Abstract
Aurora B kinase has emerged as a key regulator of mitosis and deregulation of Aurora B activity is closely related to the development and progression of human cancers. In the present study, we found that Aurora B is overexpressed in human esophageal squamous cell carcinoma (ESCC), high levels of Aurora B protein were associated with a worse overall survival rate in ESCC patients. Depleting of Aurora B blunted the malignant phenotypes in ESCC cells. Importantly, we demonstrated that a natural compound, deguelin, has a profound anti-tumor effect on ESCC via inhibiting Aurora B activity. Deguelin potently inhibited in vitro Aurora B kinase activity. The in silico docking study further indicated that deguelin was docked into the ATP-binding pocket of Aurora B. Inhibition of Aurora B activity attenuated growth of ESCC cells, resulted in G2/M cell cycle arrest, polyploidy cells formation, and apoptosis induction. Knocking down of Aurora B decreased the sensitivity of ESCC cells to deguelin. The in vivo results showed that deguelin blocked the phosphorylation of histone H3 and inhibited the growth of ESCC tumor xenografts. Overall, we identified deguelin as an effective Aurora B inhibitor, which deserves further studies in other animal models and ESCC treatment.
Abbreviations: ESCC, esophageal squamous cell carcinoma;; RNase A, ribonuclease A; OS, overall survival; shRNA, short nairpin RNA; DMSO, dimethyl sulphoxide
Keywords: Esophageal squamous cell carcinoma, Deguelin, Aurora B, Mitosis
Highlights
-
•
Aurora B is overexpressed in human esophageal squamous cell carcinoma (ESCC), high levels of Aurora B protein are associated with a worse overall survival rate in ESCC patients.
-
•
Deguelin inhibits the growth of ESCC cells both in vitro and in vivo.
-
•
Deguelin potently inhibits in vitro Aurora B kinase activity and is docked into the ATP-binding pocket of Aurora B. Deguelin acts as an effective Aurora B inhibitor.
1. Introduction
Human esophageal carcinoma is one of the most frequently diagnosed cancers, ranked as the eighth leading causes of cancer-related mortality worldwide. Esophagus squamous cell carcinoma (ESCC) is the most common histological type of esophageal carcinomas, especially with a higher incidence in developing nations (Abnet et al., 2017, Rustgi and El-Serag, 2014). Epidemiological studies have demonstrated that alcohol consumption and tobacco use are closely linked to increased ESCC risk. ESCC is notoriously aggressive in nature, spreading by a variety of pathways including direct extension, lymphatic spread and hematogenous metastasis (Abnet et al., 2017, Lagergren and Lagergren, 2013, Liang et al., 2017). Despite advances in early detection and standard treatment, ESCC is often diagnosed at an advanced stage and has a poor prognosis. The overall 5-year survival rate for late stage ESCC is < 15%, which has hardly improved during the past few decades (Liang et al., 2017). The underlying reasons for this disappointingly low survival rate remain to be greatly elucidated. Therefore, a better understanding of the molecular mechanisms of ESCC pathogenesis or identifies a novel chemical entity with activity against ESCC is expected to facilitate the development of novel therapies that can complement current traditionally therapeutic methods.
The Aurora kinases, a closely related subgroup of 3 serine/threonine kinases, including Aurora A, B, and C, are believed to play a key role in protein phosphorylation in mitosis and have been shown to contribute to the development and progression of cancer (Nguyen and Schindler, 2017, Tang et al., 2017). Aurora B is localized to the centromeres from the prophase to the metaphase-anaphase transition. Thereafter, it is localized to midzone spindle microtubules during the telophase and subsequently to midbody during cytokinesis. Small-molecule inhibitors and siRNA of Aurora B abrogate the mitotic-spindle checkpoint and cause premature mitotic exit without the completion of cytokinesis, leading to 4 N DNA-containing cells that continue to progress through the cell cycle (Carmena et al., 2009, Krenn and Musacchio, 2015, Liu et al., 2009). In malignancy, alterations of Aurora kinase have been linked with genetic instability, including mitotic errors, chromosomal aneuploidy, deregulation of cell proliferation and apoptosis, which are highly associated with tumorigenesis (Mahadevan et al., 2017, Portella et al., 2011, Schecher et al., 2017). Aurora B is overexpressed in many tumor types and has been linked to poor patient prognosis in cancers (Lens et al., 2010, Otto and Sicinski, 2017). Recently, several Aurora B inhibitors have been developed, including AZD1152, BI 811283, ZM447439, and VX-680, etc., the clinical trial data demonstrated that inhibition of Aurora B kinase displayed a generally manageable safety profile and disease stabilization in some patients (Bavetsias and Linardopoulos, 2015, Falchook et al., 2015, Tang et al., 2017, Yan et al., 2016, Ziemska and Solecka, 2016).
In the present study, we identified a natural compound, deguelin, which was isolated from the Legume family, Lonchocarpus, Derris, or Tephrosia, as an effective Aurora B inhibitor for using in ESCC therapy. We investigated the therapeutic effect of deguelin in ESCC both in vitro and in vivo. Our results could provide an option for clinical targeting anti-mitotic therapies of ESCC.
2. Materials and Methods
2.1. Cell Lines and Culture
Human esophageal squamous carcinoma cell line Eca109, KYSE180, and KYSE450 were purchased from Cell Bank of Chinese Academy of Sciences, Shanghai, China. Human esophageal carcinoma cell lines KYSE150 was kindly provided by Dr. Qian Tao from The Chinese University of Hong Kong, Hong Kong, China. Human normal esophageal epithelial cell Het-1A was purchased from American Type Culture Collection (ATCC, Manassas, VA). The cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum, 100 units/mL penicillin and 100 mg/mL streptomycin. All cell lines were incubated at 37 °C in a humidified atmosphere containing 5% CO2.
2.2. Chemicals and Cell Treatments
Tris, NaCl, and SDS for molecular biology and buffer preparation were obtained from Sigma-Aldrich (St. Louis, MO). Deguelin (> 98%) and Hesperadin (> 95%) were purchased from Sigma-Aldrich. Cell culture media and supplements were from Invitrogen (Grand Island, NY). Subconfluent cells were treated with the compound at indicated concentrations for an indicated time. Detailed treatment procedures were described in figure legends. The final concentration of DMSO in the culture media was kept < 0.1% which had no significant effect on the cell growth. Vehicle controls were prepared for all treatments.
2.3. MTS Assay
ESCC cells were seeded (2.5 × 103/well/100 mL) into 96-well plates, and proliferation was assessed by MTS assay (Promega, Madison, WI) according to the instructions provided.
2.4. Anchorage-Independent Cell Growth Assay
The anchorage-independent growth assay was conducted as described previously (Liu et al., 2016). Briefly, a total of 8000 cells were suspended between a layer of solidified basal medium Eagle/10% FBS/0.5% bottom agar with different concentrations of compound or vehicle and 1 mL basal medium Eagle/10% FBS/0.3% top agar with the same concentration of compound or vehicle in each well of six-well plates. After maintenance at 37 °C, 5% CO2 for 2 weeks, colonies were scored under a microscope using the Image-Pro Plus software (version 6.2) program (Media Cybernetics. Rockville, MD).
2.5. Immunofluorescence Staining
The immunofluorescence staining was conducted as described previously (Li et al., 2013). Briefly, after treated with deguelin for 24 h, an asynchronous population of cancer cells were washed with PBS, and fixed in 4% paraformaldehyde and permeabilized in 0.5% Triton X-100 for 30 min. Fixed cells were incubated with a α-tubulin rabbit antibody (#2144, Cell Signaling Technology, Danvers, MA) overnight at 4 °C followed by incubation with green fluorescent Alexa Fluor 488 dye-labeled anti-mouse IgG (Invitrogen, Grand Island, NY). Nuclei were stained with DAPI. Samples were viewed with a Nikon inverted fluorescence microscope (Melville, NY).
2.6. Flow Cytometry
The flow cytometry was performed as described previously (Yu et al., 2015). Briefly, ESCC cells were seeded into 6-well plates in RPMI 1640 containing 10% FBS. After culturing for 24 h, different concentrations of deguelin were added to each well and left on the cells for 24 h. After treatment, attached and floating cells were harvested. For apoptosis analysis, the cells were suspended in 1 × 106 cells/mL, and 5 μL Annexin V and Propidium Iodide staining solution were added to 300 μL of the cell suspension. After incubated 10–15 min at room temperature in the dark, stained cells were assayed and quantified using a FACSort Flow Cytometer (BD, San Jose, CA, USA). For cell cycle analysis, cells were harvested and washed with PBS for two times and then fixed in 70% ethanol overnight at 4 °C. Cells were counterstained in the dark with 50 μg/mL propidium iodide and 0.1% ribonuclease A (RNase A) in 400 μL of PBS at room temperature for 30 min. Stained cells were assayed and quantified using a FACSort Flow Cytometer (BD, San Jose, CA, USA).
2.7. Protein Preparation and Western Blotting
Cultured cells were harvested and whole cell lysates were prepared according to the method previously described (Yu et al., 2017a). Protein concentration was determined using the BCA Assay Reagent (Cat. no. 23228, Pierce, Rockford, IL). Western blotting was performed as previously described (Li et al., 2013). The following antibodies were used for immunodetection with appropriate dilutions: Aurora B (#3094, 1:1000), Phospho-Aurora B (Thr232) (#2914, 1:1000), Histone H3 (#4499,1:2000) and Phospho-Histone H3 (Ser10) (#3377,1:2000) antibodies were purchased from Cell Signaling Technology (Beverly, MA); β-actin (A5316, 1:10,000) was purchased from Sigma (St. Louis, MO); Ki67 (ab16667,1:200) was purchased from Abcam (Cambridge, UK).
