Abstract
Chronic kidney disease (CKD) is a public health problem with very high prevalence and mortality. Yet, there is a paucity of effective treatment options, partly due to insufficient knowledge of underlying pathophysiology. We combined metabolomics (GCMS) with kidney gene expression studies to identify metabolic pathways that are altered in adults with non-diabetic stage 3–4 CKD versus healthy adults. Urinary excretion rate of 27 metabolites and plasma concentration of 33 metabolites differed significantly in CKD patients versus controls (estimate range − 68% to + 113%). Pathway analysis revealed that the citric acid cycle was the most significantly affected, with urinary excretion of citrate, cis-aconitate, isocitrate, 2-oxoglutarate and succinate reduced by 40–68%. Reduction of the citric acid cycle metabolites in urine was replicated in an independent cohort. Expression of genes regulating aconitate, isocitrate, 2-oxoglutarate and succinate were significantly reduced in kidney biopsies. We observed increased urine citrate excretion (+ 74%, p = 0.00009) and plasma 2-oxoglutarate concentrations (+ 12%, p = 0.002) in CKD patients during treatment with a vitamin-D receptor agonist in a randomized trial. In conclusion, urinary excretion of citric acid cycle metabolites and renal expression of genes regulating these metabolites were reduced in non-diabetic CKD. This supports the emerging view of CKD as a state of mitochondrial dysfunction.
Keywords: Citric acid cycle, Chronic kidney disease, Metabolomics, Gene expression, Mitochondria
Highlights
-
•
Urinary excretion rate and plasma concentration of 60 metabolites differed significantly in CKD patients versus controls.
-
•
Pathway analysis revealed that the citric acid cycle was the most significantly affected.
-
•
Expression of genes regulating TCA cycle was significantly reduced in kidney biopsies.
Chronic kidney disease (CKD) is very common and carries a high risk of complications and death. Patients with advanced disease have severe fatigue, organ dysfunction beyond the kidneys, and disturbances in sugar, protein and fat metabolism. We found many metabolites that differed significantly in kidney patients versus healthy controls. Analysis revealed that the citric acid cycle was the most significantly affected metabolic pathway. The citric acid cycle is performed in the cells` mitochondria and is the central metabolic hub where fuel molecules are converted into energy. This supports the view of CKD as a state of mitochondrial dysfunction.
1. Introduction
Chronic kidney disease (CKD) is an important public health problem with a high prevalence world-wide (10–14% of the general population) and strongly increased age-standardized death rate (GBD_Collaborators 2015). Yet, there is a paucity of effective treatment options with no major breakthroughs to reduce kidney damage since the introduction of renin-angiotensin system blockers 30 years ago. Lack of relevant animal models is a major limitation to identify novel targets for therapy, and the tradition of CKD patients being excluded from many clinical trials makes drug repurposing from other indications difficult (Ramos et al., 2015, Strippoli et al., 2004). Therefore, studies with patient based pathophysiology research and clinical testing of mechanisms may be highly beneficial.
For decades, the main focus has been on glomerular dysfunction and pathology, but recently it has been suggested that the proximal tubule is important for initiation and progression of CKD (Bonventre, 2014, Takaori et al., 2016). This nephron segment has a very high content of mitochondria and is highly dependent on oxidative phosphorylation (Chevalier 2016). Likewise, muscle weakness and atrophy, fatigue, and non-renal organ dysfunction are major CKD symptoms indicating a basal defect in cell metabolism. Mitochondrial dysfunction has turned out to be an important mechanism in diabetic kidney disease (DKD) (Hallan and Sharma 2016). A recent metabolomics comparison of 24-h urine samples from subjects with DKD and healthy controls identified 13 metabolites that were reduced in DKD, 12 of which were intermediates in mitochondrial metabolic pathways, suggesting a global suppression of mitochondrial activity (Sharma et al. 2013). Interestingly, 12 of the 13 metabolites were not reduced in diabetes without CKD, suggesting that presence of CKD, and not diabetes alone, was necessary for the observed urine metabolite pattern. Independent studies of kidney biopsy tissue demonstrated reduction of mitochondrial proteins and mitochondrial biogenesis (Sharma et al. 2013).
Metabolomics, the quantitative analysis of small molecules in biological samples, has uncovered numerous abnormalities in the blood of uremic patients. These include accumulation of renally excreted gut metabolites such as p-cresol sulfate (Aronov et al., 2011, Yu et al., 2014), indoxyl sulfate (Aronov et al., 2011, Kobayashi et al., 2014, Yu et al., 2014), and trimethylamine-N-oxide (Mutsaers et al. 2013); altered metabolism of amino acids such as tryptophan (Duranton et al., 2014, Goek et al., 2013, Kobayashi et al., 2014, Rhee et al., 2013, Toyohara et al., 2010), arginine (Duranton et al., 2014, Goek et al., 2013, Nkuipou-Kenfack et al., 2014, Shah et al., 2013, Yu et al., 2014), tyrosine/phenylalanine (Duranton et al. 2014), and glycine (Yu et al. 2014); impaired organic anion transport (Shah et al., 2013, Sharma et al., 2013); and increased anaerobic metabolism(Qi et al. 2012). Although urine is an important bio-sample for metabolomic studies in CKD, there have been relatively few studies evaluating metabolomics in 24-h urine. Furthermore, metabolomics may be a useful tool to identify novel therapeutic targets for CKD and to evaluate the effects of promising interventions. Vitamin D receptor agonists (VDRAs) such as paricalcitol are associated with reduced all-cause and cardiovascular mortality (Duranton et al., 2013, Zheng et al., 2013), reduced urine albumin excretion, and may have other renoprotective and cardioprotective effects (de Borst et al. 2013). However, the mechanisms underlying these effects, as well as other metabolic effects of VDRAs in CKD, remain incompletely understood, limiting full translation to clinical care.
To further investigate the metabolic abnormalities associated with non-diabetic CKD and to explore the effects of VDRAs, we quantified plasma and urine metabolites among participants in a clinical trial of paricalcitol (de Boer et al. 2013). We used an established wide-ranging panel of metabolites that are dysregulated in human inborn errors of metabolism and compared results to healthy controls. Key differences at baseline were replicated in an independent cohort, and expression of genes relevant to the significantly altered metabolites was analyzed in kidney tissue. Finally, we examined the effect of interventional treatment with paricalcitol vs. placebo on blood and urine metabolite profiles in CKD.
2. Methods
2.1. Study Populations
First, we compared 22 non-diabetic CKD stage 3–4 patients and 10 healthy controls to identify abnormalities in plasma and urine present in CKD patients. CKD participants were recruited from nephrology clinics at three medical centers associated with the University of Washington and enrolled in a clinical trial designed to test the effects of paricalcitol on glucose metabolism (NCT01003275) (de Boer et al. 2013). Inclusion criteria for the Paricalcitol study included age ≥ 18 years; estimated GFR 15–59 mL/min/1.73m2; and fasting serum glucose 100–125 mg/dL. Exclusion criteria included a clinical diagnosis of diabetes or use of glucose-lowering medications; history of maintenance dialysis or kidney transplantation; use within past 8 weeks of prednisone, immunosuppressive medications, or other medications known to strongly affect blood glucose; change in dose of any medication within 8 weeks; and serum calcium > 10.1 mg/dL. Healthy control participants were University of Washington employees required to be ≥ 18 years of age, and the same exclusion criteria were applied. In addition, healthy control participants were required to have an estimated GFR ≥ 60 mL/min/1.73m2.
Second, major findings were replicated in the Study of Glucose and Insulin in Renal Disease (SUGAR), which included 45 non-diabetic CKD patients and 15 controls matched for age, sex and race (de Boer et al. 2016). SUGAR is a cross-sectional study of insulin and glucose metabolism in moderate to severe non-diabetic CKD. From 2011 to 2014, participants were recruited from nephrology and primary care clinics associated with the University of Washington and the neighboring institutions in Seattle, WA. Inclusion criteria included age ≥ 18 years and estimated GFR < 60 mL/min/1.73m2. Healthy control participants were individuals with GFR ≥ 60 mL/min/1.73m2, spot urine albumin-creatinine ratio < 30 mg/g, with the same distribution of age, sex and race as the enrolled participants with CKD. Exclusion criteria included a clinical diagnosis of diabetes, end-stage renal disease (ongoing or imminent maintenance hemodialysis or kidney transplantation) or use of medications known to affect glucose metabolism (e.g. corticosteroids), fasting serum glucose ≥ 126 mg/dL, and hemoglobin < 10 g/dL. All study procedures were approved by the University of Washington Institutional Review Board, and all participants provided written informed consent.
We also studied gene expression in kidney biopsies from 155 patients from the European Renal cDNA Bank cohort with biopsy-proven, non-diabetic CKD (FSGS or minimal change n = 24, hypertensive nephropathy n = 15, IgA nephropathy n = 27, minimal change disease n = 14, membranous glomerulonephritis n = 21, rapidly progressive glomerulonephritis n = 22, lupus nephritis n = 32). The control kidney biopsies were obtained from healthy kidney transplant donors (n = 31) prior to kidney donation, following the usual clinical protocols.
2.2. Paricalcitol Treatment
CKD participants enrolled in the paricalcitol cross-over intervention trial were allocated to paricalcitol for 8 weeks and placebo for 8 weeks, separated by an 8-week washout period (de Boer et al. 2013). The order of paricalcitol and placebo treatment periods was randomly assigned by the University of Washington Investigational Drug Services and was blinded to both participants and investigators. The active intervention was paricalcitol (19-nor-1,25-(OH)2-vitamin D2) 2 micrograms daily by mouth. Participants were encouraged not to change their use of non-study medications during the course of the study. Healthy control participants were not treated with paricalcitol (de Boer et al. 2013).
