Abstract
Background
Sensitization in early childhood may precede respiratory allergy in adolescence.
Methods
IgE reactivity against 132 allergen molecules was evaluated using the MeDALL microarray in sera obtained from a random sample of 786 children at the age of 4, 8 and 16 years in a population based birth cohort (BAMSE). Symptoms were analyzed by questionnaire at ages 4, 8 and 16 years. Clinically and independent relevant allergen molecules accounting for ≥ 90% of IgE reactivities in sensitized individuals and at all time-points were identified as risk molecules and used to predict respiratory allergy. The data was replicated in the Manchester Asthma and Allergy Study (MAAS) birth cohort by studying IgE reactivity with the use of a commercial IgE microarray. Sera were obtained from children at the ages of 3, 5, 8 and 11 years (N = 248) and the outcome was studied at 11 years.
Findings
In the BAMSE cohort 4 risk molecules could be identified, i.e.: Ara h 1 (peanut), Bet v 1 (birch), Fel d 1 (cat), Phl p 1 (grass). For MAAS the corresponding number of molecules was 5: Der p 1 (dust mite), Der f 2 (dust mite), Phl p 1 (grass), Phl p 5 (grass), Fel d 1 (cat). In BAMSE, early IgE reactivity to ≥ 3 of 4 allergen molecules at four years predicted incident and persistent asthma and/or rhinitis at 16 years (87% and 95%, respectively). The corresponding proportions in the MAAS cohort at 16 years were 100% and 100%, respectively, for IgE reactivity to ≥ 3 of 5 risk molecules.
Interpretations
IgE reactivity to a few allergen molecules early in life identifies children with a high risk of asthma and/or rhinitis at 16 years. These findings will be of importance for developing preventive strategies for asthma and rhinitis in children.
Keywords: Asthma, IgE, Prediction, Rhinitis, Sensitisation
Highlights
-
•
IgE reactivity to only few allergen molecules in early childhood predicts respiratory allergy in adolescence
-
•
It may be possible to develop individualized risk prediction charts for allergic respiratory diseases.
-
•
These findings could be targets for novel intervention therapies.
Birth cohorts are essential for understanding the life course of allergy. With a novel approach using a large panel of micro-arrayed allergen molecules from more than forty allergen sources, we identified a strong IgE signature against a handful of allergen molecules at ages 3–5 years that predicted respiratory allergy with > 90% probability up until adolescence in two geographically separate populations. The results suggest generalizability across populations. The findings are of clinical importance for pediatricians or physicians seeing children at a young age, who could perform early allergy diagnosis with the key allergen molecules to initiate preventive measures
1. Introduction
IgE sensitization in early childhood is associated with increased risks of asthma, rhinitis and eczema but there are no reliable parameters that can predict the transition from IgE sensitization to the development of respiratory allergy later in life (Illi et al., 2001, Illi et al., 2004, Brockow et al., 2009). This accounts for both aero- and food allergens. Allergic diseases are heterogeneous in terms of phenotypes and trajectories (Belgrave et al., 2014, Ranciere et al., 2013). In most studies, IgE antibody reactivity has been analyzed using singleplex assays with a limited number of allergen extracts (Illi et al., 2001, Illi et al., 2004, Brockow et al., 2009, Chiu et al., 2014) that cannot distinguish genuine sensitization from cross-reactivity. However, with multiplex platforms using allergen molecules spotted on a microarray, IgE to > 100 defined allergen components can be simultaneously analyzed (Hiller et al., 2002, Canonica et al., 2013, Melioli et al., 2013, Patelis et al., 2012, Hatzler et al., 2012, van Hage et al., 2017). This allows an unprecedented precision of mapping the sensitization profiles down to the level of disease-causing allergen molecules (Bousquet et al., 2016).
Birth cohorts are essential for understanding the life course of allergy (Bousquet et al., 2014), allowing to identify windows of opportunity for preventive strategies (Westman et al., 2017). The role of sensitization to specific allergen molecules or to patterns of molecules in childhood for predicting development of allergic diseases in adolescence is unknown. We hypothesized that early age IgE sensitization to specific allergen molecules would be a strong predictor of risk to develop respiratory allergy from childhood to adolescence. Since there are obvious differences in regional sensitizations we aimed to investigate two cohorts from different regions to understand heterogeneity and improve generalizability of the results.
Our goal was to investigate if IgE recognition of certain allergen molecules early in life could be linked to respiratory allergy, rhinitis and/or asthma, in childhood up to adolescence. For this purpose sera from children from the Swedish birth cohort BAMSE (Barn/Children, Allergy, Milieu, Stockholm, Epidemiological study) were analyzed using a large panel of micro-arrayed allergen molecules and the results were replicated in the Manchester Asthma and Allergy Study (MAAS) birth cohort (Custovic et al., 2002).
