMS Foundation—who perform these validations are carefully reviewed by the steering committee at the PMS Foundation before access is granted. This ensures a certain level of credibility to the validation input from the experts. At present, the identity of the users who perform these validations are logged by PMS_DN but not displayed to the end users. In future, the credentials of the users will also be displayed to authorized users with Level 2 access privileges so the quality of the input can be independently assessed by users.
4.3. PRO questionnaire
The PRO dataset comprises answers to approximately 1,300 questions that were sourced by the PMSF from existing surveys and databases, including the Autism Genetic Resource Exchange (AGRE) (Lajonchere & Consortium, 2010), the PMS survey by Dr. Katy Phelan, and Common Data Elements and questions in other specific condition surveys about phenotypes reported by PMS patients including seizures, lymphedema, sleep disorders, behavioral disorders, and developmental delays as well as cardiac and renal abnormalities. Expert researchers reviewed and edited the initial draft of questions and delivered two sets of questions: A Clinical Survey of 100 questions split into 23 topics such as Cardiovascular, Seizures, and Sleep and a Developmental Survey split into 11 topics such as Fine and Gross Motor Skills, Puberty Status, and Communication Development. Some of these questions are specific to the symptoms exhibited by PMS patients such as dysplastic toenails. There are also a number of questions that ask about more common conditions such as seizures, reflux, and behavioral patterns associated with ASD. The ASD related questions are relevant given that studies have reported the prevalence of symptoms of ASD in PMS patients (Oberman, Boccuto, Cascio, Sarasua, & Kaufmann, 2015) and gene‐linkage studies have associated SHANK3 mutations with ASD (Leblond et al., 2014; Uchino & Waga, 2013).
4.4. Data sharing
It would be desirable to promote data sharing between PMS_DN and the other PPRNs to foster collaborative research between these projects. However, the stipulations of patient privacy regulations preclude easy data sharing. Therefore, at present, only patient counts can be shared across these projects over distributed research networks such as SCILHS SHRINE and PCORnet PopMedNet. A unified questionnaire pertinent to PMS patients as well as patients diagnosed with disorders related to other PPRNs would be highly desirable. This would spare the families of patients with rare disorders from the hassle of having to repeatedly provide the same information across different questionnaires. The Research Domain Criteria (RDoC) framework from the National Institute of Mental Health (NIMH) (Insel, Cuthbert, Garvey, Heinssen, Pine, Quinn & Wang, 2010) could be useful in addressing this concern. The objective of the RDoC framework is to bring about synergy between the diverse research projects into mental and behavioral disorders and by extension, between the various surveys that are used in the research into these disorders. RDoC provides a rigid framework comprising units of analysis (from molecules to self‐report) for behavioral and developmental domains including cognitive, positive valence, negative valence, social processes, arousal, and regulatory systems. We are in the process of mapping the questions from the PRO dataset of PMS_DN to the domains of the RDoC framework as lead off work in this direction. With similar mapping initiatives from other PPRNs over time, the vision of a unified questionnaire for all rare disorders can be achieved.
5. CONCLUSION
In this paper, we have described PMS_DN, a Patient Powered Research Network that exemplifies the potential of collaborations between academic researchers and family organizations such as PMSF to drive research into a rare genetic disorder: PMS. PMS_DN addresses the paucity of patient data in rare disease research by exploiting the rich yet underutilized sources of knowledge about patient conditions: clinical notes and self‐reported outcomes. PMS_DN uses a state‐of‐the‐art NLP engine, Apache cTAKES, to extract context sensitive knowledge from rich text descriptions in patient clinical notes before making this knowledge, along with self‐reported outcomes and genetic reports of the same patient cohort, available to authorized investigators. Further, to minimize inaccuracies in the extracted knowledge, PMS_DN implements a novel knowledge validation tool that utilizes clinical expert input to eliminate residual ambiguities. PMS_DN is hosted in a cloud computing environment guaranteeing scalability while mitigating concerns regarding long term viability of the project. By integrating diverse and heterogeneous data about patient phenotypes and genotypes, PMS_DN facilitates research that can identify patient subgroups for targeted therapies based upon genomic and phenotypic profiles. The comparative analyses of integrated datasets, made possible by PMS_DN, has the potential to yield an improved understanding of the associations between genotypic profiles and PMS patient phenotypes.