2.3. Covariate Data
Demographic data and comorbidities were assessed by questionnaire. Weight and height were measured while wearing light clothing. Blood pressure was measured after resting in a seated position for at least 5 min. Serum creatinine was measured in fasting plasma using a method traceable to isotope dilution mass spectrometer and GFR was estimated with the 2009 CKD-EPI equation (Levey et al. 2009). Timed urine samples (approximately 24-hour) were collected on ice, the urine volume registered, and 10 mL samples frozen at − 80 for later analysis; albumin was measured in fresh urine immediately after collection using a turbidimetric assay and used to calculate daily albumin excretion rate.
2.4. Metabolomics
For CKD participants and healthy controls in the paricalcitol trial, plasma and urine samples were analyzed on a gas chromatography mass spectroscopy (GC–MS) platform at ClinMet, Inc. (San Diego, CA). This panel of metabolites has been established over the years to detect inborn errors of metabolism, and the focus on organic acids is also well suited for CKD since the handling of these compounds are central in kidney function and metabolism. A list of measured metabolites is given in supplemental material (Supplemental Table 3) and includes metabolites from 37 different metabolic pathways. Eleven of these pathways were represented with a sufficiently high number of metabolites so that potential perturbations could be detected with significance. All citric acid cycle metabolites except succinyl CoA and oxalacetate were measured.
Urine aliquots corresponding to 1 μmol of creatinine or 2 mL of plasma were treated with pentafluorobenzyl hydroxylamine to oximate ketoacids prior to lyophilization overnight. Subsequently, the organic acids were extracted by liquid chromatography on silica (42% 2-methyl-2-butanol in chloroform). Solvent was evaporated and the dry residue was silylated with 300 μL of Trisil-N,O-bis (trimethylsilyl) trifluoroacetamide, and finally 1 μL of the reconstituted derivatized sample were injected into a 30 m × 0.32 mm column (Agilent DB-5) in Agilent 5890 gas chromatogram, followed by elution using a 4 °C/min gradient of 70–300°C. Electron impact mass spectrometry using an Agilent 5973 mass selective detector was used to detect the metabolites. Each analyte was identified from the spectrum and the confirmed ratio of qualifying and quantifying ions. The integrated current from the quantifying ion was used to estimate concentration using standard curves with 4–6 calibration points. Peak areas were normalized to internal standards (4-nitrophenol or 2-oxocaproate) added to samples prior to derivatization. Metabolites with results below the lower detection limit in more than two-thirds of all participants were excluded from further analysis in the current study (N = 4 of 66 for urine metabolites, N = 7 of 66 plasma metabolites). A control sample of pooled healthy urines was run every 8 samples to control for day-to-day variation. Citric acid cycle metabolites had coefficient of variation (CV) of 3–10% (Supplemental Table 4). Urine metabolite mass per 1 mmol creatinine was converted to mass excreted per 24 h to reduce problems due to different muscle mass, diet, and circadian rhythm. For details of metabolomics methods in the replication cohort (SUGAR), bioinformatics analyses, and gene expression in human kidney tissue, please see the Supplemental methods.
2.5. Statistical Analysis
Because metabolite distributions were greatly skewed, we applied the natural log transformation log(x + 1) to all metabolites. For each metabolite, we compared the difference in log-metabolites between CKD participants on placebo and normal controls using t-tests that assumed unequal variances; for ease of interpretation we present the results as the percent difference between these two groups. To compare the effect of paricalcitol on CKD participants, we likewise performed paired t-tests on the difference in log-metabolites when the participant was on paricalcitol versus on placebo. We defined metabolites with statistically significant differences as those with ≥ 5% difference between CKD and control combined with a false discovery rate (FDR) ≤ 20%.(Benjamini and Hochberg 1995) Principal components analysis, first scaling each metabolite to unit variance, was applied to investigate systematic patterns of difference between the groups. Main findings were replicated in the SUGAR study with log-transformed urine metabolite concentrations regressed on CKD status, adjusting for age, race (white/non-white), sex, weight and the use of RAAS inhibitors. The p-values were obtained from a two-sided Wald test using sandwich-based robust standard errors. We calculated correlation of eGFR to urine excretion rate and inverse plasma concentrations to illustrate how much of the metabolite variation was caused by filtration. Fractional excretion (FE %) was calculated to illustrate tubular handling of metabolites. All statistical analyses were performed using R for Windows Version 3.1.2.
Metabolic pathway analysis was performed to help identify metabolic pathways potentially associated with non-diabetic CKD. All metabolites found to be significantly different in CKD patients compared to controls were included in these functional analysis performed with MetaboAnalyst 3.0. Enrichment analysis test the probability that significantly differing metabolites should belong to a pathway only by chance, and this web-based tool uses relative between-ness centrality and out-degree centrality measures to calculate compound importance. The pathway impact is calculated as the sum of the importance measures of the matched metabolites normalized by the sum of the importance measures of all metabolites in the pathway. The maximum importance of each pathway is 1 with values ≥ 0.10 indicating significant perturbation of the pathway.
For differential analysis of transcriptomic data, unpaired analyses between healthy controls (living kidney donors) and non-diabetic CKD groups were performed using Significance Analysis of Microarrays (SAM) method implemented in Multi Experiment Viewer (MeV) application (Saeed et al., 2006, Saeed et al., 2003). Genes differentially expressed between two groups with a q-value (False Discovery Rate) below 0.05 were considered significant.
3. Results
3.1. Clinical Characteristics of the Primary Study Population
In the paricalcitol study, the participants with CKD had a mean age of 66 years, and the majority were Caucasian (77%) and male (91%). Almost all participants with CKD were treated for hypertension, 41% had a history of cardiovascular disease, and acid-base balance was in the normal range for most patients. Median estimated glomerular filtration rate (eGFR) and albumin excretion rate were 40 mL/min/1.73m2 and 59 mg/day, respectively. Healthy control subjects were on average 43 years old, were also predominantly male and Caucasian, had fewer comorbidities than the participants with CKD, and had a median estimated GFR of 92 mL/min/1.73m2. Fasting glucose levels were higher in participants with CKD than healthy controls, but none had diabetes mellitus based in the ADA diagnostic criteria (Table 1).
Table 1.
Characteristics of study participants.
| Characteristic | Primary cohort (Paricalcitol trial) |
Replication cohort (SUGAR trial) |
Gene expression cohort (European Renal Biopsy Bank) |
|||
|---|---|---|---|---|---|---|
| CKD (n = 22) | Controls (n = 10) | CKD (n = 45) | Controls (n = 15) | CKD (n = 155) | Controls (n = 31) | |
| Demographics | ||||||
| Age (years) | 66 (12) | 43 (11) | 62 (14) | 56 (12) | 46 (17) | 48 (12) |
| Male sex | 20 (91%) | 7 (70%) | 24 (53%) | 9 (60%) | 85 (55%) | 16 (51%) |
| Race | ||||||
| Caucasian | 17 (77%) | 10 (100%) | 29 (64%) | 12 (80%) | 155 (100%) | 31 (100%) |
| African American | 3 (14%) | 0 (0%) | 12 (27%) | 2 (13%) | 0 (0%) | 0 (0%) |
| Asian | 2 (9%) | 0 (0%) | 4 (9%) | 1 (7%) | 0 (0%) | 0 (0%) |
| Medical history | ||||||
| Hypertension | 21 (96%) | 2 (20%) | 40 (89%) | 5 (33%) | 75 (58%) | None § |
| Cardiovascular disease | 9 (41%) | 0 (0%) | 18 (40%) | 1 (7%) | – | None§ |
| Current smoking | 3 (14%) | 2 (20%) | 9 (20%) | 1 (7%) | – | – |
| Medical treatment | ||||||
| RAAS inhibitors | 18 (82%) | 0 (0%) | 30 (67%) | 5 (33%) | 66(63%) | None§ |
| Vitamin D supplements | 10 (46%) | 5 (50%) | 28 (62%) | 7 (47%) | – | None§ |
| Physical examination | ||||||
| Weight (kg) | – | – | 88.6 (19.9) | 81.7 (20.2) | 75.3 (15.9) | Normal§ |
| Body mass index (kg/m2) | 31 (8) | 24 (3) | 30.3 (6.2) | 27.3 (5.9) | 26 (4) | Normal§ |
| Systolic BP (mmHg) | 132 (16) | 117 (10) | 134.4 (15.9) | 122.4 (14.1) | 136 (21) | Normal § |
| Diastolic BP (mmHg) | 77 (11) | 75 (9) | 80.6 (9.7) | 77.2 (9.3) | 82(14) | Normal § |
| Laboratory data | ||||||
| Median eGFR (mL/min/1.73 m2) | 40 (34–46) | 92 (80–101) | 36 (23–45) | 92 (71–99) | 66 (38–94) | 105 (85–117) |
| Median AER (mg/24 h) | 59 (21–271) | 4 (3–4) | 98 (16–279) | 6 (3 − 10) | – | Normal § |
| p-Bicarbonate (mmol/L) | 24.0 (2.6) | 27.6 (2.3) | ||||
| Fasting glucose (mg/dL) | 107 (11) | 95 (12) | 102.5 (8.9) | 97.1 (8.9) | – | Normal § |
Note: Data are presented as N (%) for categorical variables (some missing data for the replication and the kidney biopsy cohorts), means (SD) or medians (interquartile range) for continuous variables. Estimated GFR (eGFR) was calculated using serum concentrations of creatinine measured at baseline using the 2009 CKD-EPI eq. BP: blood pressure; AER: albumin excretion rate.
§ Kidney biopsy controls were kidney donors. Numerical data were not available except for age, sex and race, but by definition they do not have diabetes, CVD, hypertension, obesity, etc.