2. Methods
2.1. Study Design and Participants
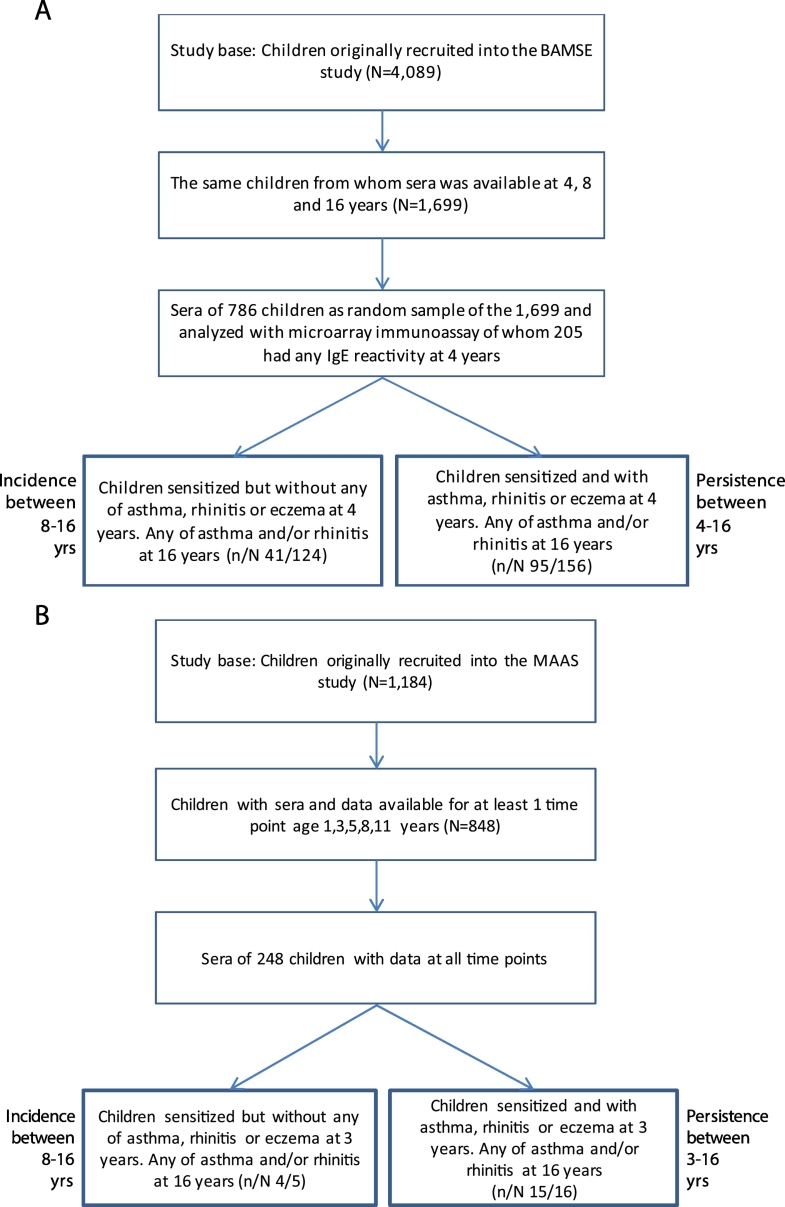
Discovery population: BAMSE is an unselected population-based birth cohort comprising 4089 children from representative areas of Stockholm (Wickman et al., 2002). Parents of all children born between 1994 and 1996 in certain areas of Stockholm were invited to participate. The BAMSE cohort comprised 4089 infants, corresponding to 75% of the eligible subjects. The children were recruited postnatally and follow-ups were performed repeatedly including collection of questionnaire data on exposure and outcome, mainly of asthma, rhinitis and eczema. A period prevalence of reported symptoms during the last 12 months was assessed. When the infants were about two months old, the parents were asked to fill in a detailed questionnaire regarding parental allergy, background factors and other relevant exposures. Follow-ups were performed at 1, 2, 4, 8, 12 and 16 years with a response rate of 78% at the latest follow-up. Allergy testing based on venous blood samples was performed in 2605; 2470 and 2547 children at four, eight and 16 years, respectively, which corresponds to 64%, 60% and 62%, of children at baseline. In 1699 children (42%) sera were available from all three time points, and 800 children were randomly picked for IgE analyses (Fig. 1A) (Asarnoj et al., 2016, Westman et al., 2015).
Fig. 1.
A. (BAMSE) Flow chart of children in the study base and study population as well as for those remaining for statistical analysis of onset and persistency of asthma, rhinitis and/or eczema at 16 years. Analyses in relation to sensitization to allergen molecules at 4 years with (persistency) and without (onset) children with any of asthma (A), rhinitis (R) or eczema (E) at 4 years. B. (MAAS). Flow chart of children in the study base and study population as well as for those remaining for statistical analysis of onset and persistency of asthma, rhinitis and/or eczema at 16 years. Analyses in relation to sensitization to allergen molecules at 3 years with (persistency) and without (onset) children with any of asthma (A), rhinitis (R) or eczema (E) at 3 years.
Replication population: MAAS is a population based-cohort of 1184 prenatally enrolled children. 248 participants attended all five follow-ups. For each follow-up the attendance rate was: at ages 3 years, (N = 248), 5 years, (N = 588), 8 years (N = 543), 11 years (N = 461) and 16 years (N = 772). Children from whom sera were available at all first three time points were included (Supplementary Fig. 1B) (Custovic et al., 2002, Prosperi et al., 2014).
Ethics committees have approved both studies and parents have provided written informed consent.
2.2. Definitions and Data Sources
Validated questionnaires were used in both studies to ascertain symptoms, physician-diagnosed illnesses and medication use.
2.2.1. Definitions of exposures and outcomes
2.2.1.1. Parental Allergy
Mother and/or father with doctor's diagnosis of asthma and asthma medication and/or doctor's diagnosis of hay fever in combination with furred pets- and/or pollen allergy at the time of the baseline questionnaire.
2.2.1.2. Breast Feeding
Four months or more of exclusive breastfeeding.
2.2.1.3. Second hand tobacco smoke
Either of the parents smoked at least one cigarette per day at the time of the first questionnaire.
2.2.1.4. Socio-economic Status
Assessed using the Nordic standard occupational classification and Swedish socio-economic classification (Occupations in Population and Housing Census 1985 (FoB 85) according to Nordic standard occupational classification (Nordisk yrkesklassificering, NYK) and Swedish socio-economic classification (Socioekonomisk indelning, SEI)1989. Available at: http://www.scb.se/statistik/_publikationer/OV9999_1982A01_ BR_X11%C3%96P8204.pdf. Accessed April 27, 2014.
2.2.1.5. Asthma
Children fulfilled the criteria of asthma at ages one and two years if they had at least three episodes of wheeze in combination with inhaled corticosteroids and/or signs of bronchial hyperreactivity without concurrent upper respiratory infection during the previous 12 months (Wickman et al., 2003). Asthma at age 4, 8, 12 and 16 years was defined as having at least four episodes of wheeze in the previous 12 months or at least one episode of wheeze in combination with occasional or regular use of prescribed inhaled corticosteroids in the last 12 months (Ballardini et al., 2012).
2.2.1.6. Rhinoconjunctivitis
Defined at all ages as “sneezing, runny or blocked nose in the last 12 months without common cold or flu, accompanied by itchy, watery eyes” according to the definition of the International study of asthma and allergies in childhood (ISAAC) (validated in Swedish language) (Asher et al., 2006).
2.2.1.7. Eczema
Defined at all ages as having dry skin in combination with a pruritic rash for two or more weeks with typical localization (face or arm/leg flexures or wrists, ankles or neck) in the past 12 months and/or a doctor's diagnosis of eczema (Bohme et al., 2002).