2.8. In Vitro Aurora Kinase Assay
The in vitro aurora kinase assay was performed as described previously (Sheng et al., 2014). Histone H3 and active Aurora kinase B were purchased from Merck Millipore (Billerica, MA). Histone H3 (1 μg) and active Aurora kinase (100 ng) were mixed with different doses of deguelin or hesperadin in a 20 μL reaction, which was conducted in 100 μM ATP and 1 × kinase buffer (Cell Signaling Technology) at 30 °C for 30 min. Reactions were stopped by boiling samples in 5 × SDS loading buffer, and proteins were analyzed by Western blot.
2.9. Lentiviral Infection
Four lentivirus plasmids targeting Aurora B (TRCN0000000776, TRCN0000000777, TRCN0000000778, TRCN0000000779) were purchased from Thermo Scientific. pLKO.1-sh-GFP (Addgene plasmid #30323), the lentiviral packaging plasmid psPAX2 (Addgene plasmid #12260) and the envelope plasmid pMD2.G (Addgene plasmid #12259) were available on Addgene (Cambridge, MA). The generation of gene stable knocking down cell lines was performed as described previously (Yu et al., 2017b). Briefly, to generate Aurora B knocking down cells, pLKO.1-sh-GFP or pLKO.1-sh-Aurora B lentivirus plasmid was co-transfected into 293 T cells with psPAX2 and pMD2.G. Viral supernatant fractions were collected at 48 h after transfection and filtered through a 0.45 μm filter followed by infection into KYSE150 cells together with 8 μg/mL polybrene. At 16 h after infection, the medium was replaced with fresh medium containing 2 μg/mL puromycin and cells were incubated for another 6 days.
2.10. Xenograft Mouse Model
All the experimentation for animals was performed following guidelines approved by the Animal Ethics Committee of Central South University. KYSE150 cells (2 × 106) in 100 μL 1640 medium were inoculated s.c. into the right flank of 6-week-old female athymic nude mice. Eight days after inoculation, mice were given an i.p. injection of deguelin at a dose of 4 mg/kg daily, whereas control mice were administered vehicle. The body weight of each mouse was recorded and tumor volume was determined by Vernier caliper twice a week. Volume was calculated following the formula of A × B2 × 0.5, wherein A is the longest diameter of tumor, B is the shortest diameter and B2 is B squared.
2.11. Molecular Modeling
To predict the binding mode of deguelin targeting Aurora B, the crystal structure of the kinase domain (PDB ID: 4C2V) was obtained from the Protein Data Bank. This structure was then prepared using the default parameters of Protein Preparation Wizard in Schrödinger Suite 2013. Hydrogen atoms were added consistent with a pH of 7, and all water molecules were removed. Finally, an ATP-binding site-based receptor grid was generated at the centroid of the ligand, barasertib, from the crystal structure, with default settings in Receptor Grid Generation in Schrödinger Suite 2013. For deguelin, 3D structures of each stereoisomer were generated and prepared in the module of LigPrep in Schrödinger Suite 2013, with other parameters kept the default. Docking was performed using the program of Glide in Schrödinger Suite 2013 with default parameters under the standard precision mode. Three poses of each stereoisomer or state of deguelin were output to observe the scores and binding modes.
2.12. Immunohistochemistry Staining
Tumor tissues obtained from euthanized xenografted mice were embedded and subjected to immunohistochemistry staining with specific antibodies against p-Histone H3-Ser10 (1:100) or Ki67 (1:200) according to the DAKO system protocol. Hematoxylin was used for counterstaining. Slides were viewed and photographed under a light microscope and analyzed using Image-Pro Plus Software (version 6.2) program (Media Cybernetics). Human ESCC tissue arrays (HEso-Squ180Sur-03) were purchased from Shanghai Outdo Biotech Co., Itd. (Shanghai, China). The arrays included 80 cases of squamous cell carcinoma with clinical stages and follow-up records for 5 years. The latest follow-up information was updated in September 2014, overall survival (OS) was defined as the time from completion of therapy to the date of death or when censored at the latest date if patients were still alive. Aurora B expression was scored according to staining intensity and the percentage of positive cells as previously described (Luo et al., 2012). The percentage of positive cells was scored as follows: 0, no positive cells; 1, ≤ 10% positive cells; 2, 10–50% positive cells; 3, > 50% positive cells. Staining intensity was scored as follows: 0, no staining; 1, faint staining; 2, moderate staining; 3, dark staining. Comprehensive score = staining percentage × intensity. Aurora B expression: < 2 low expression, ≥ 2 high expression.
2.13. Statistical Analysis
Statistical analysis was performed with SPSS 16.0 (SPSS, Inc., Chicago, IL). Survival curve was estimated using the Kaplan-Meier method. The log-rank test was used to identify statistically significant differences between survival curves. The association between clinicopathologic factors and Aurora B level was evaluated using Chi-square test. The data were expressed as means ± SD as indicated. Significant differences were determined by a Student's t-test or one-way ANOVA. A probability value of < 0.05 was used as the criterion for statistical significance.
3. Results
3.1. Elevated Aurora B Expression Predicts Poor Overall Survival in Esophageal Squamous Carcinoma Patients
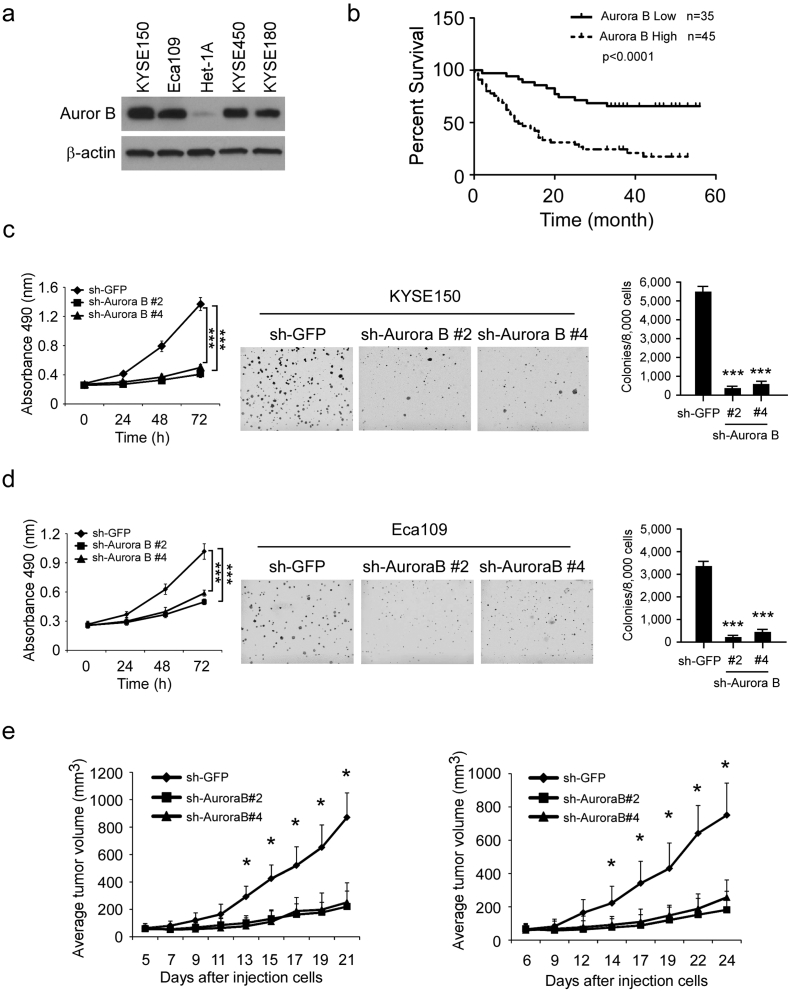
We first checked the expression of Aurora B in esophageal squamous carcinoma cells. The result showed that, compared with the normal esophageal epithelial cell Het-1A, Aurora B is overexpressed in ESCC cells (Fig. 1a). Then, the tissue arrays were used to evaluate the Aurora B expression in ESCC patient. Of 80 tumor tissue samples, 45 (56.3%) ESCC cancers had high Aurora B expression, whereas only 8 (10%) had high expression in adjacent non-tumor tissues. These results suggest that Aurora B expression is upregulated in ESCC compared to non-tumor tissues (p < 0.001, Table 1). To further explore the relationship between Aurora B expression and patient prognosis, Kaplan-Meier analysis was conducted. The result showed that patients with higher level of Aurora B expression had significantly shorter overall survival (OS) compared with patients whose tumors expressed a lower level of Aurora B (p < 0.0001, Fig. 1b). The association between Aurora B level and clinical features of patients, including age, gender, initial clinical stage, tumor stage, and lymph node status were summarized in Table 2. Aurora B was positively correlated with Lymph nodes status (p = 0.013, Table 2), which suggested that high expression of Aurora B may play a role in ESCC metastasis. In order to further determine the critical role of Aurora B in the tumorigenesis of ESCC, we generated stable knockdown Aurora B KYSE150 (Fig. S1a) and Eca109 (Fig. S1b) cells and validated several shRNAs that effectively depleted Aurora B after transfection. Two shRNA sequences were used for the further study (#2 and #4). Results showed that knocking down of Aurora B inhibited proliferation of KYSE150 (Fig. 1c left) and Eca109 cells (Fig. 1d left). Knocking down Aurora B expression also decreased the anchorage-independent growth of KYSE150 (Fig. 1c, middle and right) and Eca109 (Fig. 1d, middle and right) cells. Furthermore, we found that xenograft growth of KYSE150 (Fig. 1e left, Fig. S2a) and Eca109 cells (Fig. 1e right, Fig. S2b) in athymic nude mice were also attenuated after Aurora B knockdown. These results suggest that Aurora B possesses an important biological function in ESCC and blocking Aurora B expression significantly reduces the tumorigenic properties of ESCC cells.