3.2. Metabolites in Non-diabetic CKD vs. Healthy Controls in Primary Study Cohort
Plasma concentration and/or urinary excretion of 49 unique metabolites varied significantly between CKD patients and healthy controls after correction for multiple testing (Table 2). The largest relative differences in plasma were for 3-hydroxybutyrate, the most abundant ketone-body, and hippurate, a well characterized gut-microbial end-product, with 113% and 109% higher plasma concentrations in CKD and unchanged urine excretion, respectively. In CKD, long chain fatty acids stearate (C18:0) and palmitate (C16:0) were higher in plasma while urine excretion was lower. Furthermore, there was significantly reduced urinary excretion of central citric acid cycle – also known as the Krebs cycle or the tricarboxylic acid (TCA) cycle – metabolites (citrate − 68% and succinate − 47%) with unchanged plasma levels. In general, CKD patients had many metabolites with higher blood concentrations and lower urine excretion rates. However, correlations of estimated glomerular filtration rate (eGFR) to urine excretion rates were not significant and explained only a small proportion of the variation for most metabolites (R-squared interquartile range 0.01–0.08). The correlations of plasma levels of metabolites with eGFR were stronger, but still weak (R-squared interquartile range 0.02–0.16, see Supplemental Table 1). Differences in tubular handling of metabolites, measured as fractional excretion (FE%), were small and non-significant for most metabolites. Principal component analysis showed that the measured metabolites segregated participants with CKD from healthy controls (Supplemental Fig. 1).
Table 2.
Differences in urine excretion rate and blood metabolite concentration in the primary cohort, comparing participants with and without CKD.
| Metabolite | ID-number | Urine excretion rate |
Blood concentration |
||||
|---|---|---|---|---|---|---|---|
| % difference (95% CI) | p-Value | FDR | % difference (95% CI) | p-Value | FDR | ||
| Significant in both urine and blood | |||||||
| Stearate | HMDB00827 | − 53 (− 77, − 4) | 0.041 | 0.11 | 47 (13, 92) | 0.0068 | 0.021 |
| Palmitate | HMDB00220 | − 52 (− 75, − 8) | 0.03 | 0.092 | 55 (16, 107) | 0.0059 | 0.02 |
| Glycolate | HMDB00115 | − 45 (− 59, − 26) | 0.00026 | 0.0049 | − 19 (− 25, − 13) | 9.80E-07 | 5.80E-05 |
| 3-Hydroxyisovalerate | HMDB00754 | − 41 (− 55, − 23) | 0.00032 | 0.0049 | 25 (10, 42) | 0.0017 | 0.0093 |
| Isocitrate | HMDB00193 | − 41 (− 57, − 19) | 0.0021 | 0.016 | 24 (2, 52) | 0.033 | 0.061 |
| Homovanillate | HMDB00118 | − 37 (− 60, − 1) | 0.047 | 0.12 | − 13 (− 21, − 3) | 0.012 | 0.029 |
| L-2-Hydroxyglutarate | HMDB00694 | − 36 (− 48, − 20) | 0.0003 | 0.0049 | 6 (2, 11) | 0.0082 | 0.023 |
| Hydroxypropionate | HMDB00700 | − 29 (− 52, 4) | 0.077 | 0.18 | 18 (3, 36) | 0.021 | 0.043 |
| 2-Methylcitrate | HMDB00379 | − 28 (− 41, − 12) | 0.0027 | 0.018 | 13 (7, 19) | 0.00006 | 0.00094 |
| 3-Hydroxyglutarate | HMDB00428 | − 24 (− 42, 1) | 0.06 | 0.15 | 14 (6, 22) | 0.00077 | 0.0057 |
| Leucinate | HMDB00665 | − 5 (− 7, − 2) | 0.0029 | 0.018 | − 8 (− 14, − 1) | 0.032 | 0.061 |
| Significant in blood only | |||||||
| 3-Hydroxybutyrate | HMDB00011 | − 5 (− 27, 24) | 0.72 | 0.78 | 113 (22, 272) | 0.011 | 0.028 |
| Hippurate | HMDB00714 | 6 (− 34, 71) | 0.79 | 0.83 | 109 (55, 181) | 0.00002 | 0.00057 |
| Oleate | HMDB00207 | − 11 (− 24, 4) | 0.14 | 0.29 | 80 (25, 159) | 0.0044 | 0.017 |
| Acetoacetate | HMDB00060 | − 1 (− 20, 24) | 0.95 | 0.95 | 76 (3, 203) | 0.041 | 0.071 |
| 3-Hydroxyadipate | HMDB00345 | − 16 (− 46, 31) | 0.42 | 0.6 | 38 (17, 62) | 0.00035 | 0.0034 |
| Glycerate | HMDB00139 | − 25 (− 54, 21) | 0.22 | 0.38 | 34 (1, 78) | 0.041 | 0.071 |
| 2-Hydroxybutyrate | HMDB00008 | 3 (− 19, 31) | 0.81 | 0.83 | 34 (− 1, 82) | 0.058 | 0.095 |
| Myristate | HMDB00806 | − 1 (− 4, 1) | 0.2 | 0.36 | 29 (− 6, 77) | 0.11 | 0.16 |
| 2-Ethylhydracrylate | HMDB00396 | − 16 (− 41, 20) | 0.32 | 0.5 | 28 (11, 47) | 0.0012 | 0.0072 |
| L-Malate | HMDB00156 | − 22 (− 52, 27) | 0.3 | 0.48 | 22 (7, 40) | 0.0054 | 0.02 |
| p-Hydroxyphenylacetate | HMDB00020 | − 7 (− 28, 22) | 0.6 | 0.72 | 18 (8, 29) | 0.00071 | 0.0057 |
| 3-Methylglutaconate | HMDB00522 | − 4 (− 27, 27) | 0.78 | 0.83 | 16 (6, 27) | 0.0025 | 0.011 |
| Hydroxyphenyllactate | HMDB00755 | − 8 (− 28, 18) | 0.48 | 0.61 | 13 (2, 26) | 0.021 | 0.043 |
| 4-Hydroxyhippurate | HMDB13678 | − 28 (− 72, 84) | 0.47 | 0.61 | 10 (3, 18) | 0.008 | 0.023 |
| Phenylpyruvate | HMDB00205 | − 2 (− 5, 2) | 0.26 | 0.42 | 10 (2, 20) | 0.017 | 0.038 |
| Fumarate | HMDB00134 | − 4 (− 17, 12) | 0.62 | 0.72 | 7 (2, 12) | 0.011 | 0.028 |
| Adipate | HMDB00448 | − 1 (− 31, 42) | 0.94 | 0.95 | 6 (2, 9) | 0.001 | 0.0067 |
| Hexanoylglycine | HMDB00701 | − 23 (− 48, 14) | 0.18 | 0.35 | 6 (1, 11) | 0.017 | 0.038 |
| 3-Methyladipate | HMDB00555 | − 10 (− 31, 18) | 0.43 | 0.6 | 5 (2, 7) | 0.00022 | 0.0026 |
| Benzoate | HMDB01870 | − 5 (− 25, 20) | 0.65 | 0.73 | − 7 (− 11, − 2) | 0.0069 | 0.021 |
| Methylsuccinate | HMDB01844 | 4 (− 9, 19) | 0.52 | 0.64 | − 21 (− 35, − 5) | 0.018 | 0.04 |
| 2-Methylacetoacetate | HMDB03771 | − 5 (− 12, 2) | 0.14 | 0.29 | − 29 (− 43, − 13) | 0.0027 | 0.011 |
| Significant in urine only | |||||||
| Citrate | HMDB00094 | − 68 (− 82, − 44) | 0.00049 | 0.006 | − 11 (− 35, 22) | 0.45 | 0.56 |
| Succinate | HMDB00254 | − 47 (− 65, − 20) | 0.0048 | 0.023 | − 4 (− 13, 7) | 0.49 | 0.60 |
| 4-Hydroxybutyrate | HMDB00710 | − 46 (− 63, − 20) | 0.0049 | 0.023 | 5 (− 2, 14) | 0.16 | 0.22 |
| 2-oxoglutarate | HMDB00208 | − 42 (− 68, 5) | 0.071 | 0.17 | 9 (− 7, 28) | 0.27 | 0.36 |
| cis-Aconitate | HMDB00072 | − 40 (− 58, − 16) | 0.0051 | 0.023 | − 20 (− 41, 9) | 0.14 | 0.20 |
| Methylmalonate | HMDB00202 | − 40 (− 60, − 10) | 0.018 | 0.06 | 1 (− 4, 6) | 0.74 | 0.84 |
| 2-Methyl-3-hydroxybutyrate | HMDB00354 | − 39 (− 53, − 21) | 0.0011 | 0.011 | 3 (− 1, 8) | 0.097 | 0.15 |
| Tiglylglycine | HMDB00959 | − 37 (− 49, − 24) | 0.00005 | 0.0029 | 2 (0, 4) | 0.03 | 0.058 |
| Ethylmalonate | HMDB00622 | − 37 (− 55, − 10) | 0.015 | 0.054 | 4 (0, 9) | 0.057 | 0.095 |
| N-acetyl-l-aspartate | HMDB00812 | − 31 (− 47, − 10) | 0.0085 | 0.033 | 6 (− 16, 32) | 0.62 | 0.74 |
| Pyroglutamate | HMDB00267 | − 30 (− 50, − 2) | 0.039 | 0.11 | 16 (− 5, 43) | 0.15 | 0.20 |
| Glutarate | HMDB00661 | − 27 (− 41, − 10) | 0.0071 | 0.029 | 4 (− 3, 10) | 0.24 | 0.33 |
| Glutaconate | HMDB00620 | − 24 (− 36, − 9) | 0.0049 | 0.023 | 1 (− 2, 3) | 0.7 | 0.82 |
| Ortho-hydroxyphenylacetate | HMDB00669 | − 23 (− 37, − 6) | 0.016 | 0.057 | 1 (0, 1) | 0.1 | 0.16 |
| Azelate | HMDB00784 | − 22 (− 31, − 10) | 0.002 | 0.016 | 0 (− 4, 3) | 0.81 | 0.88 |
| 3-Hydroxymethylglutarate | HMDB00355 | − 20 (− 35, − 1) | 0.037 | 0.11 | 2 (1, 2) | 0.0021 | 0.01 |
Note: Data are relative difference (% and 95% CI) between non-diabetic CKD and healthy controls. A positive percent difference indicates that the metabolite was higher in CKD than in controls. The significance threshold is ≥ 5% difference between CKD and controls and Q-values indicating a FDR < 20%. The % difference and p-values for metabolites passing this threshold are in bold print. Analytes are ordered by magnitude of difference between cases and controls. p-Values are based on t-test on log-transformed values assuming unequal variance.