The definitions were identical for asthma and rhinitis and considered similar for eczema in the two populations. Allergic multimorbidity was defined as simultaneously having asthma and rhinitis. Incident disease was defined as disease reported at eight or 16 years without prior disease. Persistent disease was defined as disease at both 4 and 16 years for BAMSE and at 3 and 16 years for MAAS irrespective of disease at a younger age.
2.3. Allergen-specific IgE Measurements
2.3.1. Multiplex Microarray
For analysis of BAMSE samples, a customized allergen chip based on ISAC technology (Thermo Fisher Scientific, Uppsala, Sweden) that was developed in the FP7-funded European Union project MeDALL (MeDALL Chip) was used at the Medical University of Vienna (EK1641/2014)(Lupinek et al., 2014) and for MAAS, the ImmunoCAP ISAC (Thermo Fisher) was employed (Gadisseur et al., 2011). The same detection systems and cut-off levels of 0.3 ISU (ISAC standardized units) were used in both assays. In the final dataset, 132 allergen molecules from 51 allergen sources were studied in BAMSE and 99 allergen molecules from 44 allergen sources in MAAS.
2.3.2. MeDALL Allergen Chip
Allergy testing was based on venous blood samples drawn at 4, 8 and 16 years. All sera were analyzed anonymously. The MeDALL allergen-chip represents a comprehensive collection of spotted allergen molecules for the detection of allergen-specific antibody signatures (Lupinek et al., 2014). Aliquots of 30 μl serum were incubated on the microarray and after 120 min of incubation at room temperature, slides were washed, and fluorochrome-labeled IgE specific detection antibody (Thermo Fisher) was added and incubated for 30 min at room temperature. Chips were then washed, dried and analyzed using a Laser Scan Confocal microarray reader (LuxScan 10 K/A; Capital-Bio, Beijing, China). The results were evaluated using Phadia Microarray Image Analysis (MIA) software and reported in ISAC Standardized Units for IgE detection (ISU) by using a standard calibration curve. Calibration for IgE has been established for the MeDALL allergen-chip by relating antibody levels to several allergens detected by microarray with the respective results from ImmunoCAP measurements (Lupinek et al., 2014). The cut off was 0.3 ISU-E according to instructions from the manufacturer. In this study we used 132 allergen molecules for analysis corresponding to 41 allergen sources which have recently been described and validated (Lupinek et al., 2014). Allergen molecules from venoms (bee and wasp), anisakis, experimental molecules and glycosylated molecules were excluded (Cabauatan et al., 2014), as were allergen molecules not relevant for the regions. From those allergens where preparations from several different expression systems were spotted, only that preparation which gave results with highest reproducibility was used for analysis.
2.4. Statistical Analysis
Prevalence rates were expressed in percent of the total number of observations available and with 95% confidence intervals. The median IgE concentration and interquartile range (IQR) for each specific allergen component were calculated. Quintile regression was used for detection of changes of IgE over time and for tests of equality of proportions z-tests were used.
We identified clinically relevant allergen molecules responsible for sensitization in at least 90% of all sensitized individuals at each of the three ages. For this procedure IgE-levels ≥ 1 ISU were required in order to reduce misclassification of sensitization. With this approach the number of sensitizing molecules responsible for the cumulative proportion of sensitized individuals increased rather linearly from 55% to just below 90%, and to turn exponential with little additive effect on the overall sensitization rate thereafter (Fig. 1 in the supplementry appendix). In order to identify clinically relevant molecules, hereafter called key allergen molecules, a combined rank of the prevalence and median IgE-level (ISU) for each allergen molecule was constructed separately at the three ages. From the initial 132 molecules 16, 13 and 9 molecules were identified as potential risk molecules at 4, 8 and 16 years, respectively (Table 1A-C in the supplementray appendix). Over the entire time period of 12 years, 18 sensitizing key molecules could be identified in total. Because allergic children are more likely to be sensitized to multiple allergens (Wickman et al., 2014), some of which may be clinically relevant and some only cross-reacting, we wanted to identify risk allergen molecules of clinical relevancy. For this identification we required that the risk allergen molecule should be identified at all three time points (4, 8, 16 years) and should contribute to at least 1% to the cumulative proportion of sensitization.
Odds ratios (ORs) were estimated using multiple logistic regression models and 95% confidence intervals (95% CI). Fitted predicted probability curves according to the levels of specific IgE to different allergen components were plotted using the results from the logistic regression. Previously identified confounders for sensitization and disease development changed the odds ratios by < 10%. Therefore, all analyses were performed unadjusted. Positive and negative predictive values (PPV and NPV) respectively, as well as binomial exact 95% confidence intervals (95% CI) were calculated. A P-value < 0.05 was considered significant. All statistical analyses were performed with the software program STATA 13 (College Station, TX, USA).
3. Results
3.1. Demographic Data
Flow charts of the selection of children from each cohort as well as outcomes of asthma/rhinitis/eczema at 16 years are presented in Fig. 1A–B and Table 1).
Table 1.
Background characteristics at baseline and at 4, 8 and 16 years (BAMSE cohort) and at 3, 5, 8, 11 and 16 years (MAAS cohort) of children in the study base and children in the study population.