ACKNOWLEDGMENTS
The Phelan‐McDermid Syndrome Foundation, the Phelan‐McDermid Syndrome International Registry, the patients and their families, Chris Botka, and the Harvard Medical School Research Computing center. This work was partially funded through a Patient‐Centered Outcomes Research Institute (PCORI) Award (PPRN‐1306‐04814) phase I and II for development of the National Patient‐Centered Clinical Research Network, known as PCORnet; by Research Grant EDU_R_FY2015_Q2_HarvardMedicalSchool_Avillach‐NEW from Amazon Inc.; and National Institutes of Health — RFA‐HG‐13‐009 — Centers of Excellence for Big Data Computing in the Biomedical Sciences (U54) — Grant Number 1U54HG007963‐01.
CONFLICT OF INTEREST
None
DISCLAIMER
The statements presented in this article are solely the responsibility of the author(s) and do not necessarily represent the views of the Patient‐Centered Outcomes Research Institute (PCORI), its Board of Governors or Methodology Committee or other participants in PCORnet.
Kothari C, Wack M, Hassen‐Khodja C, et al. Phelan‐McDermid syndrome data network: Integrating patient reported outcomes with clinical notes and curated genetic reports. Am J Med Genet Part B. 2018;177B:613–624. 10.1002/ajmg.b.32579
REFERENCES
- Aberdeen, J. , Bayer, S. , Yeniterzi, R. , Wellner, B. , Clark, C. , Hanauer, D. , … Hirschman, L . (2010). The MITRE identification scrubber toolkit: Design, training, and assessment. International Journal of Medical Informatics, 79(12), 849–859. 10.1016/j.ijmedinf.2010.09.007 [DOI] [PubMed] [Google Scholar]
- Avillach, P. , Buelow, J. , Colletti, R. , Eichler, F. , Ginsberg, S. , Dewitt, E. M. , … Fleurence, R . (2014). Patient‐powered research networks: Building capacity for conducting patient‐centered clinical outcomes research. Journal of American Medical Informatics Association, 21(4), 583–586. 10.1136/amiajnl-2014-002758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baynam, G. , Walters, M. , Claes, P. , Kung, S. , LeSouef, P. , Dawkins, H. , … Goldblatt, J . (2015). Phenotyping: Targeting genotype's rich cousin for diagnosis. Journal of Paediatrics and Child Health, 51(4), 381–386. 10.1111/jpc.12705 [DOI] [PubMed] [Google Scholar]
- Bodenreider, O. (2004). The unified medical language system (UMLS): Integrating biomedical terminology. Nucleic Acids Research, 32(Database issue), D267–D270. 10.1093/nar/gkh061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, F. S. , & Varmus, H. A. (2015). New initiative on precision medicine. New England Journal of Medicine, 372.9(2015), 793–795. 10.1056/NEJMp1500523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusmano‐Ozog, K. , Manning, M. , & Eugene Hoyme, H . (2007). 22q13 deletion syndrome: A recognizable malformation syndrome associated with marked speech and language delay. American Journal of Medical Genetics, 145C(4), 393–398. 10.1002/ajmg.c.30155 [DOI] [PubMed] [Google Scholar]
- Deleger, L. , Molnar, K. , Savova, G. , Xia, F. , Lingren, T. , Li, Q. , … Solti, I . (2013). Large‐scale evaluation of automated clinical note de‐identification and its impact on information extraction. Journal of American Medical Informatics Association, 20(1), 84–94. 10.1136/amiajnl-2012-001012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delude, C. (2015). Deep phenotyping: The details of disease. Nature, 527, S14–S15. 10.1038/527S14a [DOI] [PubMed] [Google Scholar]
- Fleurence, R. L. , Beal, A. C. , Sheridan, S. E. , Johnson, L. B. , & Selby, J. V . (2014). Patient‐powered research networks aim to improve patient care and health research. Health Affairs, 33(7), 1212–1219. 10.1377/hlthaff.2014.0113 [DOI] [PubMed] [Google Scholar]