3.3. Pathway Analysis
To assist in the biological interpretation of our data, we used metabolite enrichment analysis, which is based on several metabolite libraries consisting of ∼ 1000 entries, to explore whether significantly differing metabolites belong to a common pathway. There was an enrichment of significantly differing compounds in several metabolic pathways (Table 3), including the citric acid cycle (7 significant metabolites while only 0.27 was expected by chance, FDR < 0.001), phenylalanine with subsequent tyrosine and dopamine metabolism (7 metabolites vs. 0.6 expected, FDR < 0.001), short-chain fatty acids butanoate and propanoate metabolism (5 metabolites vs 0.5, FDR = 0.002)), and synthesis and degradation of ketone bodies (2 vs 0.08, FDR = 0.028). Among these pathways, the citric acid cycle and ketone body metabolism had the highest pathway impact scores (0.25 and 0.70, respectively) indicating substantially disturbed pathways due to high numbers and central location of differing metabolites (see note below Table 3). Five of the early citric acid cycle metabolites had significantly reduced urine excretion (citrate, cis-aconitate, isocitrate, 2-oxoglutarate and succinate) but were not significantly different in plasma, while two late citric acid cycle metabolites were significantly increased in plasma only (fumarate and malate).
Table 3.
Altered metabolic pathways in non-diabetic CKD versus controls.
| Pathway | Enrichment analysis |
Topology analysis |
|||||
|---|---|---|---|---|---|---|---|
| Total | Expected | Hits | Raw p | Holm adjust | FDR | Impact | |
| Citric acid cycle (TCA cycle) | 20 | 0.27 | 7 | 1.15E − 07 | 9.19E − 06 | 0.0000 | 0.25 |
| Phenylalanine metabolism | 45 | 0.60 | 7 | 1.17E − 06 | 9.24E − 05 | 0.0000 | 0.14 |
| Glyoxylate & dicarboxylate metab. | 50 | 0.66 | 7 | 2.46E − 06 | 1.92E − 04 | 0.0001 | 0.10 |
| Propanoate metabolism | 35 | 0.47 | 5 | 7.35E − 05 | 5.66E − 03 | 0.0015 | 0.03 |
| Butanoate metabolism | 40 | 0.53 | 5 | 1.42E − 04 | 1.08E − 02 | 0.0023 | 0.11 |
| Tyrosine metabolism | 76 | 1.01 | 6 | 3.83E − 04 | 2.87E − 02 | 0.0051 | 0.11 |
| Ketone bodies metabolism | 6 | 0.08 | 2 | 2.49E − 03 | 1.84E − 01 | 0.0284 | 0.70 |
| Alanine, aspartate, & glutamate metab. | 24 | 0.32 | 3 | 3.58E − 03 | 2.61E − 01 | 0.0358 | 0.00 |
| Fatty acid biosynthesis | 49 | 0.65 | 3 | 2.6E − 02 | 1E + 00 | 0.2310 | 0.00 |
| Valine, leucine, & isoleucine metab. | 40 | 0.53 | 2 | 9.77 E − 02 | 1E + 00 | 0.7820 | 0.00 |
Note: Enrichment analysis test if the compounds significantly differing between CKD patients and controls are found more often in a specific metabolic pathway than expected by chance. Pathways are ranked according to their statistical significance, i.e. citric acid cycle is on top since our finding of 7 differing metabolites in this pathway is extremely unlikely to happen by chance. In addition, topology analysis, which takes into account the structure of a given pathway to evaluate the relative importance of the differing compounds based on their relative locations, shows that the 7 differing TCA metabolites represent changes in key positions of the network and will trigger more severe impact on the pathway than changes on marginal or relatively isolated positions. Impact scores range 0 to 1, and scores ≥ 0.1 suggest significant pathway alteration.
3.4. Replication of Citric Acid Cycle (TCA) Metabolite Differences
We also evaluated the association of CKD status with urine metabolite excretion in an independent study of non-diabetic CKD stage 3–5, the Study of Glucose and Insulin in Renal Disease (SUGAR) (de Boer et al. 2013). In SUGAR, participants with CKD (median eGFR 37 mL/min/1.73m2) were matched to control subjects without CKD on age, sex, and race, and the cohort had greater racial and gender diversity than the paricalcitol trial (Table 1). Pathway analyses based on all 140 metabolites measured in SUGAR showed that the citric acid cycle was again the most strongly affected pathway (p = 0.0006 after adjustment for multiple testing and impact score of 0.43). All five TCA cycle intermediates with reduced urine excretion in the paricalcitol cohort (isocitric, citric, succinic, 2-oxoglutarate and cis-aconitate) were also significantly reduced in 24-hour urine samples from the SUGAR cohort after adjusting for age, race, sex, and weight (Table 4).
Table 4.
Differences in citric acid (TCA) cycle metabolites in 24-hour urine samples in the replication cohort (SUGAR study), comparing participants with non-diabetic CKD to matched controls.
| Metabolite | HMDB | % difference (95% CI) | p-value |
|---|---|---|---|
| Citrate | HMDB00094 | − 81 (− 89, − 65) | 7.3E-08 |
| Aconitate | HMDB00072 | − 61 (− 73, − 42) | 2.3E-06 |
| Isocitrate | HMDB00193 | − 61 (− 72, − 45) | 1.0E-07 |
| 2-oxoglutarate | HMDB00208 | − 71 (− 81, − 54) | 1.1E-07 |
| Succinate | HMDB00254 | − 41 (− 59, − 15) | 4.5E-03 |
| Fumarate | HMDB00134 | − 37 (− 56, − 11) | 9.3E-03 |
Note: Log-transformed urine metabolite concentrations were regressed on CKD status, adjusting for age, race (white/non-white), sex, weight, RAAS medication and baseline CVD. The p-values were obtained from a two-sided Wald test using sandwich-based robust standard errors.
3.5. Gene Expression for Citric Acid (TCA) Cycle Enzymes
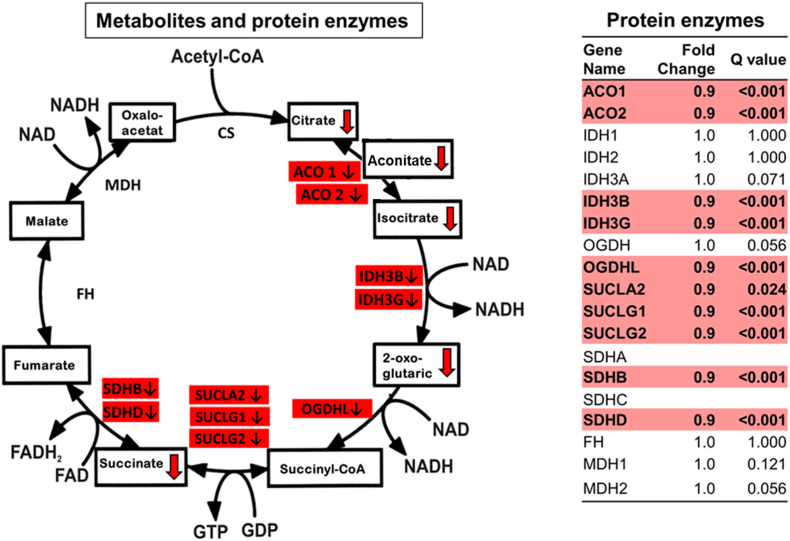
As many of the metabolites from the TCA cycle were consistently reduced in the 24-hour urine collections of subjects with CKD we considered whether gene expression for enzymes relevant to the TCA cycle could also be affected. We therefore evaluated the renal gene expression of citric acid cycle enzymes in 155 non-diabetic CKD patients and 31 healthy kidney donors from the European Renal cDNA Bank (ERCB) with age, blood pressure and eGFR comparable to our primary cohort (Table 1) (Yasuda et al. 2006). There were significantly reduced mRNA levels for ten citric acid cycle enzymes, compared with controls (Fig. 1). Aconitase 1 and 2 (ACO1, ACO2), oxoglutarate dehydrogenase-like (OGDHL), succinate-CoA ligase (SUCLA2, SUCLG1 and SUCLG2), succinate dehydrogenase subunits B and D (SDHB, SDHD), and isocitrate dehydrogenase subunits 3B and 3G (IDH3B, IDH3G) mRNA levels were significantly reduced in the tubulointerstitial compartment. The glomerular compartment displayed very similar findings (Supplemental Fig. 2). This pattern of reduction in gene expression corresponded to the observed reduction of urinary metabolites from the proximal half of the citric acid cycle (e.g. citrate, aconitate, isocitrate, 2-oxoglutarate and succinate, Table 2 and Fig. 1).
Fig. 1.