| The BAMSE cohort (study base) (N = 4089) |
BAMSE study population (N = 786) |
The MAAS cohort (study base) (N = 1184) |
MAAS study population (N = 848) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | 95% CI | n | % | 95% CI | N | % | 95% CI | n | % | 95% CI | |
| At baseline | ||||||||||||
| Female | 2024 | 49.5 | 48.0–51.0 | 395 | 50.3 | 46.7–53.8 | 542 | 45.8 | 42.9–48.6 | 389 | 45.9 | 42.5–49.2 |
| Parental allergy | 1200 | 29.7 | 28.3–31.1 | 260 | 33.3 | 30.0–36.7 | 444 | 39.1 | 36.3–42.0 | 340 | 41.5 | 38.1–44.9 |
| Breastfed ≥ 4 months | 3116 | 79.5 | 78.2–80.8 | 616 | 79.4 | 76.4–82.2 | 435 | 39.0 | 36.1–41.9 | 339 | 41.5 | 38.2–44.9 |
| Parents smoking | 855 | 21.0 | 19.8–22.3 | 161 | 20.6 | 17.8–23.6 | 328 | 32.0 | 29.1–34.9 | 250 | 31.3 | 28.1–34.5 |
| Young mother at birth (≤ 25 years) | 319 | 7.8 | 7.0–8.7 | 58 | 7.4 | 5.7–9.4 | 154 | 13.8 | 11.8–15.9 | 101 | 12.6 | 10.3–14.9 |
| White collar parent | 695 | 17.3 | 16.1–18.5 | 114 | 14.6 | 12.2–17.3 | 464 | 61.5 | 58.1–65.0 | 357 | 61.4 | 57.5–65.4 |
| Pet (cat and/or dog) at home | 590 | 14.4 | 13.4–15.5 | 115 | 14.6 | 12.2–17.3 | 389 | 33.5 | 30.8–36.3 | 284 | 34.1 | 30.9–37.3 |
| Older siblings | 1980 | 48.4 | 46.9–50.0 | 393 | 50.0 | 46.4–53.6 | 599 | 55.7 | 52.7–58.6 | 464 | 56.0 | 52.6–59.4 |
| At 4 years | At 3 years | |||||||||||
| Asthma | 537 | 14.5 | 13.4–15.7 | 135 | 17.4 | 14.8–20.2 | 155 | 14.4 | 12.3–16.6 | 25 | 10.1 | 6.3–13.9 |
| Rhinitis | 386 | 10.5 | 9.5–11.5 | 89 | 11.4 | 9.3–13.9 | 43 | 3.9 | 1.9–5.8 | 13 | 5.2 | 2.4–8.0 |
| Asthma and/or Rhinitis (AR) | 787 | 21.5 | 20.2–22.9 | 188 | 24.3 | 21.3–27.5 | 184 | 17.1 | 14.9–19.4 | 33 | 13.3 | 9.0–17.6 |
| At 5 years | ||||||||||||
| Asthma | 135 | 17.4 | 14.8–20.2 | 123 | 21.3 | 17.9–24.6 | ||||||
| Rhinitis | 89 | 11.4 | 9.3–13.9 | 156 | 27.2 | 22.6–31.9 | ||||||
| Asthma and/or Rhinitis (AR) | 188 | 24.3 | 21.3–27.5 | 231 | 40.2 | 36.2–44.2 | ||||||
| At 8 years | At 8 years | |||||||||||
| Asthma | 337 | 10.0 | 9.0–11.0 | 94 | 12.0 | 9.8–14.5 | 182 | 17.8 | 15.4–20.1 | 87 | 16.2 | 13.0–19.3 |
| Rhinitis | 453 | 13.3 | 12.2–14.5 | 129 | 16.5 | 13.9–19.3 | 297 | 28.9 | 26.1–31.7 | 139 | 25.9 | 22.2–29.6 |
| Asthma and/or Rhinitis (AR) | 642 | 19.0 | 17.7–20.4 | 176 | 22.6 | 19.7–25.7 | 385 | 37.6 | 34.6–40.5 | 187 | 34.8 | 30.8–38.9 |
| At 11 years | ||||||||||||
| Asthma | 190 | 20.8 | 18.2–23.4 | 99 | 21.7 | 16.9–24.4 | ||||||
| Rhinitis | 314 | 33.8 | 30.2–37.3 | 151 | 32.8 | 28.5–37.1 | ||||||
| Asthma and/or Rhinitis (AR) | 390 | 42.4 | 39.2–45.6 | 187 | 41.0 | 36.5–45.5 | ||||||
| At 16 years | At 16 years | |||||||||||
| Asthma | 342 | 10.8 | 9.8–12.0 | 93 | 12.0 | 9.8–14.5 | 133 | 17.9 | 15.1–20.6 | 68 | 17.7 | 13.9–21.5 |
| Rhinitis | 789 | 25.4 | 23.9–26.9 | 211 | 27.6 | 24.4–30.9 | 309 | 41.2 | 37.7–44.7 | 154 | 40.0 | 35.1–44.9 |
| Asthma and/or Rhinitis (AR) | 918 | 29.6 | 28.0–31.2 | 242 | 31.6 | 28.3–35.0 | 350 | 46.8 | 43.2–50.4 | 177 | 46.0 | 41.0–51.0 |
3.2. Evolution of IgE Reactivity to Allergen Molecules
In BAMSE, 199 (25.3%), 319 (40.6%) and 409 (52.0%) children at 4, 8 and 16 years of age showed IgE reactivity to at least one of the 132 molecules (Fig. 2A). The corresponding numbers in MAAS were 46 (18.6%), 172 (29.4%), 184 (33.9%) and 194 (42.1%) children at 3, 5, 8 and 11 years for at least one of the 99 molecules (Fig. 2B). For children with resolving sensitization, i.e. positive at one time point, but negative (< 0.3 ISU) at the next follow-up for the same allergen molecule, IgE-levels at the first time point was below 1 ISU in 87.7% and 92.9% at 8 and 16 years, respectively.
Fig. 2.
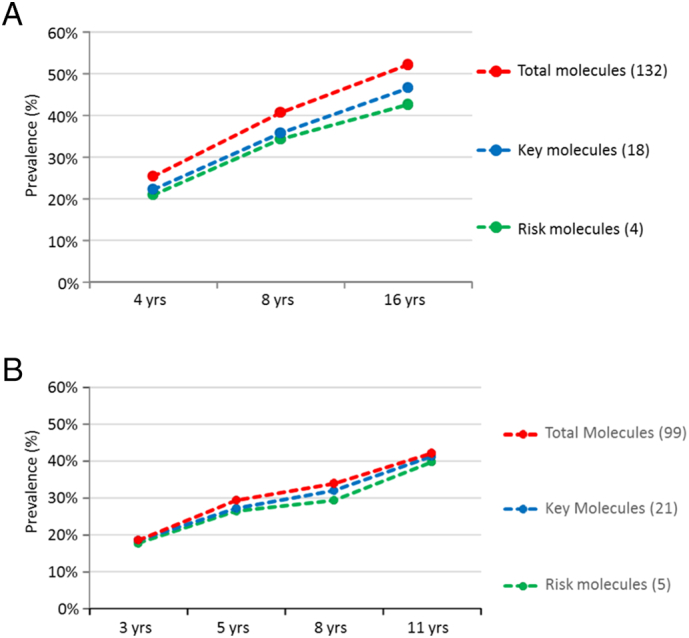
A. (BAMSE). Prevalence of IgE reactivity (%, y-axis) in the population of 786 children at ages 4, 8, 16 years (x-axis) using different sets of molecules at all ages (132), as well as key (18) and risk molecules (4) identified at 4 years. Prevalence indicates IgE reactivity to any of the molecules in each of the three groups. Four risk molecules were identified: Ara h 1 (peanut); Bet v 1 (birch pollen); Fel d 1 (cat) and Phl p 1 (timothy grass pollen). B. (MAAS). Prevalence of IgE reactivity (%, y-axis) in the population of 848 children at ages 3, 5, 8, 11 years (x-axis) using different sets of molecules at all ages (99), as well as key (21) and risk molecules (5) identified at 3 years. Prevalence indicates IgE reactivity to any of the molecules in each of the three groups. Five risk molecules were identified: Der p 1 (Dermatophagoides pteronyssinus); Der f 2 (Dermatophagoides farinae), Phl p 1 and Phl p 5 (timothy grass pollen), and Fel d 1 (cat).