- Frank, L. , Forsythe, L. , Ellis, L. , Schrandt, S. , Sheridan, S. , Gerson, J. , … Daugherty, S . (2015). Conceptual and practical foundations of patient engagement in research at the patient‐centered outcomes research institute. Quality of Life Research, 24(5), 1033–1041. 10.1007/s11136-014-0893-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier, J. , Spiegelman, D. , Piton, A. , Lafrenière, R. G. , St‐Onge, J. , Lapointe, L. , … Rouleau, G. A . (2008). Novel de novo SHANK3 mutation in autistic patients. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 150B(3), 421–424. 10.1002/ajmg.b.30822 [DOI] [PubMed] [Google Scholar]
- Girard, S. L. , Gauthier, J. , Noreau, A. , Xiong, L. , Zhou, S. , Jouan, L. , … Rouleau, G. A . (2011). Increased exonic de novo mutation rate in individuals with schizophrenia. Nature Genetics, 43(9), 860–863. 10.1038/ng.886 [DOI] [PubMed] [Google Scholar]
- Hripcsak, G. , & Rothschild, A. (2005). Agreement, the F‐measure, and reliability in information retrieval. Journal of the American Medical Informatics Association, 12(3), 296–298. 10.1197/jamia.M1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel, T. , Cuthbert, B. , Garvey, M. , Heinssen, R. , Pine, D. S. , Quinn, K. , … Wang, P . (2010). Research domain criteria (RDoC): Toward a new classification framework for research on mental disorders. American Journal of Psychiatry, 167(7), 748–751. 10.1176/appi.ajp.2010.09091379 [DOI] [PubMed] [Google Scholar]
- Iossifov, I. , Ronemus, M. , Levy, D. , Wang, Z. , Hakker, I. , Rosenbaum, J. , … Wigler, M . (2012). De novo gene disruptions in children on the autistic spectrum. Neuron, 74(2), 285–299. 10.1016/j.neuron.2012.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner, B. (2015). Psychiatric genetics, neurogenetics, and neurodegeneration. Frontiers in Genetics, 5, 467 10.3389/fgene.2014.00467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohane, I. S. (2015). Ten things we have to do to achieve precision medicine. Science, 349(6243), 37–38. [DOI] [PubMed] [Google Scholar]
- Kohane, I. S. , Drazen, J. M. , & Campion, E. W . (2012). A glimpse of the next 100 years in medicine. New England Journal of Medicine, 367, 2538–2539. 10.1056/NEJMe1213371 [DOI] [PubMed] [Google Scholar]
- Kohane, I. S. , Churchill, S. E. , & Murphy, S. N . (2012). A translational engine at the national scale: Informatics for integrating biology and the bedside. Journal of the American Medical Informatics Association, 19(2), 181–185. 10.1136/amiajnl-2011-000492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolevzon, A. , Bush, L. , Ting Wang, A. , Halpern, D. , Frank, Y. , Grodberg, D. , … Buxbaum, J. D . (2014). A pilot controlled trial of insulin‐like growth factor‐1 in children with Phelan‐McDermid syndrome. Molecular Autism, 5, 54 https://doi.org/0.1186/2040-2392-5-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, A. , Frigge, M. L. , Masson, G. , Besenbacher, S. , Sulem, P. , Magnusson, G. , … Stefansson, K . (2012). Rate of de novo mutations and the importance of father's age to disease risk. Nature, 488(7412), 471–475. 10.1038/nature11396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajonchere, C. M. , & Consortium, A. G. R. E. (2010). Changing the landscape of autism research: The autism genetic resource exchange. Neuron, 68(2), 187–191. 10.1016/j.neuron.2010.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblond, C. S. , Nava, C. , Polge, A. , Gauthier, J. , Huguet, G. , Lumbroso, S. , … Bourgeron, T . (2014). Meta‐analysis of SHANK mutations in autism spectrum disorders: A gradient of severity in cognitive impairments. PLoS Genetics. 10(9), e1004580 10.1371/journal.pgen.1004580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester, M. , Boateng, S. , Studeny, J. , & Coustasse, A . (2016). Personal health records: Beneficial or burdensome for patients and healthcare providers? Perspectives in Health Information Management 13(Spring), 1h. PMCID: PMC4832132 [PMC free article] [PubMed] [Google Scholar]