Fig. 1.
High expression of Aurora B predicts poor overall survival in ESCC patients. (a) Aurora B is highly expressed in human esophageal squamous carcinoma cells. Western blot analysis was performed to examine Aurora B expression in several ESCC cell lines and normal Het-1A cells. (b) Overall survival rates of ESCC patients with high (n = 45) or low (n = 35) expression levels of Aurora B were estimated with the Kaplan-Meier method by log-rank test (p < 0.0001). (c, d) Knocking down Aurora B expression in KYSE150 cells (c) and Eca109 cells (d) reduces their tumorigenic properties. MTS assay (left) and soft agar assay (middle and right) were performed as described in “Materials and Methods” (***p < 0.001 vs. sh-GFP group). (e) Knockdown of Aurora B reduces in vivo tumor growth of KYSE150 (left) and Eca109 (right) cells. Tumor growth curve of mice injected with KYSE150 and Eca109 was shown. Data are represented as means ± S.D. of each group (*p < 0.05 vs. vehicle-treated group).
Table 1.
Protein expression of Aurora B in ESCC tissues and adjacent normal tissues.
| Tissue sample | No. of patients | Aurora B |
p-value | |
|---|---|---|---|---|
| Low | High | |||
| Tumor | 80 | 35 | 45 | < 0.0001⁎ |
| Adjacent | 80 | 72 | 8 | |
Chi-square test.
p < 0.0001 indicates a significant upregulation of Aurora B in tumor tissue vs. adjacent tissue.
Table 2.
Relationships between the expression of Aurora B and clinical pathological characteristics in 80 patients with ESCC.
| Expression of Aurora B |
p – value | |||
|---|---|---|---|---|
| Characteristics | All cases | Low(n = 35) | High(n = 45) | |
| Gender | 0.999 | |||
| Male | 64 | 28 | 36 | |
| Female | 16 | 7 | 9 | |
| Age(years) | 0.806 | |||
| ≤ 60 | 24 | 11 | 13 | |
| > 60 | 56 | 24 | 32 | |
| Initial clinical stage | ||||
| ≤ IIa | 40 | 19 | 21 | 0.499 |
| > IIa | 40 | 16 | 24 | |
| Tumor stages | 0.088 | |||
| T1 + T2 | 22 | 13 | 9 | |
| T3 + T4 | 58 | 22 | 36 | |
| Lymph nodes status | 0.013⁎ | |||
| N0 (negative) | 40 | 23 | 17 | |
| N1 or above (positive) | 40 | 12 | 28 | |
Chi-square test.
p < 0.05 indicates a significant upregulation of Aurora B in patients with lymphatic metastasis vs. no lymphatic metastasis.
3.2. Deguelin Effectively Suppresses the Proliferation of Human Esophageal Squamous Carcinoma Cells
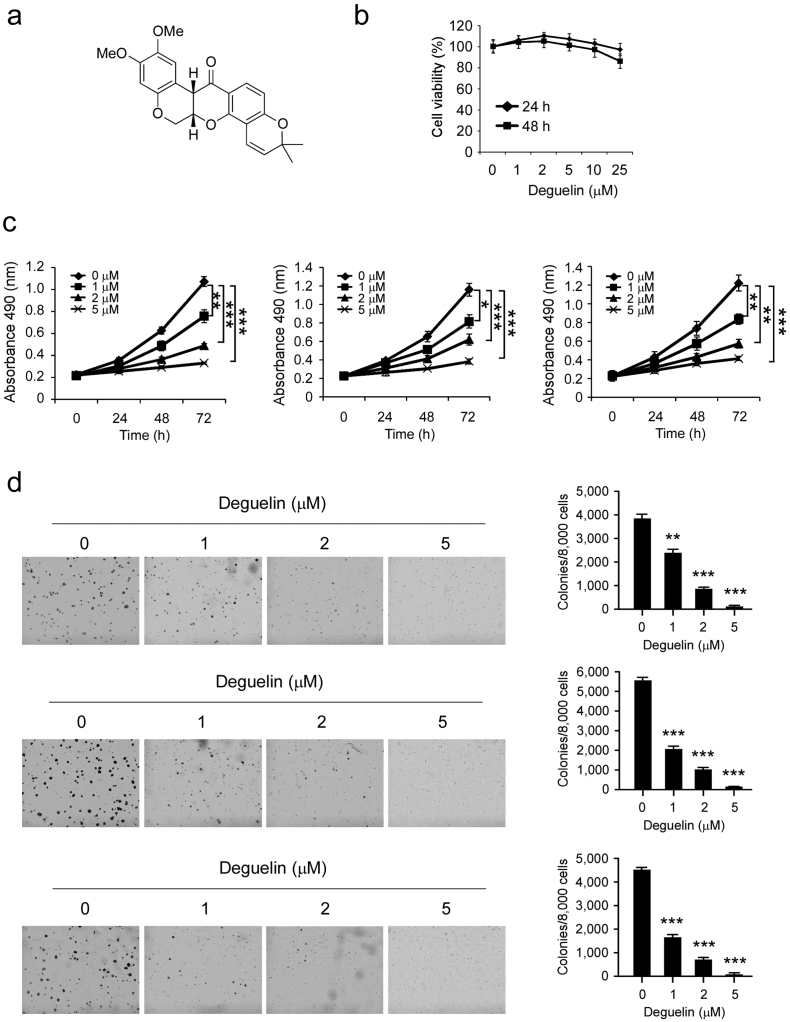
Deguelin (Fig. 2a, MW. 394.42) has shown potential chemopreventive or chemotherapeutical activities against several types of human cancers. In order to investigate the anti-tumor effect of deguelin on human esophageal squamous carcinoma, we first determined whether deguelin exerted any cytotoxic effect on normal esophageal epithelial cells. Het-1A cells were treated with different doses of deguelin for 24 or 48 h and measured by MTS assay. The result showed that deguelin had no cytotoxicity against Het-1A cells at concentrations < 25 μM (Fig. 2b). We then examined the effects of deguelin on the anchorage-dependent growth of KYSE180, KYSE150, and Eca109 cells. The results indicated that deguelin strongly inhibited ESCC cells proliferation in a dose-dependent manner (Fig. 2c). We further investigated the effects of deguelin on anchorage-independent growth of these three ESCC cells via soft agar assay. Results showed that deguelin markedly suppresses colony formation of all tested ESCC cells at different concentrations (Fig. 2d). Our results clearly demonstrate that deguelin has a potent antitumor effect on ESCC cells.
Fig. 2.
Inhibitory effects of deguelin on the growth of ESCC cells. (a) The chemical structure of deguelin. (b) Cytotoxicity of deguelin was measured in normal esophageal epithelial cells by MTS assay. Het-1A cells were treated with various concentrations of deguelin for 24 or 48 h. Data are shown as means ± S.D. from triplicate experiments. (c) The viability of ESCC cells was measured by MTS assay in KYSE180 (left), KYSE150 (middle) and Eca109 (right) cancer cells. All ESCC cells were treated for 24, 48 and 72 h with various concentrations of deguelin as indicated. Data are shown as mean values ± S.D. from 3 independent experiments conducted with triplicate samples. Statistical significance was determined by the Student's t-test (*p < 0.05, **p < 0.01, ***p < 0.001 vs. DMSO-treated group). (d) A colony formation assay was performed as described in the “Materials and Methods” section. Data shown are the colony formation ability of KYSE180 (top), KYSE150 (middle) and Eca109 (bottom) cells treated with different concentrations of deguelin compared with the DMSO group. The average colony number was calculated from three separate experiments (*p < 0.05, **p < 0.01, ***p < 0.001 vs. DMSO-treated group).
3.3. Deguelin Induces Cell Cycle Arrest and Polyploidy in Human Esophageal Squamous Carcinoma Cells
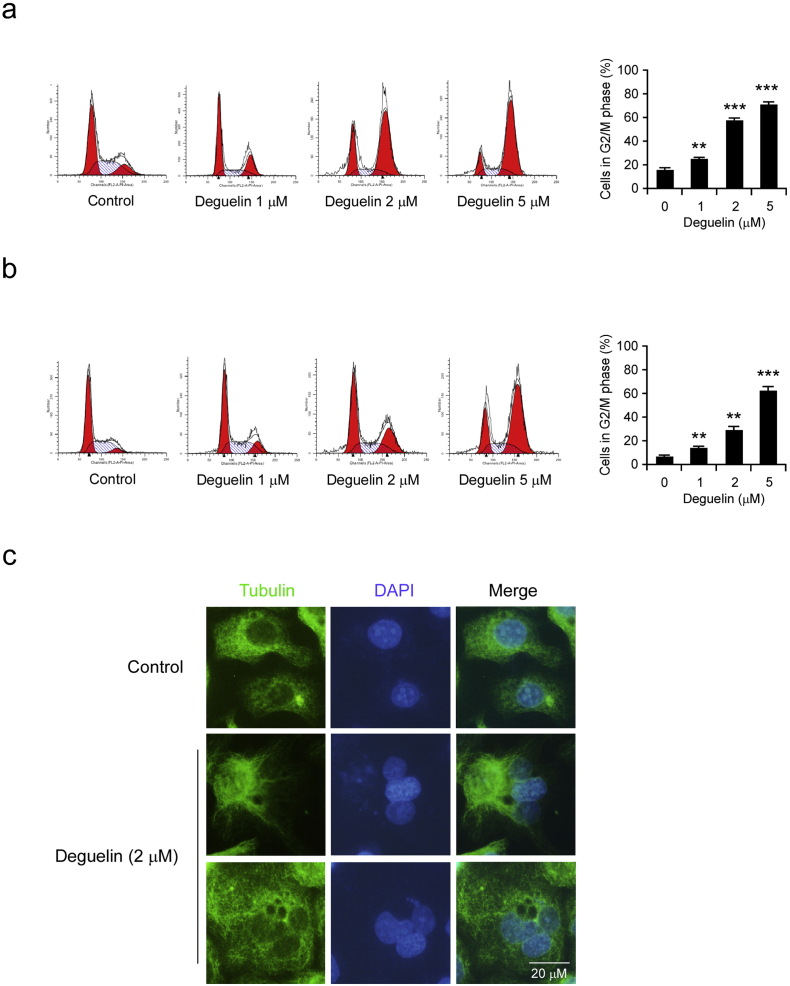
To better understand the mechanism and the activity of deguelin, we evaluated cell cycle distributions after deguelin treatment. The Flow Cytometry data displayed that treatment with deguelin for 24 h caused a dramatic increase in the number of KYSE150 (Fig. 3a) and Eca109 cells (Fig. 3b) occupying the G2/M phase. The G2/M cell cycle arrest indicated that failure in cytokinesis and abnormal exit from mitosis were induced by deguelin treatment, which could result in polyploid cells. We then studied the morphological phenotypes of ESCC cell with deguelin treatment by immunofluorescence staining. The results showed that treatment of KYSE150 cells with 2 μM deguelin caused the induction of polyploidy cells, whereas no polyploidy cells were observed in control cells (Fig. 3c). These results suggest that the inhibition of G2/M cell cycle progression is induced after deguelin treatment in ESCC cells.