3.3. Identification of IgE to Key and Risk Allergen Molecules
Eighteen different key molecules were identified in BAMSE (Tables 1A–C in the supplementary appendix) and 21 in MAAS (Tables 1 D–G in the supplementary appendix) at the statistical analyses. These molecules belonged to both inhalant and food allergen sources.
In BAMSE, 4 risk molecules were identified among the 18 key molecules: Ara h 1 (peanut); Bet v 1 (birch pollen); Fel d 1 (cat) and Phl p 1 (timothy grass pollen). At 4, 8 and 16 years 83%, 84% and 82% of all sensitized children could be identified by the 4 BAMSE risk molecules (Fig. 2A).
In MAAS, 5 risk molecules among the 21 key molecules were identified: Der p 1 and Der f 2 (house dust mites; Dermatophagoides pteronyssinus/farinae); Phl p 1 and Phl p 5 (timothy grass pollen), and Fel d 1 (cat). At 3, 5, 8 and 11 years 95%, 90%, 86% and 94% of all sensitized children could be identified by the 5 MAAS risk molecules (Fig. 2B).
3.4. IgE to Risk Molecules in Asthma and/or Rhinitis Trajectories
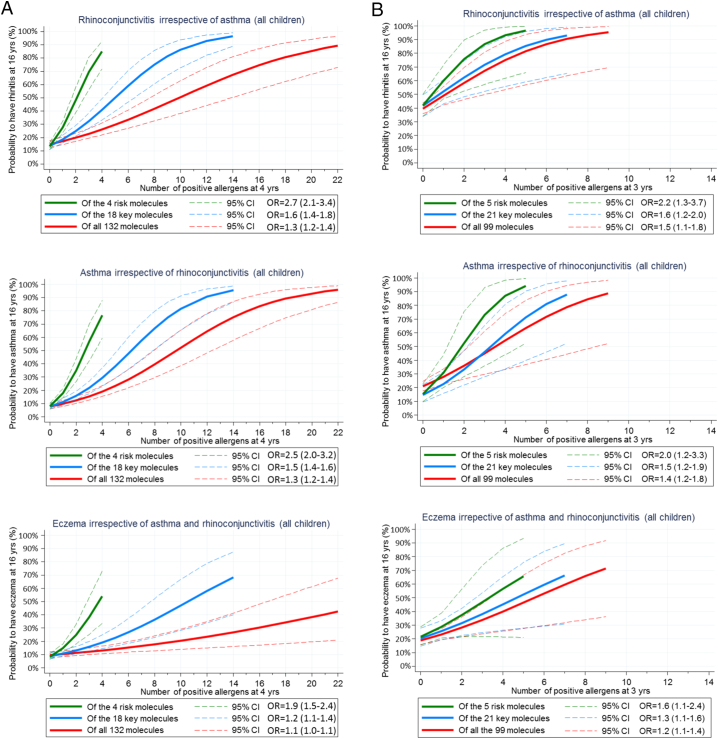
In BAMSE, sensitization to any of the 4 BAMSE risk molecules at 4 years was the strongest predictor of the risk of developing rhinitis, asthma and to some extent eczema until the age of 16 years. Children sensitized to 1, 2, 3 or 4 risk molecules showed a probability of 30%, 50%, 70% and 85% of rhinoconjuctivitis at 16 years, respectively. For key and full panel allergen molecules, children needed to be sensitized to 10 and 19 molecules, respectively, for having an 85% probability of having rhinoconjunctivis at 16 years (Fig. 3A). Results were similar for asthma and, to less extent, for eczema.
Fig. 3.
A. (BAMSE): Probability of rhinitis, asthma or eczema at 16 years in relation to the number of sensitizing risk (green), key (blue) and full panel (red) allergen molecules at 4 years. Probabilities and confidence intervals (hatched lines) were obtained from logistic regression models. Logistic models did not include respiratory allergic disease at 4 or 8 years. B. (MAAS): Probability of rhinitis, asthma or eczema at 11 years in relation to number of sensitizing risk (green), key (blue) and full panel (red) allergen molecules at 3 years. Probabilities and confidence intervals (hatched lines) were obtained from logistic regression models. Logistic models did not include respiratory allergic disease at 3 or 5 years.
In MAAS, children sensitized to any of the 5 MAAS risk molecules at 3 years had a significantly higher risk of developing rhinitis, asthma and, to some extent, eczema at 16 years than those sensitized to any of the 21 key molecules or to any of the total 99 molecules (Fig. 3B). If IgE reactivity was present to 3 of 5 risk molecules at 3 years, the probability at 16 years was 87% for rhinitis and 76% for asthma.
As sensitization at 4 years in BAMSE and at 3 years in MAAS did not affect eczema alone at 16 years (OR = 1.0), further investigation on the impact of early sensitization on allergic multimorbidity was performed for rhinitis and asthma only.