The urinary excretion of citric acid (TCA) cycle metabolites and the renal expression of genes that regulate these metabolites were significantly reduced among participants with versus without non-diabetic CKD. Urine excretion of citric acid cycle metabolites in the proximal part of the pathway were reduced in samples from patients with non-diabetic CKD, as was the mRNA expression of the enzymes catalyzing the proximal steps of the citric acid cycle in the tubulointerstitial compartment in biopsies of patients with non-diabetic CKD.
3.6. Regulators of Mitochondrial Biogenesis in Non-diabetic CKD
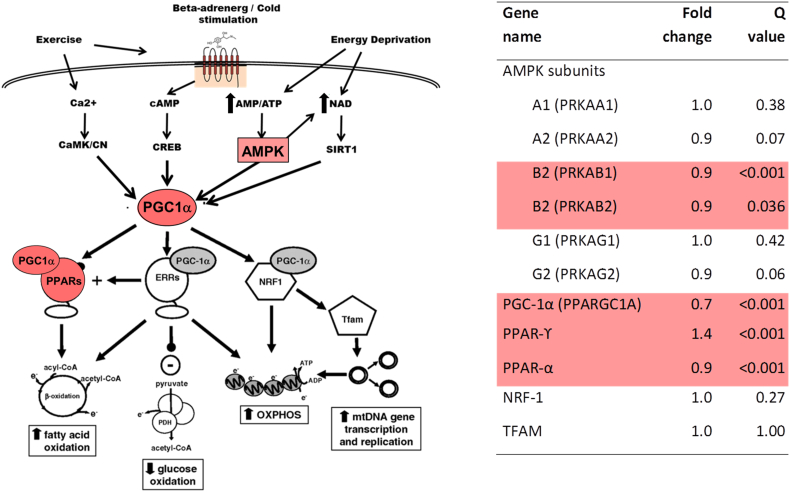
Since citric acid (TCA) cycle is an intra-mitochondrial process, we examined mRNA levels for several key regulators of mitochondrial biogenesis in the same dataset. We found significant reductions of various subunits of the AMP-activated kinase (AMPK), peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) and peroxisome proliferator-activated receptor alpha (PPAR-α) in non-diabetic CKD kidneys compared with controls (Fig. 2). In contrast, peroxisome proliferator-activated receptor gamma (PPAR- γ) was increased, while nuclear respiratory factor-1 (NRF-1) and transcription factor A, mitochondrial (TFAM) were unchanged. A simplified overview of mitochondrial stimulators, regulators, and their effects is also given (Fig. 2).
Fig. 2.
Expression of mRNA regulating mitochondrial biogenesis was significantly different comparing participants with versus without non-diabetic CKD. A. Increased AMPK activity induces activation of PGC-1α, a major regulator of mitochondrial biogenesis. Activated PGC-1α sets in motion several transcriptional programs to stimulate replication of mitochondrial DNA, as well as expression of mitochondrial enzymes such as those involved in fatty acid oxidation, citric acid cycle or the electron transport chain. B. Compared with healthy controls, subjects with non-diabetic CKD showed reduced mRNA levels for two subunits of the AMPK protein (PRKAB1, PRKAB2), as well as PGC-1α, while PPARγ mRNA was increased.
PRKAA1 to PRKAG2: AMPK subunits; PARGC1A: PPARγ coactivator 1α (PGC-1α); AMPK: AMP-dependent kinase; PPARγ: Peroxisome proliferator-activated receptor γ; ERR α: Estrogen related receptor α; NRF: Nuclear respiratory factor; TFAM: Transcription factor A, mitochondrial.
3.7. Urine and Plasma Metabolites in CKD During Paricalcitol Treatment
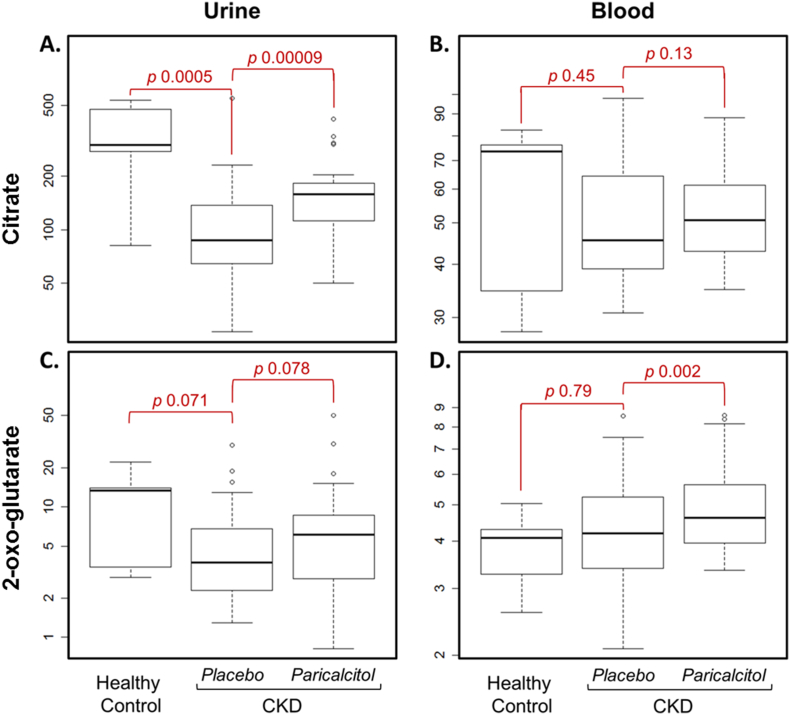
Finally, we compared citric acid (TCA) cycle metabolites in plasma and urine at the end of 8 weeks of paricalcitol treatment to those measured at the end of 8 weeks of placebo treatment (Table 5). Among 22 trial participants with CKD, we found that total renal citrate excretion and plasma citrate concentration was higher during paricalcitol treatment (+ 74%, p < 0.0001 and + 5%, p = 0.13, respectively). Furthermore, higher 2-oxoglutarate excretion and plasma concentrations were also observed during treatment (+ 34%, p = 0.08 and + 12%, p = 0.002, respectively). There were no changes in the fractional excretion of TCA metabolites (Table 5) or other metabolites (Supplementary Table 2). Distribution of citrate and 2-oxoglutarate in urine and blood is shown for healthy controls, CKD patients without paricalcitol and CKD patients with paricalcitol (Fig. 3). CKD patients also had a small but significantly increase in serum bicarbonate during treatment (24.0 vs 25.5 mmol/L, p = 0.002). There was a significant decrease of eGFR after 8 weeks of paricalcitol treatment compared with placebo (33.2 vs 36.5 mL/min/1.73m2, p = 0.01).
Table 5.
Effect of paricalcitol treatment on citric acid cycle (TCA) metabolites in non-diabetic CKD patients.
| Metabolites | Urine excretion rate |
Blood concentration |
Fractional Excretion |
|||
|---|---|---|---|---|---|---|
| % difference (95% CI) | p-value | % difference (95% CI) | p-value | % difference (95% CI) | p-value | |
| Citrate | 74 (37, 120) | < 0.0001 | 5 (− 2, 13) | 0.13 | 1 (− 2, 3) | 0.49 |
| Aconitate | 19 (− 12, 62) | 0.25 | 2 (− 5, 10) | 0.60 | 1 (− 7.7) | 0.89 |
| Isocitrate | 28 (2, 61) | 0.03 | 3 (− 3, 10) | 0.30 | 1 (− 4, 7) | 0.66 |
| 2-oxoglutarate | 34 (− 4, 85) | 0.08 | 12 (5, 19) | 0.002 | 0 (− 1, 2) | 0.66 |
| Succinyl CoA | NA | NA | NA | NA | NA | NA |
| Succinate | 29 (− 8, 83) | 0.14 | 4 (− 1, 10) | 0.10 | 0 (− 2, 2) | 0.66 |
| Fumarate | 17 (2, 35) | 0.03 | 1 (− 1, 4) | 0.34 | 0 (− 1, 1) | 0.97 |
| Malate | 25 (− 4, 64) | 0.09 | 2 (− 5, 10) | 0.59 | 0 (− 1, 2) | 0.81 |
| Oxaloacetate | NA | NA | NA | NA | NA | NA |
| Pyruvate | 8 (− 19, 46) | 0.58 | 7 (− 6, 23) | 0.28 | 0 (0, 0) | 0.89 |
Note: Cell contents are percent differences (95% confidence intervals) comparing values obtained during treatment with paricalcitol to values obtained during treatment with placebo for each trial participant (N = 22).
Fig. 3.
Urine and blood concentrations of citrate and 2-oxoglutarate in CKD were increased by paricalcitol in non-diabetic CKD. Urine excretion of citrate is reduced in CKD, compared with healthy controls, and is increased significantly after paricalcitol treatment (panel A). Plasma citrate shows a similar trend, however did not reach significance (panel B). Whisker plots depict maximum, upper quartile, median, lower quartile and the minimum values of each metabolite.
4. Discussion
We identified 49 significantly altered metabolites in plasma and urine representing several perturbed metabolic pathways, most strikingly the citric acid (TCA) cycle, in non-diabetic CKD patients as compared to healthy controls. Lower urinary excretion of citric acid cycle metabolites were replicated in an independent cohort, and the expression of genes for ten citric acid cycle enzymes were significantly reduced in kidney biopsies of patients with non-diabetic CKD. In participants with CKD, the urinary excretion of citrate and the plasma levels of 2-oxogluarate, which are central citric acid cycle metabolites, were significantly higher during treatment with paricalcitol. These results suggested that non-diabetic CKD is characterized by reduction of citric acid cycle metabolites and enzymes.