- Macedoni‐Lukšič, M. , Krgović, D. , Zagradišnik, B. , & Kokalj‐Vokač, N . (2013). Deletion of the last exon of SHANK3 gene produces the full Phelan–McDermid phenotype: A case report. Gene, 524(2), 386–389. 10.1016/j.gene.2013.03.141 [DOI] [PubMed] [Google Scholar]
- Maxonus, I. , Irnberger, E. , & Rittinger, O . (2012). Intranasal insulin may influence motor activities and behaviour in Phelan McDermid syndrome. Neuropediatrics, 43, PS15_ 02 10.1055/s-0032-1307111 [DOI] [Google Scholar]
- McMurry, A. J. , Fitch, B. , Savova, G. , Kohane, I. S. , & Reis, B. Y . (2013). Improved de‐identification of physician notes through integrative modeling of both public and private medical text. BMC Medical Informatics and Decision Making, 13, 112 10.1186/1472-6947-13-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga, K. H. , Newton, Y. , Jain, M. , Altemose, N. , Willard, H. F. , & Kent, W. J . (2014). Centromere reference models for human chromosomes X and Y satellite arrays. Genome Research, 24(4), 697–707. 10.1101/gr.159624.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, S. N. , Weber, G. , Mendis, M. , Gainer, V. , Chueh, H. C. , Churchill, S. , … Kohane, I. S . (2010). Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). Journal of the American Medical Informatics Association, 17(2), 124–130. 10.1136/jamia.2009.000893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network and Pathway Analysis Subgroup of the Psychiatric Genomics Consortium . (2015). Psychiatric genome‐wide association study analyses implicate neuronal, immune and histone pathways. Nature Neurosciences, 18(6), 926 10.1038/nn.3922 [DOI] [PubMed] [Google Scholar]
- Oberman, L. M. , Boccuto, L. , Cascio, L. , Sarasua, S. , & Kaufmann, W. E . (2015). Autism spectrum disorder in Phelan McDermid syndrome: Initial characterization and genotype‐phenotype correlations. Orphanet Journal of Rare Diseases, 10, 105 10.1186/s13023-015-0323-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak, B. J. , Deriziotis, P. , Lee, C. , Vives, L. , Schwartz, J. J. , Girirajan, S. , … Eichler, E. E . (2011). Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature Genetics, 43(6), 585–589. 10.1038/ng.835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, C. J. , Pho, N. , McDuffie, M. , Easton‐Marks, J. , Kothari, C. , Kohane, I. S. , & Avillach, P . (2016). A database of human exposomes and phenomes from the US National Health and Nutrition Examination Survey. Nature Scientific Data, 3, 160096 10.1038/sdata.2016.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perakslis, E. D. , van Dam, J. , & Szalma, S . (2010). How informatics can potentiate precompetitive open‐source collaboration to jump‐start drug discovery and development. Clinical Pharmacology and Therapeutics, 87(5), 614–616. 10.1038/clpt.2010.21 [DOI] [PubMed] [Google Scholar]
- Phelan, K. , & McDermid, H. (2012). The 22q13. 3 deletion syndrome (Phelan‐McDermid syndrome). Molecular Syndromology, 2(3–5), 186–201. 10.1159/000334260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan, M. C. (2008). Deletion 22q13. 3 syndrome. Orphanet Journal of Rare Diseases, 3, 14 10.1186/1750-1172-3-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, P. N. , Mungall, C. J. , & Haendel, M. (2015). Capturing phenotypes for precision medicine. Cold Spring Harbor Molecular Case Studies, 1, a000372 10.1101/mcs.a000372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, P. N. , Köhler, S. , Bauer, S. , Seelow, D. , Horn, D. , & Mundlos, S . (2008). The human phenotype ontology: A tool for annotating and analyzing human hereditary disease. The American Journal of Human Genetics, 83(5), 610–615. 10.1016/j.ajhg.2008.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, F. B. (1963). Communications to the editor. Bulletin of the Medical Library Association, 51(1), 114–116. [PMC free article] [PubMed] [Google Scholar]