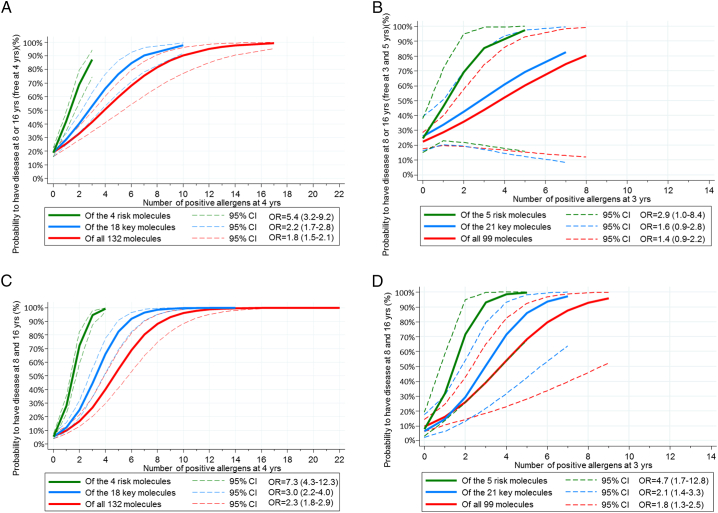
3.5. IgE to Risk Molecules for Allergic Multimorbid Disease
For both rhinitis and asthma, i.e. multimorbid disease, the risk of incident (BAMSE Fig. 4, MAAS Fig. 4B) or persistent (BAMSE Fig. 4C, MAAS Fig. 4D) disease was most significant using the 4-risk molecule approach in BAMSE and the 5-risk molecule approach in MAAS. In BAMSE no children with incident asthma and/or rhinitis at 16 years were found to be sensitized to all 4 risk molecules at 4 years. However, for IgE reactivity to 2 or 3 risk molecules at 4 years the probability of incident asthma and or rhinitis was 69% and 87%, respectively. For key and full panel allergen molecules children needed to be sensitized to 7 and 10 molecules, respectively, to have a 87% probability of having multimorbid respiratory disease at 16 years. For MAAS the corresponding proportions for IgE reactivity at 3–5 years and incident respiratory disease at 8 or 16 was for 2 or 3 risk molecules 57.4% and 66.7% and for 4 of the 5 risk molecules 100%.
Fig. 4.
A. (BAMSE) Probability of allergic multimorbidity (asthma and rhinitis) reported at 8 or 16, but not at 4 years of age. B. (MAAS). Probability of allergic multimorbidity (asthma and rhinitis) reported at 8 or 16, but not at 3 or 5 years of age. C. (BAMSE). Probability of having allergic multimorbidity (asthma and rhinitis), i.e. disease at both 8 and 16 years irrespective of disease at 4 years of age. D. (MAAS). Probability of having allergic multimorbidity (asthma and rhinitis), i.e. disease reported at both 8 and 16, irrespective of disease at 3–5 yrs.
For persistent multimorbid disease in BAMSE, children sensitized to 1, 2, 3 or 4 risk molecules showed a probability of 28%, 72%, 95% and 98%.
For key and full panel allergen molecules children needed to be sensitized to 8 and 12 molecules, respectively, to have a 98% probability of having multimorbid disease at 16 years. The corresponding probabilities for disease at 16 years in relation to IgE reactivity to at least 3 of the 5 risk molecules in MAAS at 3 years were 90% and 100% if sensitized to all 5 (Fig. 4C–D).
The positive predictive value (PPV) for asthma or rhinitis in BAMSE ranged from 73.1% to 94.7% with increasing number of risk molecules at 4 years, p < 0.001 (Table 2A). For the 3 year olds in MAAS 100% PPV was achieved for asthma and/or rhinitis (Table 2B).
Table 2A.
BAMSE: Persistent asthma and/or rhinitis (with any of asthma or rhinitis at 4 yrs. and any of these two at 16 years).
| Number of risk molecules at 4 yrs | Freq. % | TP | FP | TN | FN | Sensitivity, % (95% CI) | Specificity, % (95% CI) | PPV, % (95% CI) | NPV, % (95% CI) |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 69.5 | ||||||||
| 1 | 13.0 | 19 | 7 | 84 | 55 | 25.7 (16.2–37.2) | 92.3 (84.8–96.9) | 73.1 (52.2–88.4) | 60.4 (51.8–68.6) |
| 2 | 8.0 | 14 | 2 | 84 | 55 | 20.3 (11.6–31.7) | 97.7 (91.9–99.7) | 87.5 (61.7–98.4) | 60.4 (51.8–68.6) |
| 3–4 | 9.5 | 18 | 1 | 84 | 55 | 24.7 (15.3–36.1) | 98.8 (93.6–100.0) | 94.7 (74.0–99.9) | 60.4 (51.8–68.6) |
TP, True Positive; FP, False Positive; TN, True Negative; FN, False Negative; PPV, Positive Predictive Value; NPV, Negative Predictive Value.
Table 2B.
MAAS: Persistent asthma and/or rhinitis (with any of asthma or rhinitis at 3 yrs. and any of these two at 16 years).
| Number of risk molecules at 3 yrs | Freq.(%) | TP | FP | TN | FN | Sensitivity (%) |
Specificity (%) |
PPV |
NPV |
|---|---|---|---|---|---|---|---|---|---|
| (95% CI) | (95% CI) | (95% CI) | (95% CI) | ||||||
| 0 | 57.9 | ||||||||
| 1 | 21.1 | 4 | 0 | 4 | 7 | 36.4 (12.4–68.4) | 100 (39.6–100) | 100 (100 − 100) | 36.4 (12.4–68.4) |
| 2 | 15.8 | 3 | 0 | 4 | 7 | 30.0 (8.1–64.6) | 100 (39.6–100) | 100 (100–100) | 36.4 (12.4–68.4) |
| 3–5 | 5.3 | 1 | 0 | 4 | 7 | 12.5 (0.7–53.3) | 100 (39.6–100) | 100 (100–100) | 36.4 (12.4–68.4) |
TP, True Positive; FP, False Positive; TN, True Negative; FN, False Negative; PPV, Positive Predictive Value; NPV, Negative Predictive Value.
4. Discussion
This is the largest study to date where children from a population-based birth cohort (BAMSE) have been analyzed with 132 allergen molecules from early childhood to adolescence, corresponding to > 300,000 single IgE tests to individual allergen molecules. The study was replicated in a comparable birth cohort (MAAS) using 99 of the BAMSE allergen molecules. The results reveal that the detection of IgE against a handful of allergen molecules in early childhood provides a strong molecular signature to predict respiratory allergy until adolescence in two geographically different populations. We were able to predict incident or persistent asthma and rhinitis with > 90% probability in both studies by IgE reactivity to the risk molecules at age 3–5 years, albeit with broad confidence intervals. Interestingly, the risk for incident or persistent multimorbid disease was even more significant than for single disease. To our knowledge, our study is the first to show this association in two different cohorts with different exposure backgrounds.