Fig. 3.
Deguelin induces G2–M phase cell-cycle arrest and polyploidy in ESCC cells. (a) KYSE150 cells were treated with DMSO or deguelin (1, 2 or 5 μM) for 24 h, cells were collected and cell cycle was then analyzed using propidium iodide staining and flow cytometry. (b) Eca109 cells were treated with DMSO or deguelin (1, 2 or 5 μM) for 24 h, cells were collected and cell cycle was then analyzed using propidium iodide staining and flow cytometry. Data represent mean ± SD from three independent experiments (**p < 0.01, ***p < 0.001 vs. DMSO-treated group). (c) KYSE150 cells were treated with DMSO or deguelin (2 μM) for 24 h and then were evaluated by immunofluorescence assay by staining with anti-α-tubulin (green) and DAPI (blue). Scale bar, 20 μm.
3.4. Deguelin Promotes Apoptosis in KYSE150 and Eca109 Cells
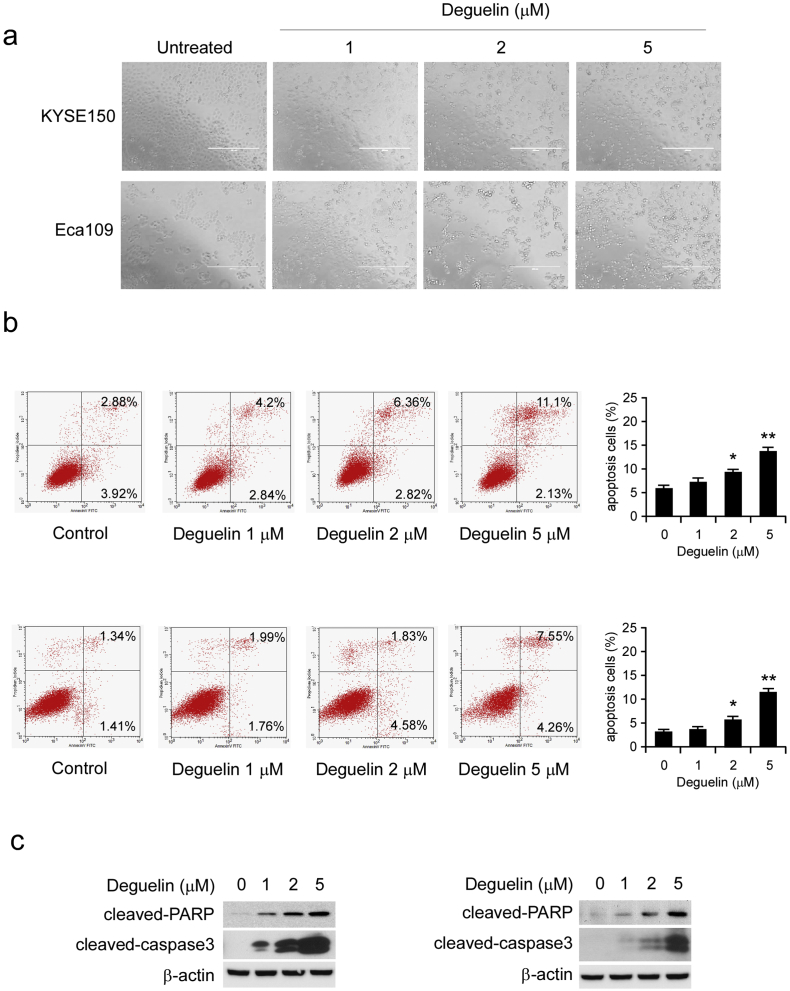
Treatment of KYSE150 and Eca109 cells with different concentrations of deguelin for 24 h dramatically induced cells morphological change. We found that a significant proportion of cells became detachment and round-up, the numbers of round-up cells increased proportionally to deguelin concentrations, which consistent with the phenotype and characteristics of G2/M phase arrest cells (Fig. 4a). Disruption in G2/M cell cycle progression could result in polyploid cells, and ultimately, leads to apoptosis. We next conducted Annexin V/PI staining to measure the effect of deguelin on the induction of apoptosis in KYSE150 and Eca109 cells. The results demonstrated that the application of 2 μM or 5 μM of deguelin for 24 h triggered a significant apoptosis (Fig. 4b). Additionally, consistent with the results of flow cytometry, the protein levels of apoptotic markers, such as cleaved-PARP and cleaved-caspase 3, were substantially upregulated upon deguelin treatment (Fig. 4c). All of these data indicate that increased apoptosis is involved in deguelin-mediated ESCC suppression.
Fig. 4.
Deguelin induces apoptosis in ESCC cells. (a) KYSE150 and Eca109 cells were treated without (DMSO) or with deguelin (1, 2 and 5 μM) for 24 h, bright field pictures were taken to show cell morphology. (b) KYSE150 (upper) and Eca109 (lower) cells were treated without (DMSO) or with deguelin (1, 2 and 5 μM) for 24 h and stained with propidium iodide and Annexin V-FITC, then analyzed by flow cytometry to determine the apoptotic cells. Data represent mean ± SD from three independent experiments (*p < 0.05, **p < 0.01 vs. DMSO-treated group). (c) KYSE150 (left) and Eca109 (right) cells were treated with deguelin (0, 1, 2 and 5 μM) for 24 h, and then the expression of apoptotic markers was detected by immunoblotting.
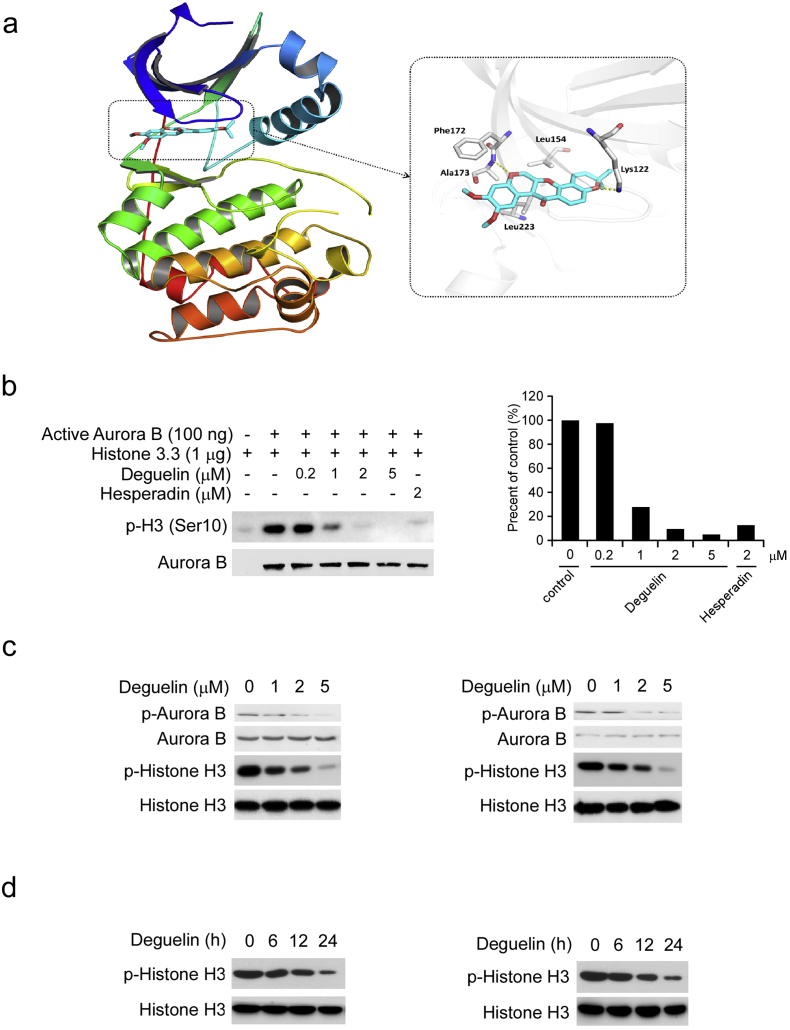
3.5. Deguelin is Associated with Inhibition of Aurora Kinase Signaling Pathways in ESCC Cells
Aurora kinase B inhibition leads to failure in cytokinesis and abnormal exit from mitosis, which could result in polyploidy cells, G2/M cell-cycle arrest, and ultimately, apoptosis or cell death. Our primary data strongly implied that deguelin disrupted the Aurora kinase B pathway in ESCC cells. In order to validate our hypothesis, the molecular docking was performed to study the binding mode of deguelin targeting Aurora B. The in silico docking study indicated that deguelin was docked into the ATP-binding pocket of Aurora B. The docked pose suggested that the flat scaffold of this compound fit well with the pocket by forming hydrophobic interactions with Phe172, Leu223, Leu154, etc. (Fig. 5a). We then used an in vitro kinase assay with a recombinant active Aurora B protein and various concentrations of deguelin to determine whether deguelin could inhibit Aurora B kinase activity in vitro directly. The findings revealed that the phosphorylation of histone H3 on Ser10, an Aurora B substrate, was strongly inhibited by deguelin (Fig. 5b). However, deguelin can't effectively suppress the Aurora A kinases activity in vitro (Fig. S4). These results indicated that deguelin substantially inhibited the kinase activity of Aurora B in a dose-dependent manner from 1 to 5 μM. In addition, we found that autophosphorylation of Aurora B at Thr232 was significantly decreased in the presence of deguelin in KYSE150 and Eca109 cells (Fig. 5c). Phosphorylation of a highly conserved serine residue (Ser10) in histone H3 is thought to be crucial for entry into mitosis. Our results showed that deguelin suppressed histone H3 phosphorylation on Ser10 in KYSE150 and Eca109 cells in a dose- and time-dependent manner (Fig. 5c, d). These results suggest that Aurora B kinase is effective targets of deguelin ex vivo and in vitro.