When GFR decreases, serum metabolite concentrations will increase until a new steady state is achieved with elimination again equaling production. Excretion based on 24-hour urine samples is therefore not affected by reduced GFR per se, but can theoretically be influenced by dietary changes, renal reabsorption, extra-renal excretion and metabolism. Citrate metabolism is well studied over decades, and blood and urine levels are generally held to reflect citric acid (TCA) cycle activity since there is no known extra-renal elimination and minimal influence by oral intake (Bashir et al., 2012, Hamm, 1990). However, renal excretion can be substantially modulated by acid-base state. Acidosis will increase tubular reabsorption of citrate with subsequent full metabolization to carbon dioxide in proximal tubular cells, but the small difference in plasma bicarbonate between well-regulated CKD stage 3 patients and healthy controls in our study can only explain a small part of the observed decrease in citrate excretion (Brennan et al., 1988, Caudarella et al., 2003, Hamm, 1990, Simpson, 1983). Although other factors decreasing urinary citrate excretion are known (starvation, potassium depletion, high-protein intake, hyperparathyroidism) (Caudarella et al. 2003), these are, except for the latter, not likely to be of importance for most CKD stage 3 patients. Therefore, our findings of substantially reduced excretion of the majority of citric acid cycle metabolites and reduced gene expression in CKD patients is best explained by reduced local or systemic citric acid cycle activity. A recent study demonstrated reduced urinary excretion of two citric acid cycle metabolites and 11 other metabolites associated with mitochondrial function in participants with diabetic kidney disease compared to healthy control subjects (Sharma et al. 2013).
Diminished citric acid (TCA) activity may be due to a reduction in overall mitochondrial biogenesis, reduced expression of the genes encoding citric acid enzymes, or reduced citric acid cycle substrate availability. Regarding the first point, we observed reduced mRNA levels of AMPK and PGC-1α, key regulators of mitochondrial biogenesis, in kidney tissues from non-diabetic CKD patients. This is the first study to report the reduction of gene expression of several members of the AMPK system in kidney biopsies from patients with CKD. Other recently published studies have also reported reduced kidney PGC-1α in CKD, both diabetic and non-diabetic (Kang et al., 2015, Sharma et al., 2013). Diabetic kidney disease patients had reduced mitochondrial protein in kidney biopsies and reduced mitochondrial mRNA in the urine suggesting global mitochondrial suppression (Sharma et al. 2013). Significantly reduced mitochondrial volume density and other abnormalities were recently found in muscle biopsies and blood leukocytes from CKD patients (Gamboa et al. 2016), and reduced mtDNA copy number was found in diabetic as well as non-diabetic CKD stage 3–4 indicating that these changes start well before end-stage renal disease (Gamboa et al. 2016). Combined, these data suggest that CKD may be associated with a general mitochondrial dysfunction regardless of diabetes status. Secondly, we observed significantly reduced expression of genes for citric acid cycle enzymes in both the glomerular and tubulointerstitial compartments of human kidneys with non-diabetic CKD. The reduction of isocitrate dehydrogenase 3 (IDH3B and IDH3G) in the tubulointerstitial compartment is especially interesting since this is considered to be the major control point in the citric acid cycle (Berg et al. 2002). The second citric acid cycle control point in animal cells, oxoglutaric dehydrogenase (OGDH), was also significantly reduced. Thirdly, it is well known that CKD patients have reduced gluconeogenesis and increased risk of hypoglycemia (Moen et al. 2009). Posphoenol-pyruvate carboxykinase (PEPCK) is the key regulator of gluconeogenesis, but an equally important function could be removal of citric acid cycle intermediates for conversion to pyruvate and fueling of the citric acid cycle (cataplerosis) (Hanson 2009). Reduced PEPCK mRNA abundance and protein expression have been demonstrated in CKD rats (Burki et al. 2015) as well as non-diabetic CKD patients from the ERCB cohort used in our study (fourfold reduction, top 1% of under-expressed genes) (Athey et al., 2012, University-of-Michigan, 2016). This could contribute to reduced substrate availability and reduced citric cycle activity.
Fatty acid oxidation and ketone body metabolism are other important mitochondrial processes which are highly integrated with the citric acid (TCA) cycle. Prior studies have suggested impaired long-chain fatty acid beta-oxidation in both chronic and end-stage kidney disease.(Stadler et al. 2015) Carnitine deficiency or inhibition of carnitine palmitoyl transferase-1 (Smogorzewski et al. 1988), which lead to impaired transport of long chain fatty acids into mitochondria for beta-oxidation, may contribute (Matera et al. 2003). Impaired oxidation of long-chain fatty acids is proposed to cause preferential consumption of shorter chain fatty acids because these can translocate into mitochondria independent of carnitine to undergo beta-oxidation, generating adipate (Mortensen 1992). The result is reduced concentration of low-molecular-weight triacylglycerols and accumulation of high-molecular-weight triacylglycerols (Rhee et al. 2010). Consistent with this theory, we and others find higher concentrations of long-chain fatty acids (palmitate, oleate, stearate and myristate) (Fouque et al., 2006, Shah et al., 2013, Wanner et al., 1988) and adipate (Rhee et al., 2010, Toyohara et al., 2010) in plasma (Table 2). In addition, we find significant alteration in metabolism of two of the most abundant short-chain fatty acids (SCFA; propanoate and butanoate), with marked reduction in urinary excretion of several distal propanoate metabolites (hydroxypropionate, methylcitrate, tiglyglycine, methylmalonate). This could be consistent with reduced propanoate availability due to reduced gut-microbial production or increased consumption. The gut-microbiome is increasingly found to influence inflammatory processes and to produce uremic toxins (Felizardo et al. 2016), and SCFA is hypothesized to be kidney protective by reducing inflammation and improving mitochondrial function (Andrade-Oliveira et al. 2015). A signature of defective fatty acid beta-oxidation was also recently reported in genome-wide transcriptomic analyses of fibrotic human kidneys (Kang et al. 2015). Furthermore, we observed strong perturbations in the ketone body metabolism with doubling of plasma beta hydroxyl butyrate, the main ketone body in humans, and it's precursor acetoacetate. Others have found four-fold increased levels of the rate-limiting enzyme in ketogenesis (HMGCS2) in DKD (Zhang et al. 2011). Our data indicate that this could be a general metabolic disturbance in CKD and could reflect a shift in metabolism to burn acetyl-coA. Overall, one could speculate whether these widespread mitochondrial dysfunctions are part of the well-known clinical protein-energy wasting syndrome in CKD and other uremic symptoms (Carrero et al. 2013).
The renoprotective mechanisms of paricalcitol are not well understood. Increased urine excretion of citrate and other metabolites could be explained by improved glomerular function, but we observed a small decrease in GFR estimated from serum creatinine during treatment. Similar results have been found previously and have been attributed to an increased creatinine production with no effect on the measured GFR (Agarwal et al. 2011). The findings are therefore more consistent with paricalcitol having a general effect on TCA cycle activity. 1-25-OH-vitamin D was recently shown to improve mitochondrial activity in human skeletal muscle cells (Ryan et al., 2016, Sinha et al., 2013). Vitamin D has a wide range of effects on several organs and even cancer cells, many of which are not well understood, and some of these could be caused be a redirection of metabolism towards oxidative phosphorylation (Christakos et al. 2016). We could speculate that paricalcitol improve basic cellular functions in podocytes and proximal tubular cell, which are the mainstays against urine albumin losses (Muller-Deile and Schiffer 2014). Alternatively, paricalcitol may increase urine citrate by reducing proximal tubular reabsorption of citrate, for example by altering the acid-base status (Hamm 1990). However, a substantial pH decrease (pH 7.40–pH 7.20, i.e. a 60% increased H + concentration) is needed to give a moderately increased citrate reabsorption (+ 20–40%) (Brennan et al., 1988, Caudarella et al., 2003, Hamm, 1990, Simpson, 1983), so the small differences in bicarbonate in our study during paricalcitol treatment of CKD patients (7% change in H + concentration) can only explain a small fraction of the change in citrate excretion. 2-oxoglutarate have an essential role in metabolic control with important effects on mitochondrial potential, tissue respiration, ROS (reactive oxygen species) production, nitrogen metabolism, and glutamate signaling (Bunik and Fernie 2009). Supplementation of 2-oxoglutarate has been found to extend C. elegans lifespan by 50%, probably by inhibiting ATP synthase similar to dietary restriction (Chin et al. 2014). Notably, administering 2-oxoglutarate has been shown to increase bone density, possibly by promoting proline hydroxylation, a key component of type I collagen backbone of the bone matrix (Tatara et al. 2005). As such, the paricalcitol-induced increase in plasma 2-oxoglutarate may relate to beneficial effects of VDRAs on bone health. More largescale clinical trials and experimental studies to look into mechanisms are needed.
Strengths of this study are use of an established, quantitative metabolite panel, simultaneous metabolite quantification in urine and plasma, the use of 24-hour urine to quantify urinary excretion, replication of key findings in an independent cohort in a different laboratory, evaluation of gene transcription to complement metabolite measurements, and application of metabolomics to a CKD-relevant treatment (paricalcitol). The main study limitations are the small sample size, the cross-sectional design, lack of complete matching of CKD participants to healthy controls in the primary study, and lack of replication for paricalcitol treatment effects. Our focus on a pre-specified set of metabolites naturally precludes a total evaluation of metabolic abnormalities in CKD, but the metabolomics platforms used survey a variety of biochemical pathways relevant to human errors of metabolism. The large observed differences, the consistency with previous findings, and the replication of key results in an independent population provide reassurance that our results are not due to confounding factors, although further studies should be performed in a larger population with greater ability to account for relevant covariates.
In conclusion, we observed a pattern of significantly altered metabolites consistent with reduced citric acid cycle activity, reduced fatty acid oxidation and increased ketone body metabolism. This could suggest generally decreased mitochondrial function in non-diabetic CKD similar to recent findings in patients with diabetic CKD.