- Savova, G. K. , Masanz, J. J. , Ogren, P. V. , Zheng, J. , Sohn, S. , Kipper‐Schuler, K. C. , & Chute, C. G . (2010). Mayo clinical text analysis and knowledge extraction system (ctakes): Architecture, component evaluation and applications. Journal of the American Medical Informatics Association, 17, 507–513. 10.1136/jamia.2009.001560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheufele, E. , Aronzon, D. , Coopersmith, R. , McDuffie, M. T. , Kapoor, M. , Uhrich, C. A. , … Palchuk, M. B . (2014). TranSMART: An open source knowledge management and high content data analytics platform. AMIA Joint Summits Translational Science Proceedings, 2014, 96–101. PMCID:pmc4333702 [PMC free article] [PubMed] [Google Scholar]
- Schneider, V. A. , Chen, H. , Clausen, C. , Meric, P. A. , Zhou, Z. , Bouk, N. , … Church, D. M . (2013). Clone DB: An integrated NCBI resource for clone‐associated data. Nucleic Acids Research 41(Database issue), D1070–D1078. 10.1093/nar/gks1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, S. , & Klein, G. (2008). SNOMED CT—Advances in concept mapping, retrieval, and ontological foundations. Selected contributions to the semantic mining conference on SNOMED CT (SMCS 2006). BMC Medical Informatics Decision Making, 8(Suppl 1), S1 10.1186/1472-6947-8-S1-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, A. , Shaffer, L. G. , & Hastings, R. J . (2013). Cytogenetic nomenclature: Changes in the ISCN 2013 compared to the edition. Cytogenetic and Genome Research, 141, 1–6. 10.1159/000353118 [DOI] [PubMed] [Google Scholar]
- Smith, R. (2007). An overview of the Tesseract OCR engine. Proceedings of the 9th IEEE International Conference on Document Analysis and Recognition (ICDAR), 2, 629–633. [Google Scholar]
- Szalma, S. , Koka, V. , Khasanova, T. , & Perakslis, E. D . (2010). Effective knowledge management in translational medicine. Journal of Translational Medicine, 8, 68 10.1186/1479-5876-8-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchino, S. , & Waga, C. (2013). SHANK3 as an autism spectrum disorder‐associated gene. Brain and Development, 35(2), 106–110. 10.1016/j.braindev.2012.05.013 [DOI] [PubMed] [Google Scholar]
- Uzuner, O. , Luo, Y. , & Szolovits, P . (2007). Evaluating the state‐of‐the‐art in automatic de‐identification. Journal of the American Medical Informatics Association, 14(5), 550–563. 10.1197/jamia.M2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers, L. E. , de Ligt, J. , Gilissen, C. , Janssen, I. , Steehouwer, M. , de Vries, P. , … Veltman. J. A . (2010). A de novo paradigm for mental retardation. Nature Genetics, 42(12), 1109–1112. 10.1038/ng.712 [DOI] [PubMed] [Google Scholar]
- Weber, G. M. , Murphy, S. N. , McMurry, A. J. , MacFadden, D. , Nigrin, D. J. , Churchill, S. , & Kohane, I. S . (2009). The shared health research information network (SHRINE): A prototype federated query tool for clinical data repositories. Journal of the American Medical Informatics Association, 16(5), 624–630. 10.1197/jamia.M3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, H. (2013). Neuropsychiatric disorders: Blurring diagnostic boundaries: Common genetic risk variants in major psychiatric disorders. Nature Reviews Neurology, 9, 181 10.1038/nrneurol.2013.54 [DOI] [PubMed] [Google Scholar]
- Xu, B. , Ionita‐Laza, I. , Roos, J. L. , Boone, B. , Woodrick, S. , Sun, Y. , … Karayiorgou, M . (2012). De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nature Genetics, 44(12), 1365–1369. 10.1038/ng.2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwanenberg, R. J. , Bocca, G. , Ruiter, S. A. J. , Dillingh, J. H. , Flapper, B. C. T. , van den Heuvel, E. R. , & van Ravenswaaij‐Arts, C. M. A . (2016). Is there an effect of intranasal insulin on development and behaviour in Phelan‐McDermid syndrome? A randomized, double‐blind, placebo‐controlled trial. European Journal of Human Genetics, 24(12), 1696–1701. 10.1038/ejhg.2016.109 [DOI] [PMC free article] [PubMed] [Google Scholar]