Fig. 5.
Deguelin inhibits Aurora B kinase activity both in vitro and ex vivo. (a) Computational predicted binding mode of deguelin with Aurora B. Left, The ligand is shown in sticks with red oxygen atoms and cyan carbon atoms. Left, binding pocket. The kinase domain of Aurora B is shown as a rainbow colored cartoon. Right, binding mode. The pocket is colored in gray, of which the key residues are labeled and shown as sticks. Yellow dashed lines represent hydrogen bonds. The figure is generated by PyMOL. (b) Deguelin inhibits Aurora B in vitro kinase activity in a dose-dependent manner. An inactive histone 3.3 protein was used as the substrate for an in vitro kinase assay with active Aurora B and 100 μM ATP as indicated. Proteins were resolved by SDS-PAGE and detected by Western blotting. (c) KYSE150 (left) and Eca109 (right) cells were treated with deguelin (0, 1, 2 and 5 μM) for 24 h, whole cell extract and histone proteins were subjected to Western blot analysis. (d) KYSE150 (left) and Eca109 (right) cells were treated with 5 μM deguelin for various time points as indicated, and histone proteins were subjected to Western blot analysis.
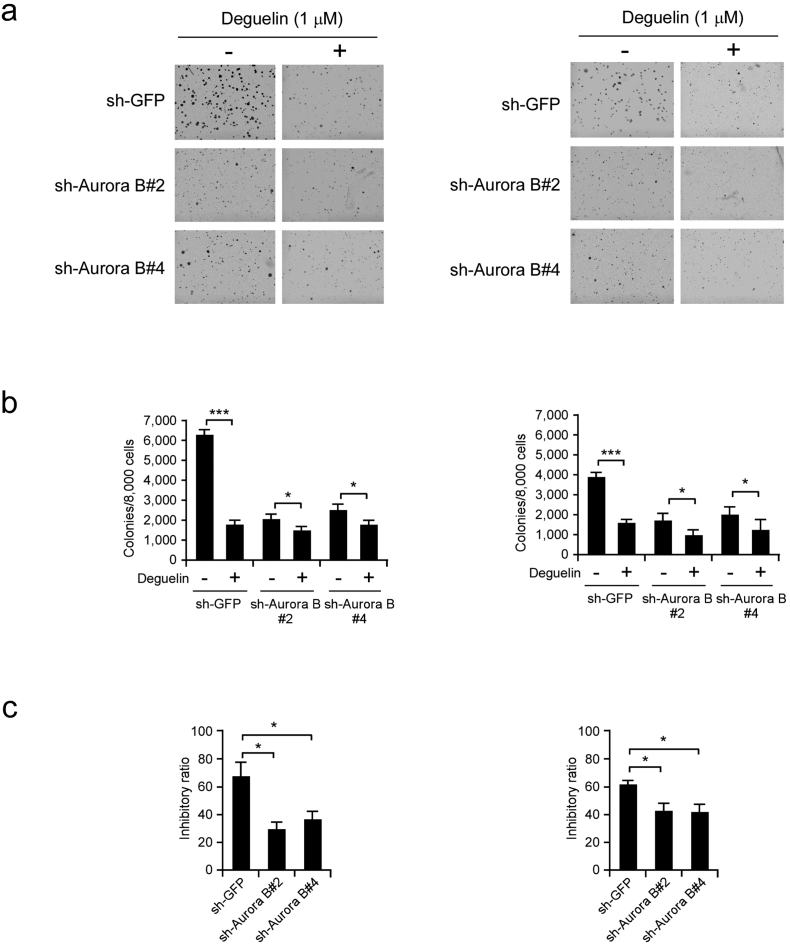
3.6. Knockdown of Aurora B Decreases the Sensitivity of KYSE150 and Eca109 Cells to Deguelin
Because deguelin was shown to specifically target Aurora B, we then examined whether knockdown of Aurora B expression could influence the sensitivity of ESCC cells to deguelin. The soft agar assay data showed that sh-Aurora B transfected cells formed fewer colonies than sh-GFP transfected KYSE150 (Fig. 6a, left) and Eca109 cells (Fig. 6a, right). Deguelin (1 μM) inhibited anchorage-independent growth of KYSE150 (Fig. 6b, left) and Eca109 (Fig. 6b, right) cells which transfected with sh-GFP by about 70% and 60%, respectively. However, in Aurora B shRNA transfected cells, the inhibition ratio was < 30% in KYSE150 sh-Aurora B cells (Fig. 6c, left), and 45% in Eca109 sh-AuroraB cells (Fig. 6c, right). Our results indicated that ESCC cells transfected with Aurora B shRNAs were more resistant to deguelin treatment. These results suggest that Aurora B kinase plays an important role in the sensitivity of KYSE150 and Eca109 cells to the anticancer effects of deguelin.
Fig. 6.
Knockdown of Aurora B decreases the sensitivity of ESCC cells to deguelin. (a, b) Anchorage-independent growth of KYSE150 (left) and Eca109 (right) cells transfected with sh-GFP or sh-Aurora B, treated or not treated with deguelin (1 μM). The average colony number was calculated from three separate experiments (*p < 0.05, ***p < 0.001 vs DMSO-treated group). (c) The inhibitory ratio of deguelin on colony formation ability of KYSE150 (left) and Eca109 (right) cells transfected with sh-GFP or sh-Aurora B (*p < 0.05 vs. sh-GFP group).
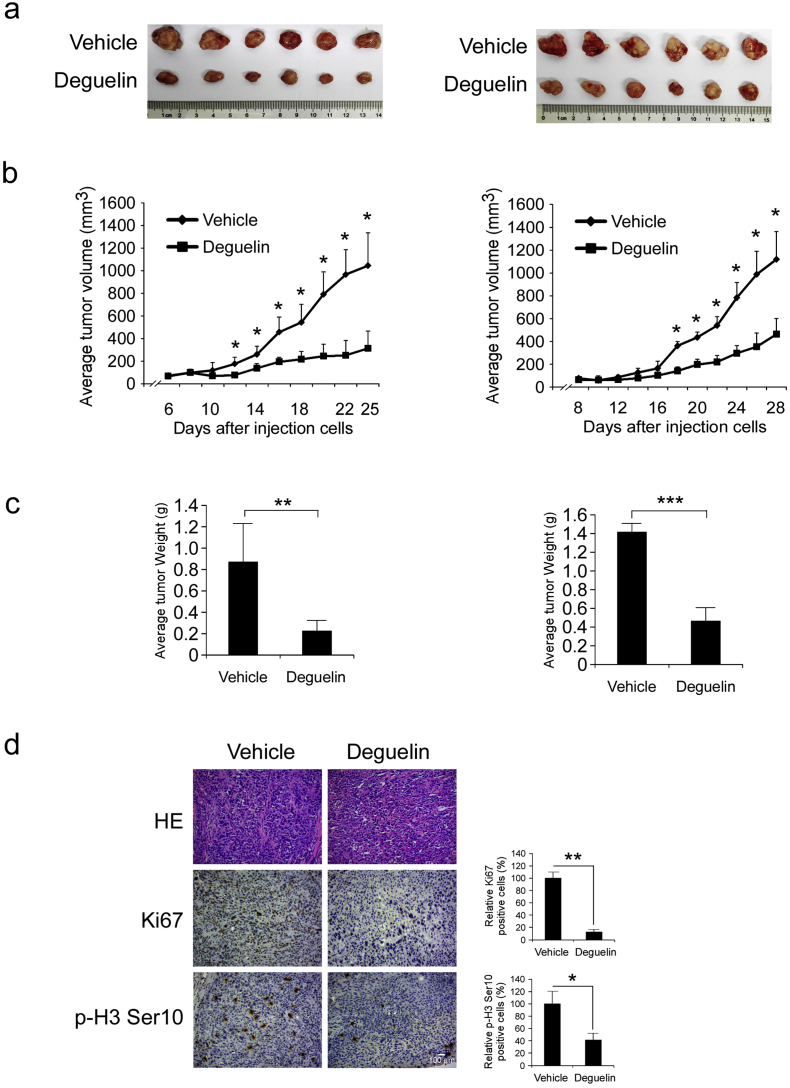
3.7. Deguelin Inhibits Xenograft Growth of KYSE150 and Eca109 Cells in Athymic Nude Mice
To determine the chemotherapeutic effect of deguelin in vivo, we used an athymic nude xenograft mouse model of KYSE150 and Eca109 cells. Data showed that deguelin markedly reduced tumor size (Fig. 7a, b) and tumor weight (Fig. 7c) as expected. Moreover, no obvious loss of body weight was observed (Fig. S3a, b). Immunohistochemical analysis of tumor sections from the KYSE150 animal model showed that the positive staining of Ki67 and phosphorylation of histone H3 Ser10 were significantly suppressed in the deguelin-treated group compared with the vehicle-treated group (Fig. 7d). These results indicate that deguelin suppresses tumor growth in vivo.