We used an investigator driven strategy, with a priori defined criteria for key and risk allergen molecules associated with allergic disease. Using this approach, it was possible to obtain a detailed pattern of the IgE recognition repertoire and link the molecular responses to clinical symptoms in both cohorts. We identified key allergen molecules accounting for 90% of IgE reactivities at each of the studied time points (18/132 in BAMSE and 21/99 in MAAS) and risk molecules (4 in BAMSE and 5 in MAAS) associated with development of allergic disease. These risk molecules were the major allergens from birch and timothy grass pollen, as well as those from cat and peanut in the Swedish setting, whereas the major allergens of dust mites, grass and cat were recognized in the UK environment.
4.1. Strengths and Limitations
A major strength of both birth cohorts is their design to primarily study the development of allergic disease in childhood up to adolescence using questionnaire and serological data serially available in a large number of children at similar ages both for detection of IgE reactivity and outcome. In addition, nearly the same panel of allergen molecules was used for both cohorts. Furthermore, using cohorts from two different geographical locations, with a total number of 1034 children generating similar conclusions, suggests the results to be generalizable across populations. A limitation could be that IgE measurements on the chip are performed under conditions of low amounts of allergen immobilized to the solid phase, which may be affected by allergen-specific IgG antibodies (Lupinek et al., 2016). However, the MeDALL allergen-chip has been carefully evaluated with respect to specificity and sensitivity and was found to be more sensitive and specific for detecting IgE sensitization than allergen-extract-based skin prick testing and conventional serology (Skrindo et al., 2015). Some risk molecules reflected region-specific differences in exposure. The major role of birch pollen in Sweden is quite obvious due to the large diffusion of birch trees, while for dust mites previous reports showed low prevalence of sensitization to these species (Wickman et al., 2014, Rönmark et al., 2003). In other regions, sensitization to one of these or other allergen molecules might appear and disappear depending on exposure.
4.2. Clinical Implication of Risk Allergen Molecules for Prediction of Onset and Persistency of Allergic Airways Disease in Preschool Children
Sensitization to common food and inhalant allergen sources during the first year of life has been identified as a risk factor for development of asthma and rhinitis at 6 years and for asthma in adults, but it has been unclear how many and what allergen molecules in these allergen sources are involved, thus making the use of IgE sensitization difficult to predict the development and persistence of respiratory allergic disease. Furthermore, previous studies have not assessed IgE to a combination of allergens in relation to development or prediction of disease (Brockow et al., 2009, Rhodes et al., 2001).
Our study is the first to show that indeed IgE reactivity to only few allergen molecules in early childhood predicts respiratory allergy in adolescence. The major allergens of cat (Fel d 1) and timothy grass pollen (Phl p 1) were identified as risk allergens in both the BAMSE and MAAS cohort. In fact, we have recently shown that Fel d 1 is not only a marker of present but also a prognostic marker of future cat allergy (Asarnoj et al., 2015). Furthermore, Phl p 1 has been identified as a molecule in the beginning of grass pollen allergy (Hatzler et al., 2012). The results of the present study also confirm that the major birch pollen allergen (Bet v 1) is a risk allergen in Sweden while the major allergens of dust mites (Der p 1, and Der f 2) are risk allergens in the UK and in Germany (Posa et al., 2017). However, it was unexpected to find the peanut storage protein Ara h 1 as a risk allergen in BAMSE since it represents a typical food allergen. However, it has been suggested that sensitization toward food allergens may also occur via the skin or the respiratory system (Roberts and Lack, 2003, Brough et al., 2013). The results of the two birth cohorts, where IgE reactivities to allergen molecules from more than forty allergen sources were analyzed, indicate that a few allergen molecules are sufficient to provide a strong molecular signature to predict the development and persistence of allergic respiratory disease. Previous efforts to predict asthma development during childhood have shown limited capacity to predict which children with wheeze in early childhood will develop asthma at a later age (Smit et al., 2015). Based on our results it may therefore be possible to develop individualized risk prediction charts for allergic respiratory diseases. Our results indicate the possibility to develop allergen-specific forms of preventive immunotherapy. Using approaches based on recombinant allergen derivatives or synthetic allergen-derived peptide vaccines that are composed of the identified risk allergen molecules could be designed (Valenta et al., 2012, Zieglmayer et al., 2016, Valenta et al., 2017, Campana et al., 2017).
The predictive values of IgE reactivity to increasing numbers of risk molecules for incidence and persistence of asthma and/or rhinitis were similar in the two cohorts indicating the generalizability of our data. Since other risk factors could not be found in the analysis and the risk molecules are linked to the environments in Sweden and the UK, our data suggest that at early age there is a window of plasticity and with an opportunity to modulate the immune system to escape disease development based on few allergen molecules. A very limited proportion of children with IgE reactivity to a specific allergen molecule at a certain time point had IgE below the cutoff at the next follow up. Almost all of these children had at the preceding time point an allergen-specific IgE level below 1 ISU. This was shown for the majority of food allergens molecules. The results are in line with previous reports where children with low allergen-specific IgE levels have an increased tendency to outgrow their allergies (Boyano-Martinez et al., 2002). However, our data imply that for the majority of children with IgE reactivity to common allergens few become desensitized.
In conclusion, IgE reactivity early in life to a few risk allergen molecules, from birch and timothy grass pollen as well as from cat and peanut, to 95% predicts persistence of allergic airway disease up to adolescence in a Swedish setting. Replication of data from the MAAS birth cohort revealed similar results with a partially overlapping set of few risk molecules related to differences in allergen exposure. Thus, only a few allergen molecules are of importance for predicting onset and persistency of respiratory allergic diseases and should be the focus for preventive strategies and targets for novel therapies. The findings are of clinical importance for pediatricians or physicians seeing children at a young age, who could perform early allergy diagnosis with the key allergen molecules to initiate preventive measures.
Contributors
MW, CL, RV and MvH designed the study. CL and RV measured IgE reactivity to allergen molecules in BAMSE. NA and AA performed the statistical analyses in BAMSE and DB in MAAS. Leadership and coordination for the specific cohorts and projects were provided by MW, GP, AS, IK, AB, EM, and JB and JMA for MeDALL (BAMSE) and AC for MAAS. MW, CL, AA, CH, RV and MvH drafted the report with input from all authors. All authors approved the final version of the paper.