Funding Sources
This study was supported by an investigator-initiated research grant from Abbvie, with additional support from the University of Washington Institute of Translational Health Sciences (UL1TR000423), as well as NIDDK grants R01DK087726, R01DK087726-S1. Dr. Afkarian's effort was supported by grant K23DK089017 from National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and the Norman S. Coplon Extramural Grant from Satellite Healthcare. Ms. Sharma was an employee of ClinMet, Inc. and is presently a consultant with University of Texas Health San Antonio.
Dr. de Boer's effort was also supported by NIDDK grants R01DK088762, and R01DK099199. Dr. Sharma's effort was supported by NIDDK DP3 DK094352. Dr. Hallan was supported by grants from NTNU, St Olav Hospital and Norwegian Research Council. Dr. Saito's effort was supported by NIDDK DP3 DK094352.
None of the funders had any role in study design, data collection, data analysis, interpretation, or writing of the manuscript.
Conflict of Interest
K.S. is the founder of Clinical Metabolomics (ClinMet), Inc. All other authors have declared that no conflict of interest exists.
Author Contributions
Study conception and design: Hallan, Afkarian, K. Sharma, de Boer
Acquisition of data: deBoer, K. Sharma, Kretzler, Ju, Darshi, S. Sharma, Raftery, Barding,
Analysis and interpretation of data: Hallan, Afkarian, Zelnick, Kestenbaum, S·Sharma, Saito, Darshi, Barding, Raftery, Ju, Kretzler, K. Sharma, de Boer
Drafting of manuscript: Hallan, Afkarian
Critical revision: Hallan, Afkarian, Zelnick, Kestenbaum, S·Sharma, Saito, Darshi, Barding, Raftery, Ju, Kretzler, K. Sharma, de Boer.
Acknowledgments
Acknowledgements
Authors thank the Applied Systems Biology Core of the O'Brien Renal Center at the University of Michigan and Compendia Bioscience, part of Life Techologies, for making the Nephroseq (formerly Nephromine) datasets and analysis engine publically available to the scientific community.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2017.10.027.
Appendix A. Supplementary Data
Supplementary material
References
- Agarwal R., Hynson J.E., Hecht T.J., Light R.P., Sinha A.D. Short-term vitamin D receptor activation increases serum creatinine due to increased production with no effect on the glomerular filtration rate. Kidney Int. 2011;80:1073–1079. doi: 10.1038/ki.2011.207. [DOI] [PubMed] [Google Scholar]
- Andrade-Oliveira V., Amano M.T., Correa-Costa M., Castoldi A., Felizardo R.J., de Almeida D.C., Bassi E.J., Moraes-Vieira P.M., Hiyane M.I., Rodas A.C. Gut bacteria products prevent AKI induced by ischemia-reperfusion. J. Am. Soc. Nephrol. 2015;26:1877–1888. doi: 10.1681/ASN.2014030288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronov P.A., Luo F.J., Plummer N.S., Quan Z., Holmes S., Hostetter T.H., Meyer T.W. Colonic contribution to uremic solutes. J. Am. Soc. Nephrol. 2011;22:1769–1776. doi: 10.1681/ASN.2010121220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athey B.D., Cavalcoli J.D., Jagadish H.V., Omenn G.S., Mirel B., Kretzler M., Burant C., Isokpehi R.D., DeLisi C., Ncibi faculty, t The NIH National Center for integrative biomedical informatics (NCIBI) J. Am. Med. Inform. Assoc. 2012;19:166–170. doi: 10.1136/amiajnl-2011-000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir S., Khan N.A., Gilani A.-H. Physiology of Renal Handling of Citrate. In: Talati J.J., Tiselius H.-G., Albala D.M., Ye Z., editors. Urolithiasis: Basic Science and Clinical Practice. Springer London; London: 2012. pp. 183–186. [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- Berg J., T J., Stryer L. 5th edn. Freeman; New York: 2002. Biochemistry. [Google Scholar]
- de Boer I.H., Sachs M., Hoofnagle A.N., Utzschneider K.M., Kahn S.E., Kestenbaum B., Himmelfarb J. Paricalcitol does not improve glucose metabolism in patients with stage 3-4 chronic kidney disease. Kidney Int. 2013;83:323–330. doi: 10.1038/ki.2012.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer I.H., Zelnick L., Afkarian M., Ayers E., Curtin L., Himmelfarb J., Ikizler T.A., Kahn S.E., Kestenbaum B., Utzschneider K. Impaired glucose and insulin homeostasis in moderate-severe CKD. J. Am. Soc. Nephrol. 2016;27:2861–2871. doi: 10.1681/ASN.2015070756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonventre J.V. Maladaptive proximal tubule repair: cell cycle arrest. Nephron Clin. Pract. 2014;127:61–64. doi: 10.1159/000363673. [DOI] [PubMed] [Google Scholar]
- de Borst M.H., Hajhosseiny R., Tamez H., Wenger J., Thadhani R., Goldsmith D.J. Active vitamin D treatment for reduction of residual proteinuria: a systematic review. J. Am. Soc. Nephrol. 2013;24:1863–1871. doi: 10.1681/ASN.2013030203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan S., Hering-Smith K., Hamm L.L. Effect of pH on citrate reabsorption in the proximal convoluted tubule. Am. J. Phys. 1988;255:F301–306. doi: 10.1152/ajprenal.1988.255.2.F301. [DOI] [PubMed] [Google Scholar]
- Bunik V.I., Fernie A.R. Metabolic control exerted by the 2-oxoglutarate dehydrogenase reaction: a cross-kingdom comparison of the crossroad between energy production and nitrogen assimilation. Biochem. J. 2009;422:405–421. doi: 10.1042/BJ20090722. [DOI] [PubMed] [Google Scholar]
- Burki R., Mohebbi N., Bettoni C., Wang X., Serra A.L., Wagner C.A. Impaired expression of key molecules of ammoniagenesis underlies renal acidosis in a rat model of chronic kidney disease. Nephrol. Dial. Transplant. 2015;30:770–781. doi: 10.1093/ndt/gfu384. [DOI] [PubMed] [Google Scholar]
- Carrero J.J., Stenvinkel P., Cuppari L., Ikizler T.A., Kalantar-Zadeh K., Kaysen G., Mitch W.E., Price S.R., Wanner C., Wang A.Y. Etiology of the protein-energy wasting syndrome in chronic kidney disease: a consensus statement from the International Society of Renal Nutrition and Metabolism (ISRNM) J. Ren. Nutr. 2013;23:77–90. doi: 10.1053/j.jrn.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Caudarella R., Vescini F., Buffa A., Stefoni S. Citrate and mineral metabolism: kidney stones and bone disease. Front. Biosci. 2003;8:s1084–1106. doi: 10.2741/1119. [DOI] [PubMed] [Google Scholar]
- Chevalier R.L. The proximal tubule is the primary target of injury and progression of kidney disease: role of the glomerulotubular junction. Am. J. Physiol. Ren. Physiol. 2016;311:F145–161. doi: 10.1152/ajprenal.00164.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin R.M., Fu X., Pai M.Y., Vergnes L., Hwang H., Deng G., Diep S., Lomenick B., Meli V.S., Monsalve G.C. The metabolite alpha-ketoglutarate extends lifespan by inhibiting ATP synthase and TOR. Nature. 2014;510:397–401. doi: 10.1038/nature13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christakos S., Dhawan P., Verstuyf A., Verlinden L., Carmeliet G. Vitamin D: metabolism, molecular mechanism of action, and pleiotropic effects. Physiol. Rev. 2016;96:365–408. doi: 10.1152/physrev.00014.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duranton F., Rodriguez-Ortiz M.E., Duny Y., Rodriguez M., Daures J.P., Argiles A. Vitamin D treatment and mortality in chronic kidney disease: a systematic review and meta-analysis. Am. J. Nephrol. 2013;37:239–248. doi: 10.1159/000346846. [DOI] [PubMed] [Google Scholar]
- Duranton F., Lundin U., Gayrard N., Mischak H., Aparicio M., Mourad G., Daures J.P., Weinberger K.M., Argiles A. Plasma and urinary amino acid metabolomic profiling in patients with different levels of kidney function. Clin. J. Am. Soc. Nephrol. 2014;9:37–45. doi: 10.2215/CJN.06000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felizardo R.J.F., Castoldi A., Andrade-Oliveira V., Camara N.O.S. The microbiota and chronic kidney diseases: a double-edged sword. Clin. Trans. Immunol. 2016;5 doi: 10.1038/cti.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouque D., Holt S., Guebre-Egziabher F., Nakamura K., Vianey-Saban C., Hadj-Aissa A., Hoppel C.L., Kopple J.D. Relationship between serum carnitine, acylcarnitines, and renal function in patients with chronic renal disease. J. Ren. Nutr. 2006;16:125–131. doi: 10.1053/j.jrn.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Gamboa J.L., Billings F.T.t., Bojanowski M.T., Gilliam L.A., Yu C., Roshanravan B., Roberts L.J., II, Himmelfarb J., Ikizler T.A., Brown N.J. Mitochondrial dysfunction and oxidative stress in patients with chronic kidney disease. Phys. Rep. 2016;4 doi: 10.14814/phy2.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBD_Collaborators Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;385:117–171. doi: 10.1016/S0140-6736(14)61682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goek O.N., Prehn C., Sekula P., Romisch-Margl W., Doring A., Gieger C., Heier M., Koenig W., Wang-Sattler R., Illig T. Metabolites associate with kidney function decline and incident chronic kidney disease in the general population. Nephrol. Dial. Transplant. 2013;28:2131–2138. doi: 10.1093/ndt/gft217. [DOI] [PubMed] [Google Scholar]