Fig. 7.
Deguelin inhibits tumor growth in xenograft mouse models. (a, b and c) Deguelin significantly inhibited tumor growth in KYSE150 (left) and Eca109 (right) xenograft mouse models. At the treatment endpoint, mice were sacrificed and tumors were removed, weighed and photographed (a). The tumor volume of mice from the vehicle- and the deguelin-treated group was measured (b). The tumor weight from vehicle- and deguelin-treated group was measured (c). Statistical significance was as follows: * p < 0.05, ** p < 0.01, *** p < 0.001 vs. vehicle-treated group. (d) Immunohistochemical staining examination of Ki67 and p-H3-Ser10 in the KYSE150 tumor sections from the mice of the vehicle- or deguelin-treated group. All panels are of the same magnification (* p < 0.05, ** p < 0.01 vs. vehicle-treated group). Scale bar, 100 μm.
4. Discussion
Abnormal activation of mitosis plays a crucial role in the process of human tumorigenesis. In the past few decades, targeting anti-mitotic therapies is a highly successful strategy for human anticancer treatment. Microtubule dynamics targeted agents, including the vinca alkaloids and taxanes, are highly active against a variety of human cancer types. However, development of drug resistance and highly toxic to normal dividing cells remain persistent problems (Hardin et al., 2017, Kavallaris, 2010, Louage et al., 2017). Therefore, identification of novel nonmicrotubule proteins therapeutic targets is still an urgent demand for anticancer treatment. Recently, the Aurora kinases have gained prominence as essential regulators of somatic cell division. Aurora B kinase is essential for chromosome condensation, kinetochore function, cytokinesis, and the proper function of the spindle assembly checkpoint when spindle tension is perturbed. Aurora B was first demonstrated to be overexpressed in colon cancer in 1998 (Bischoff et al., 1998). Cumulative evidence indicated that the amplification/overexpression and/or hyperactivation of Aurora B kinase is a frequent finding in a panel of human malignancies, such as breast (Larsen et al., 2015, Zhang et al., 2015), liver (Liu et al., 2017), lung (Al-Khafaji et al., 2017, Helfrich et al., 2016), prostate (Schecher et al., 2017, Zekri et al., 2017) cancer and leukemia (Floc'h et al., 2017, Song et al., 2017). The evidence linking Aurora B overexpression and malignancy has generated significant interest in the development of small-molecule inhibitors against this protein.
In this report, the natural compound deguelin potently and dose-dependently suppressed Aurora B kinase activity in vitro, indicating that this compound is an effective Aurora B inhibitor. The docking study indicated that deguelin was docked into the ATP-binding pocket of Aurora B and formed hydrophobic interactions with Phe172, Leu223, Leu154, etc. In addition, deguelin had no obvious effect on Aurora A activity at the same concentration (Fig. S4). Previous studies have demonstrated that the predominant response of tumor cells to Aurora B inhibitor treatment was aborted cell division without a protracted mitotic arrest, inducing the formation of polyploidy cell and resulting in polyploidy-specific lethality or senescence (Payton et al., 2010, Sadaie et al., 2015). Our results showed that deguelin inhibited the anchorage-dependent and -independent growth of human ESCC cells. Treatment with deguelin induced accumulation of G2/M cells, polyploidy cell formation, and apoptosis, which is consistent with Aurora B inhibition. In addition, knockdown of Aurora B decreases the sensitivity of ESCC cells to deguelin, which agrees with our observations that Aurora B plays an important role in the anticancer activity of deguelin. Moreover, our in vivo study also demonstrated that deguelin significantly decreased tumor growth and down-regulated phosphorylated Histone-H3 staining without obvious cytotoxic effect.
Esophageal carcinoma is often refractory to current therapeutic approaches and has a poor prognosis. The incidence of esophageal carcinoma is closely linked to geographic region and racial background. During the past few decades, the incidence of adenocarcinoma which arises in the esophagogastric junction or distal esophagus has increased considerably in western countries, whereas in China and other Asian nations, such as Japan, over 90% of all esophageal carcinomas are squamous-cell carcinomas (Abnet et al., 2017, Lagergren and Lagergren, 2013, Rice et al., 2017). Although ESCC can be treated with various techniques, including chemotherapy, radiotherapy, and endoscopy, the surgical resection has remained the mainstay of treatment. Currently, attention has focused on the role of targeted agents in the treatment of various types of cancer. Studies of targeted agents for ESCC have just begun. Although the combination of EGFR targeted therapy agent cetuximab and radiotherapy in ESCC patients obtained a clinical complete response rate of 70%, more evidence is needed to prove that adding cetuximab to preoperative chemoradiotherapy is feasible without increasing postoperative mortality (Enzinger et al., 2016). Previous studies indicated that Aurora A is highly expressed in ESCC (Tamotsu et al., 2015, Tong et al., 2004), whereas the status of Aurora B in ESCC is not clear. In the present study, we found that Aurora B is overexpressed in ESCC cell lines, and the high levels of Aurora B protein were associated with worse overall survival rate. Moreover, knockdown Aurora B in human ESCC cells reduced tumorigenic properties, including in vitro cell growth and in vivo xenografted tumor formation. All of this evidence suggested that the development of Aurora kinases targeted therapy may serve as a selective strategy for clinical ESCC treatment. Recently, a growing number of inhibitors of Aurora kinases have been developed for both hematopoietic malignancies and solid tumors, either at preclinical or clinical stages (Ashton et al., 2016, Dar et al., 2010, Floc'h et al., 2017). Previous studies have demonstrated that the Aurora B specific inhibitor, AZD1152, significantly suppressed human solid tumors, including prostate (Zekri et al., 2017), lung (Helfrich et al., 2016), breast (Larsen et al., 2015) cancer and nasopharyngeal carcinoma (Li et al., 2015). However, the antitumor effect of Aurora B kinase inhibitors against ESCC is still elusive. Our study clearly showed that deguelin-induced ESCC growth inhibition was dependent on deguelin-mediated Aurora B activity suppression because down-regulation of Aurora B significantly decreased the sensitivity of ESCC cells to deguelin treatment. Our results are in agreement with those recent publications that the Aurora B specific inhibitor also exerts the potent anti-tumor effect in solid tumors.
The combination of Aurora kinase inhibitors with standard chemotherapeutics or radiotherapeutics are currently in progress, such as AZD1152 plus radiotherapy in various solid cancer models. In addition, Azzariti et al. found that AZD1152-HQPA enhanced oxaliplatin and gemcitabine effectiveness in the colon and pancreatic cancer, respectively (Azzariti et al., 2011). However, little is known about the mechanism of action of Aurora kinase inhibitors in combination with radio- and chemotherapy at the molecular level. Azzariti et al. suggested that the potential mechanism for AZD1152-HQPA enhanced chemotherapy effectiveness is due to the DNA fragmentation associated with apoptosis occur in a cell population that has undergone endoreduplication to become > 4 N (Azzariti et al., 2011). Additionally, the M phase and G2 phase are the most sensitive stage to radiotherapy or DNA damage agents. Therefore, chemotherapy and fractionated radiotherapy may work better in concurrent of the Aurora B kinase inhibitors by partial synchronization of cancer cells in the most radiosensitive G2/M phase (Pawlik and Keyomarsi, 2004).
In summary, our in vitro and in vivo results suggest that deguelin is a bioavailable and potent Aurora B kinase inhibitor for ESCC treatment. Importantly, neoadjuvant chemotherapy or radiotherapy followed by surgery has emerged as a new standard treatment for ESCC currently (Markar et al., 2015, Matsuda et al., 2016, Pasquali et al., 2017, Sudo et al., 2014). Thus, a single treatment with deguelin or combination with conventional chemotherapy/radiotherapy deserves further investigation and development for clinic translation.
The following are the supplementary data related to this article.
Supplementary Fig. 1.
Validation of the Aurora B knockdown by shRNAs in human ESCC cells. (a, b) KYSE150 (a) and Eca109 (b) cells were transduced with lentiviral short hairpin RNAs (shRNAs) targeting GFP control (shGFP) and Aurora B (#1 to #4). The Aurora B expression was confirmed by Western blot analysis. β-actin was used as a loading control.
Supplementary Fig. 2.
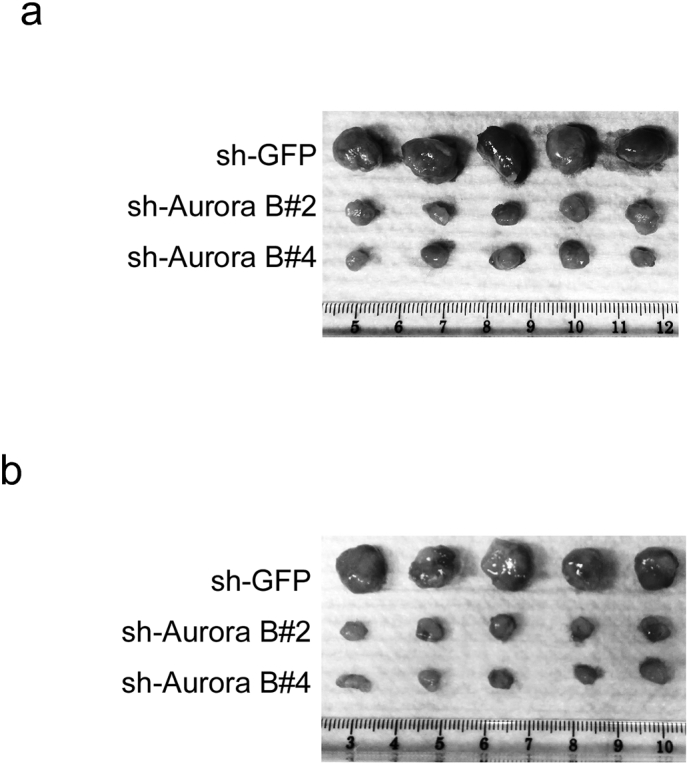
Knockdown of Aurora B inhibits tumor growth in xenograft mouse models. (a) Photograph of tumors dissected from mice injected with KYSE150-shGFP or KYSE150-shAurora B cells. (b) Photograph of tumors dissected from mice injected with Eca109-shGFP or Eca109-shAurora B cells.
Supplementary Fig. 3.
Body weight of mice from the vehicle- and deguelin-treated group. (a) The body weight of KYSE150 cell-transplanted mice. (b) The body weight of Eca109 cell-transplanted mice.
Supplementary Fig. 4.
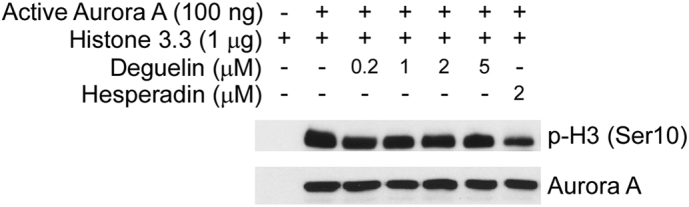
The effect of deguelin on Aurora A in vitro kinase activity. An inactive histone 3.3 protein was used as the substrate for an in vitro kinase assay with active Aurora A and 100 μM ATP as indicated. Proteins were resolved by SDS-PAGE and detected by Western blotting.
Funding Sources
This work was supported by the National Natural Science Foundation of China (No.81401548, No.81572280, No.81371690, No.81502121, and No.81201171) and New Xiangya Talent Project of the Third Xiangya Hospital of Central South University (No.JY2015011).
Conflicts of Interest
The authors declare no conflicts of interest.
Author Contributions
Conception and design: H.-D. Liu, W. Li, X.-F. Yu.
Development of methodology: H.-D. Liu, W. Li, X.-F. Yu.
Acquisition of data: H.-D. Liu, W. Li, X.-F. Yu, Q. Liang, W.-B. Liu, L. Zhou.
Analysis and interpretation of data: H.-D. Liu, W. Li, X.-F. Yu.
Writing, review, and/or revision of the manuscript: H.-D. Liu, W. Li, X.-F. Yu, Q. Liang.
Administrative, technical, or material support: H.-D. Liu, W. Li, X.-F. Yu,
Study supervision: H.-D. Liu, W. Li.
Acknowledgments
Acknowledgement
None.
Contributor Information
Wei Li, Email: Weililx@163.com.
Haidan Liu, Email: haidanliu@csu.edu.cn.
References
- Abnet C.C., Arnold M., Wei W.Q. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology. 2017 doi: 10.1053/j.gastro.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khafaji A.S., Davies M.P., Risk J.M., Marcus M.W., Koffa M., Gosney J.R., Shaw R.J., Field J.K., Liloglou T. Aurora B expression modulates paclitaxel response in non-small cell lung cancer. Br. J. Cancer. 2017;116:592–599. doi: 10.1038/bjc.2016.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton S., Song Y.H., Nolan J., Cadogan E., Murray J., Odedra R., Foster J., Hall P.A., Low S., Taylor P. Aurora kinase inhibitor nanoparticles target tumors with favorable therapeutic index in vivo. Sci. Transl. Med. 2016;8:325ra317. doi: 10.1126/scitranslmed.aad2355. [DOI] [PubMed] [Google Scholar]
- Azzariti A., Bocci G., Porcelli L., Fioravanti A., Sini P., Simone G.M., Quatrale A.E., Chiarappa P., Mangia A., Sebastian S. Aurora B kinase inhibitor AZD1152: determinants of action and ability to enhance chemotherapeutics effectiveness in pancreatic and colon cancer. Br. J. Cancer. 2011;104:769–780. doi: 10.1038/bjc.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavetsias V., Linardopoulos S. Aurora kinase inhibitors: current status and outlook. Front. Oncol. 2015;5:278. doi: 10.3389/fonc.2015.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff J.R., Anderson L., Zhu Y., Mossie K., Ng L., Souza B., Schryver B., Flanagan P., Clairvoyant F., Ginther C. A homologue of Drosophila Aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M., Ruchaud S., Earnshaw W.C. Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr. Opin. Cell Biol. 2009;21:796–805. doi: 10.1016/j.ceb.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar A.A., Goff L.W., Majid S., Berlin J., El-Rifai W. Aurora kinase inhibitors—rising stars in cancer therapeutics? Mol. Cancer Ther. 2010;9:268–278. doi: 10.1158/1535-7163.MCT-09-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enzinger P.C., Burtness B.A., Niedzwiecki D., Ye X., Douglas K., Ilson D.H., Villaflor V.M., Cohen S.J., Mayer R.J., Venook A. CALGB 80403 (alliance)/E1206: a randomized phase II study of three chemotherapy regimens plus Cetuximab in metastatic esophageal and Gastroesophageal junction cancers. J. Clin. Oncol. 2016;34:2736–2742. doi: 10.1200/JCO.2015.65.5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falchook G.S., Bastida C.C., Kurzrock R. Aurora kinase inhibitors in oncology clinical trials: current state of the progress. Semin. Oncol. 2015;42:832–848. doi: 10.1053/j.seminoncol.2015.09.022. [DOI] [PubMed] [Google Scholar]
- Floc'h N., Ashton S., Taylor P., Trueman D., Harris E., Odedra R., Maratea K., Derbyshire N., Caddy J., Jacobs V.N. Optimizing therapeutic effect of Aurora B inhibition in acute myeloid leukemia with AZD2811 nanoparticles. Mol. Cancer Ther. 2017;16:1031–1040. doi: 10.1158/1535-7163.MCT-16-0580. [DOI] [PubMed] [Google Scholar]
- Hardin C., Shum E., Singh A.P., Perez-Soler R., Cheng H. Emerging treatment using tubulin inhibitors in advanced non-small cell lung cancer. Expert. Opin. Pharmacother. 2017;18:701–716. doi: 10.1080/14656566.2017.1316374. [DOI] [PubMed] [Google Scholar]
- Helfrich B.A., Kim J., Gao D., Chan D.C., Zhang Z., Tan A.C., Bunn P.A., Jr. Barasertib (AZD1152), a small molecule aurora B inhibitor, inhibits the growth of SCLC cell lines in vitro and in vivo. Mol. Cancer Ther. 2016;15:2314–2322. doi: 10.1158/1535-7163.MCT-16-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavallaris M. Microtubules and resistance to tubulin-binding agents. Nat. Rev. Cancer. 2010;10:194–204. doi: 10.1038/nrc2803. [DOI] [PubMed] [Google Scholar]
- Krenn V., Musacchio A. The aurora B kinase in chromosome bi-orientation and spindle checkpoint signaling. Front. Oncol. 2015;5:225. doi: 10.3389/fonc.2015.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagergren J., Lagergren P. Recent developments in esophageal adenocarcinoma. CA Cancer J. Clin. 2013;63:232–248. doi: 10.3322/caac.21185. [DOI] [PubMed] [Google Scholar]
- Larsen S.L., Yde C.W., Laenkholm A.V., Rasmussen B.B., Duun-Henriksen A.K., Bak M., Lykkesfeldt A.E., Kirkegaard T. Aurora kinase B is important for antiestrogen resistant cell growth and a potential biomarker for tamoxifen resistant breast cancer. BMC Cancer. 2015;15:239. doi: 10.1186/s12885-015-1210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lens S.M., Voest E.E., Medema R.H. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat. Rev. Cancer. 2010;10:825–841. doi: 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- Li W., Peng C., Lee M.H., Lim D., Zhu F., Fu Y., Yang G., Sheng Y., Xiao L., Dong X. TRAF4 is a critical molecule for Akt activation in lung cancer. Cancer Res. 2013;73:6938–6950. doi: 10.1158/0008-5472.CAN-13-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Hong M.J., Chow J.P., Man W.Y., Mak J.P., Ma H.T., Poon R.Y. Co-inhibition of polo-like kinase 1 and aurora kinases promotes mitotic catastrophe. Oncotarget. 2015;6:9327–9340. doi: 10.18632/oncotarget.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Fan J.H., Qiao Y.L. Epidemiology, etiology, and prevention of esophageal squamous cell carcinoma in China. Cancer Biol. Med. 2017;14:33–41. doi: 10.20892/j.issn.2095-3941.2016.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M.J., Lampson M.A., Lens S.M. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Li W., Yu X., Gao F., Duan Z., Ma X., Tan S., Yuan Y., Liu L., Wang J. EZH2-mediated puma gene repression regulates non-small cell lung cancer cell proliferation and cisplatin-induced apoptosis. Oncotarget. 2016;7:56338–56354. doi: 10.18632/oncotarget.10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Wang G., Wang X., Che Z., Dong W., Guo X., Wang Z., Chen P., Hou D., Zhang Q. Targeting high Aurora kinases expression as an innovative therapy for hepatocellular carcinoma. Oncotarget. 2017;8:27953–27965. doi: 10.18632/oncotarget.15853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louage B., De Wever O., Hennink W.E., De Geest B.G. Developments and future clinical outlook of taxane nanomedicines. J. Control. Release. 2017;253:137–152. doi: 10.1016/j.jconrel.2017.03.027. [DOI] [PubMed] [Google Scholar]