- Hallan S.I., Sharma K. The role of mitochondria in diabetic kidney disease. Curr. Diab. Rep. 2016;16:61. doi: 10.1007/s11892-016-0748-0. [DOI] [PubMed] [Google Scholar]
- Hamm L.L. Renal handling of citrate. Kidney Int. 1990;38:728–735. doi: 10.1038/ki.1990.265. [DOI] [PubMed] [Google Scholar]
- Hanson R.W. Thematic minireview series: a perspective on the biology of phosphoenolpyruvate carboxykinase 55 years after its discovery. J. Biol. Chem. 2009;284:27021–27023. doi: 10.1074/jbc.R109.040519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.M., Ahn S.H., Choi P., Ko Y.A., Han S.H., Chinga F., Park A.S., Tao J., Sharma K., Pullman J. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 2015 doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Yoshida T., Fujisawa T., Matsumura Y., Ozawa T., Yanai H., Iwasawa A., Kamachi T., Fujiwara K., Kohno M. A metabolomics-based approach for predicting stages of chronic kidney disease. Biochem. Biophys. Res. Commun. 2014;445:412–416. doi: 10.1016/j.bbrc.2014.02.021. [DOI] [PubMed] [Google Scholar]
- Levey A.S., Stevens L.A., Schmid C.H., Zhang Y.L., Castro A.F., III, Feldman H.I., Kusek J.W., Eggers P., Van Lente F., Greene T. A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera M., Bellinghieri G., Costantino G., Santoro D., Calvani M., Savica V. History of L-carnitine: implications for renal disease. J. Ren. Nutr. 2003;13:2–14. doi: 10.1053/jren.2003.50010. [DOI] [PubMed] [Google Scholar]
- Moen M.F., Zhan M., Hsu V.D., Walker L.D., Einhorn L.M., Seliger S.L., Fink J.C. Frequency of hypoglycemia and its significance in chronic kidney disease. Clin. J. Am. Soc. Nephrol. 2009;4:1121–1127. doi: 10.2215/CJN.00800209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen P.B. Formation and degradation of dicarboxylic acids in relation to alterations in fatty acid oxidation in rats. Biochim. Biophys. Acta. 1992;1124:71–79. doi: 10.1016/0005-2760(92)90128-i. [DOI] [PubMed] [Google Scholar]
- Muller-Deile J., Schiffer M. The podocyte power-plant disaster and its contribution to glomerulopathy. Front Endocrinol. (Lausanne) 2014;5:209. doi: 10.3389/fendo.2014.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutsaers H.A., Engelke U.F., Wilmer M.J., Wetzels J.F., Wevers R.A., van den Heuvel L.P., Hoenderop J.G., Masereeuw R. Optimized metabolomic approach to identify uremic solutes in plasma of stage 3-4 chronic kidney disease patients. PLoS One. 2013;8:e71199. doi: 10.1371/journal.pone.0071199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkuipou-Kenfack E., Duranton F., Gayrard N., Argiles A., Lundin U., Weinberger K.M., Dakna M., Delles C., Mullen W., Husi H. Assessment of metabolomic and proteomic biomarkers in detection and prognosis of progression of renal function in chronic kidney disease. PLoS One. 2014;9 doi: 10.1371/journal.pone.0096955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S., Ouyang X., Wang L., Peng W., Wen J., Dai Y. A pilot metabolic profiling study in serum of patients with chronic kidney disease based on (1) H-NMR-spectroscopy. Clin. Trans. Sci. 2012;5:379–385. doi: 10.1111/j.1752-8062.2012.00437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A.M., Gonzalez-Guerrero C., Sanz A., Sanchez-Nino M.D., Rodriguez-Osorio L., Martin-Cleary C., Fernandez-Fernandez B., Ruiz-Ortega M., Ortiz A. Designing drugs that combat kidney damage. Expert Opin. Drug Discovery. 2015;10:541–556. doi: 10.1517/17460441.2015.1033394. [DOI] [PubMed] [Google Scholar]
- Rhee E.P., Souza A., Farrell L., Pollak M.R., Lewis G.D., Steele D.J., Thadhani R., Clish C.B., Greka A., Gerszten R.E. Metabolite profiling identifies markers of uremia. J. Am. Soc. Nephrol. 2010;21:1041–1051. doi: 10.1681/ASN.2009111132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee E.P., Clish C.B., Ghorbani A., Larson M.G., Elmariah S., McCabe E., Yang Q., Cheng S., Pierce K., Deik A. A combined epidemiologic and metabolomic approach improves CKD prediction. J. Am. Soc. Nephrol. 2013;24:1330–1338. doi: 10.1681/ASN.2012101006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan Z.C., Craig T.A., Folmes C.D., Wang X., Lanza I.R., Schaible N.S., Salisbury J.L., Nair K.S., Terzic A., Sieck G.C. 1alpha,25-Dihydroxyvitamin D3 regulates mitochondrial oxygen consumption and dynamics in human skeletal muscle cells. J. Biol. Chem. 2016;291:1514–1528. doi: 10.1074/jbc.M115.684399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Saeed A.I., Bhagabati N.K., Braisted J.C., Liang W., Sharov V., Howe E.A., Li J., Thiagarajan M., White J.A., Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Shah V.O., Townsend R.R., Feldman H.I., Pappan K.L., Kensicki E., Vander Jagt D.L. Plasma metabolomic profiles in different stages of CKD. Clin. J. Am. Soc. Nephrol. 2013;8:363–370. doi: 10.2215/CJN.05540512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K., Karl B., Mathew A.V., Gangoiti J.A., Wassel C.L., Saito R., Pu M., Sharma S., You Y.H., Wang L. Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. J. Am. Soc. Nephrol. 2013;24:1901–1912. doi: 10.1681/ASN.2013020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson D.P. Citrate excretion: a window on renal metabolism. Am. J. Phys. 1983;244:F223–234. doi: 10.1152/ajprenal.1983.244.3.F223. [DOI] [PubMed] [Google Scholar]
- Sinha A., Hollingsworth K.G., Ball S., Cheetham T. Improving the vitamin D status of vitamin D deficient adults is associated with improved mitochondrial oxidative function in skeletal muscle. J. Clin. Endocrinol. Metab. 2013;98:E509–513. doi: 10.1210/jc.2012-3592. [DOI] [PubMed] [Google Scholar]
- Smogorzewski M., Perna A.F., Borum P.R., Massry S.G. Fatty acid oxidation in the myocardium: effects of parathyroid hormone and CRF. Kidney Int. 1988;34:797–803. doi: 10.1038/ki.1988.252. [DOI] [PubMed] [Google Scholar]
- Stadler K., Goldberg I.J., Susztak K. The evolving understanding of the contribution of lipid metabolism to diabetic kidney disease. Curr. Diab. Rep. 2015;15:40. doi: 10.1007/s11892-015-0611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strippoli G.F., Craig J.C., Schena F.P. The number, quality, and coverage of randomized controlled trials in nephrology. J. Am. Soc. Nephrol. 2004;15:411–419. doi: 10.1097/01.asn.0000100125.21491.46. [DOI] [PubMed] [Google Scholar]
- Takaori K., Nakamura J., Yamamoto S., Nakata H., Sato Y., Takase M., Nameta M., Yamamoto T., Economides A.N., Kohno K. Severity and frequency of proximal tubule injury determines renal prognosis. J. Am. Soc. Nephrol. 2016;27:2393–2406. doi: 10.1681/ASN.2015060647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatara M.R., Brodzki A., Krupski W., Sliwa E., Silmanowicz P., Majcher P., Pierzynowski S.G., Studzinski T. Effects of alpha-ketoglutarate on bone homeostasis and plasma amino acids in turkeys. Poult. Sci. 2005;84:1604–1609. doi: 10.1093/ps/84.10.1604. [DOI] [PubMed] [Google Scholar]
- Toyohara T., Akiyama Y., Suzuki T., Takeuchi Y., Mishima E., Tanemoto M., Momose A., Toki N., Sato H., Nakayama M. Metabolomic profiling of uremic solutes in CKD patients. Hypertens. Res. 2010;33:944–952. doi: 10.1038/hr.2010.113. [DOI] [PubMed] [Google Scholar]
- University-of-Michigan Nephroseq (formerly Nephromine) 2016. https://wwwnephroseqorg/resource/mainhtml Assessed May 29 2016 at.
- Wanner C., Schollmeyer P., Horl W.H. Serum carnitine levels and carnitine esters of patients after kidney transplantation: role of immunosuppression. Metab. Clin. Exp. 1988;37:263–267. doi: 10.1016/0026-0495(88)90106-0. [DOI] [PubMed] [Google Scholar]
- Yasuda Y., Cohen C.D., Henger A., Kretzler M., European Renal C, D.N.A.B.C. Gene expression profiling analysis in nephrology: towards molecular definition of renal disease. Clin. Exp. Nephrol. 2006;10:91–98. doi: 10.1007/s10157-006-0421-z. [DOI] [PubMed] [Google Scholar]
- Yu B., Zheng Y., Nettleton J.A., Alexander D., Coresh J., Boerwinkle E. Serum metabolomic profiling and incident CKD among African Americans. Clin. J. Am. Soc. Nephrol. 2014;9:1410–1417. doi: 10.2215/CJN.11971113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Yang H., Kong X., Wang K., Mao X., Yan X., Wang Y., Liu S., Zhang X., Li J. Proteomics analysis reveals diabetic kidney as a ketogenic organ in type 2 diabetes. Am. J. Physiol. Endocrinol. Metab. 2011;300:E287–295. doi: 10.1152/ajpendo.00308.2010. [DOI] [PubMed] [Google Scholar]
- Zheng Z., Shi H., Jia J., Li D., Lin S. Vitamin D supplementation and mortality risk in chronic kidney disease: a meta-analysis of 20 observational studies. BMC Nephrol. 2013;14:199. doi: 10.1186/1471-2369-14-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material