- Luo W., Fang W., Li S., Yao K. Aberrant expression of nuclear vimentin and related epithelial-mesenchymal transition markers in nasopharyngeal carcinoma. Int. J. Cancer. 2012;131:1863–1873. doi: 10.1002/ijc.27467. [DOI] [PubMed] [Google Scholar]
- Mahadevan D., Islam S., Qi W., Morales C., Cooke L., Spier C., Weterings E. Disruption of aneuploidy and senescence induced by aurora inhibition promotes intrinsic apoptosis in double hit or double expressor diffuse large B-cell lymphomas. Mol. Cancer Ther. 2017;(10):2083–2093. doi: 10.1158/1535-7163.MCT-17-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markar S., Gronnier C., Duhamel A., Pasquer A., Thereaux J., du Rieu M.C., Lefevre J.H., Turner K., Luc G., Mariette C. Salvage surgery after Chemoradiotherapy in the Management of Esophageal Cancer: is it a viable therapeutic option? J. Clin. Oncol. 2015;33:3866–3873. doi: 10.1200/JCO.2014.59.9092. [DOI] [PubMed] [Google Scholar]
- Matsuda S., Takeuchi H., Kawakubo H., Ando N., Kitagawa Y. Current advancement in multidisciplinary treatment for Resectable cStage II/III esophageal squamous cell carcinoma in Japan. Ann. Thorac. Cardiovasc. Surg. 2016;22:275–283. doi: 10.5761/atcs.ra.16-00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A.L., Schindler K. Specialize and divide (twice): functions of three aurora kinase homologs in mammalian oocyte meiotic maturation. Trends Genet. 2017;33:349–363. doi: 10.1016/j.tig.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto T., Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat. Rev. Cancer. 2017;17:93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali S., Yim G., Vohra R.S., Mocellin S., Nyanhongo D., Marriott P., Geh J.I., Griffiths E.A. Survival after Neoadjuvant and adjuvant treatments compared to surgery alone for Resectable esophageal carcinoma: a network meta-analysis. Ann. Surg. 2017;265:481–491. doi: 10.1097/SLA.0000000000001905. [DOI] [PubMed] [Google Scholar]
- Pawlik T.M., Keyomarsi K. Role of cell cycle in mediating sensitivity to radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2004;59:928–942. doi: 10.1016/j.ijrobp.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Payton M., Bush T.L., Chung G., Ziegler B., Eden P., McElroy P., Ross S., Cee V.J., Deak H.L., Hodous B.L. Preclinical evaluation of AMG 900, a novel potent and highly selective pan-aurora kinase inhibitor with activity in taxane-resistant tumor cell lines. Cancer Res. 2010;70:9846–9854. doi: 10.1158/0008-5472.CAN-10-3001. [DOI] [PubMed] [Google Scholar]
- Portella G., Passaro C., Chieffi P. Aurora B: a new prognostic marker and therapeutic target in cancer. Curr. Med. Chem. 2011;18:482–496. doi: 10.2174/092986711794480203. [DOI] [PubMed] [Google Scholar]
- Rice T.W., Gress D.M., Patil D.T., Hofstetter W.L., Kelsen D.P., Blackstone E.H. Cancer of the esophagus and esophagogastric junction-major changes in the American joint committee on cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017;67:304–317. doi: 10.3322/caac.21399. [DOI] [PubMed] [Google Scholar]
- Rustgi A.K., El-Serag H.B. Esophageal carcinoma. N. Engl. J. Med. 2014;371:2499–2509. doi: 10.1056/NEJMra1314530. [DOI] [PubMed] [Google Scholar]
- Sadaie M., Dillon C., Narita M., Young A.R., Cairney C.J., Godwin L.S., Torrance C.J., Bennett D.C., Keith W.N., Narita M. Cell-based screen for altered nuclear phenotypes reveals senescence progression in polyploid cells after Aurora kinase B inhibition. Mol. Biol. Cell. 2015;26:2971–2985. doi: 10.1091/mbc.E15-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schecher S., Walter B., Falkenstein M., Macher-Goeppinger S., Stenzel P., Krumpelmann K., Hadaschik B., Perner S., Kristiansen G., Duensing S. Cyclin K dependent regulation of Aurora B affects apoptosis and proliferation by induction of mitotic catastrophe in prostate cancer. Int. J. Cancer. 2017;141:1643–1653. doi: 10.1002/ijc.30864. [DOI] [PubMed] [Google Scholar]
- Sheng Y., Li W., Zhu F., Liu K., Chen H., Yao K., Reddy K., Lim D.Y., Oi N., Li H. 3,6,2′,4′,5′-Pentahydroxyflavone, an orally bioavailable multiple protein kinase inhibitor, overcomes gefitinib resistance in non-small cell lung cancer. J. Biol. Chem. 2014;289:28192–28201. doi: 10.1074/jbc.M114.593475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Ma R., Yang X., Pang S. The Deubiquitinating enzyme USP14 regulates leukemic chemotherapy drugs-induced cell apoptosis by suppressing Ubiquitination of aurora kinase B. Cell. Physiol. Biochem. 2017;42:965–973. doi: 10.1159/000478679. [DOI] [PubMed] [Google Scholar]
- Sudo K., Xiao L., Wadhwa R., Shiozaki H., Elimova E., Taketa T., Blum M.A., Lee J.H., Bhutani M.S., Weston B. Importance of surveillance and success of salvage strategies after definitive chemoradiation in patients with esophageal cancer. J. Clin. Oncol. 2014;32:3400–3405. doi: 10.1200/JCO.2014.56.7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamotsu K., Okumura H., Uchikado Y., Kita Y., Sasaki K., Omoto I., Owaki T., Arigami T., Uenosono Y., Nakajo A. Correlation of Aurora-a expression with the effect of chemoradiation therapy on esophageal squamous cell carcinoma. BMC Cancer. 2015;15:323. doi: 10.1186/s12885-015-1329-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A., Gao K., Chu L., Zhang R., Yang J., Zheng J. Aurora kinases: novel therapy targets in cancers. Oncotarget. 2017;8:23937–23954. doi: 10.18632/oncotarget.14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong T., Zhong Y., Kong J., Dong L., Song Y., Fu M., Liu Z., Wang M., Guo L., Lu S. Overexpression of Aurora-a contributes to malignant development of human esophageal squamous cell carcinoma. Clin. Cancer Res. 2004;10:7304–7310. doi: 10.1158/1078-0432.CCR-04-0806. [DOI] [PubMed] [Google Scholar]
- Yan M., Wang C., He B., Yang M., Tong M., Long Z., Liu B., Peng F., Xu L., Zhang Y. Aurora-a kinase: a potent oncogene and target for cancer therapy. Med. Res. Rev. 2016;36:1036–1079. doi: 10.1002/med.21399. [DOI] [PubMed] [Google Scholar]
- Yu X., Deng Q., Li W., Xiao L., Luo X., Liu X., Yang L., Peng S., Ding Z., Feng T. Neoalbaconol induces cell death through necroptosis by regulating RIPK-dependent autocrine TNFalpha and ROS production. Oncotarget. 2015;6:1995–2008. doi: 10.18632/oncotarget.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li W., Deng Q., You S., Liu H., Peng S., Liu X., Lu J., Luo X., Yang L. Neoalbaconol inhibits angiogenesis and tumor growth by suppressing EGFR-mediated VEGF production. Mol. Carcinog. 2017;56:1414–1426. doi: 10.1002/mc.22602. [DOI] [PubMed] [Google Scholar]
- Yu X., Li W., Xia Z., Xie L., Ma X., Liang Q., Liu L., Wang J., Zhou X., Yang Y. Targeting MCL-1 sensitizes human esophageal squamous cell carcinoma cells to cisplatin-induced apoptosis. BMC Cancer. 2017;17:449. doi: 10.1186/s12885-017-3442-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zekri A., Mesbahi Y., Ghanizadeh-Vesali S., Alimoghaddam K., Ghavamzadeh A., Ghaffari S.H. Reactive oxygen species generation and increase in mitochondrial copy number: new insight into the potential mechanism of cytotoxicity induced by aurora kinase inhibitor, AZD1152-HQPA. Anti-Cancer Drugs. 2017;28:841–851. doi: 10.1097/CAD.0000000000000523. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Jiang C., Li H., Lv F., Li X., Qian X., Fu L., Xu B., Guo X. Elevated Aurora B expression contributes to chemoresistance and poor prognosis in breast cancer. Int. J. Clin. Exp. Pathol. 2015;8:751–757. [PMC free article] [PubMed] [Google Scholar]
- Ziemska J., Solecka J. Tyrosine kinase, aurora kinase and leucine aminopeptidase as attractive drug targets in anticancer therapy - characterisation of their inhibitors. Rocz. Panstw. Zakl. Hig. 2016;67:329–342. [PubMed] [Google Scholar]