Declaration of Interests
CL has recieved personal fees from Thermo Fischer Scientific. AA has received personal fees from Thermo Fisher Scientific and MEDA. RV has recieved personal fees from Biomay AG, Vienna, Austria and Viravaxx AG, Vienna, Austria. MvH has received personal fees from Thermo Fisher Scientific, Biomay AG, Vienna, Austria and Hycor Biomedical LLC, CA, US. The other authors declare no competing interests.
Funding
This study was supported by research grants from the Swedish Asthma and Allergy Foundation, Stockholm County Council (ALF Medicin), Swedish Research Council, the Swedish Heart-Lung Foundation, the Swedish Cancer and Allergy Foundation, the King Gustaf V 80th Birthday Foundation, the Hesselman Foundation, The Konsul Th C Bergh Foundation, the Magnus Bergvall Foundation, Working Life and Welfare, grant F4605 of the Austrian Science Fund (FWF) and by the European Commission's Seventh Framework 29 Program MeDALL under grant agreement No. 261357. Danielle Belgrave is supported through the Medical Research Council Grant MR/M015181/1.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2017.11.009.
Appendix A. Supplementary data
Supplementary table 1 A-G and supplementary figure 1.
References
- Asarnoj A., Hamsten C., Waden K. Sensitization to cat and dog allergen molecules in childhood and prediction of symptoms of cat and dog allergy in adolescence: a BAMSE/MeDALL study. J. Allergy Clin. Immunol. 2016;137(3):813–821. doi: 10.1016/j.jaci.2015.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher M.I., Montefort S., Bjorksten B. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC phases one and three repeat multicountry cross-sectional surveys. Lancet. 2006;368(9537):733–743. doi: 10.1016/S0140-6736(06)69283-0. [DOI] [PubMed] [Google Scholar]
- Ballardini N., Kull I., Lind T. Development and comorbidity of eczema, asthma and rhinitis to age 12: data from the BAMSE birth cohort. Allergy. 2012;67(4):537–544. doi: 10.1111/j.1398-9995.2012.02786.x. [DOI] [PubMed] [Google Scholar]
- Belgrave D.C., Granell R., Simpson A. Developmental profiles of eczema, wheeze, and rhinitis: two population-based birth cohort studies. PLoS Med. 2014;11(10):e1001748. doi: 10.1371/journal.pmed.1001748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohme M., Lannero E., Wickman M., Nordvall S.L., Wahlgren C.F. Atopic dermatitis and concomitant disease patterns in children up to two years of age. Acta Derm. Venereol. 2002;82(2):98–103. doi: 10.1080/00015550252948112. [DOI] [PubMed] [Google Scholar]
- Bousquet J., Gern J.E., Martinez F.D. Birth cohorts in asthma and allergic diseases: report of a NIAID/NHLBI/MeDALL joint workshop. J. Allergy Clin. Immunol. 2014;133(6):1535–1546. doi: 10.1016/j.jaci.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet J., Anto J.M., Akdis M. Paving the way of systems biology and precision medicine in allergic diseases: the MeDALL success story: mechanisms of the development of ALLergy; EU FP7-CP-IP; project no: 261357; 2010-2015. Allergy. 2016;71(11):1513–1525. doi: 10.1111/all.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyano-Martinez T., Garcia-Ara C., Diaz-Pena J.M., Martin-Esteban M. Prediction of tolerance on the basis of quantification of egg white-specific IgE antibodies in children with egg allergy. J. Allergy Clin. Immunol. 2002;110(2):304–309. doi: 10.1067/mai.2002.126081. [DOI] [PubMed] [Google Scholar]
- Brockow I., Zutavern A., Hoffmann U. Early allergic sensitizations and their relevance to atopic diseases in children aged 6 years: results of the GINI study. J Investig Allergol Clin Immunol. 2009;19(3):180–187. [PubMed] [Google Scholar]
- Brough H.A., Santos A.F., Makinson K. Peanut protein in household dust is related to household peanut consumption and is biologically active. J. Allergy Clin. Immunol. 2013;132(3):630–638. doi: 10.1016/j.jaci.2013.02.034. [DOI] [PubMed] [Google Scholar]
- Cabauatan C.R., Lupinek C., Scheiblhofer S. Allergen microarray detects high prevalence of asymptomatic IgE sensitizations to tropical pollen-derived carbohydrates. J. Allergy Clin. Immunol. 2014;133(3):910–914. doi: 10.1016/j.jaci.2013.10.004. (e5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campana R., Huang H.J., Freidl R. Recombinant allergen and peptide-based approaches for allergy prevention by oral tolerance. Semin. Immunol. 2017;30:67–80. doi: 10.1016/j.smim.2017.08.017. [DOI] [PubMed] [Google Scholar]
- Canonica G.W., Ansotegui I.J., Pawankar R. A WAO - ARIA - GA(2)LEN consensus document on molecular-based allergy diagnostics. World Aller. Organ. J. 2013;6(1):17. doi: 10.1186/1939-4551-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C.Y., Huang Y.L., Tsai M.H. Sensitization to food and inhalant allergens in relation to atopic diseases in early childhood: a birth cohort study. PLoS One. 2014;9(7) doi: 10.1371/journal.pone.0102809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custovic A., Simpson B.M., Murray C.S. The National Asthma Campaign Manchester Asthma and Allergy Study. Pediatr. Allergy Immunol. 2002;13(Suppl. 15):32–37. doi: 10.1034/j.1399-3038.13.s.15.3.x. [DOI] [PubMed] [Google Scholar]
- Gadisseur R., Chapelle J.P., Cavalier E.A. New tool in the field of in-vitro diagnosis of allergy: preliminary results in the comparison of ImmunoCAP(c) 250 with the ImmunoCAP(c) ISAC. Clin. Chem. Lab. Med. 2011;49(2):277–280. doi: 10.1515/CCLM.2011.052. [DOI] [PubMed] [Google Scholar]
- Hatzler L., Panetta V., Lau S. Molecular spreading and predictive value of preclinical IgE response to Phleum pratense in children with hay fever. J. Allergy Clin. Immunol. 2012;130(4):894–901. doi: 10.1016/j.jaci.2012.05.053. (e5) [DOI] [PubMed] [Google Scholar]
- Hiller R., Laffer S., Harwanegg C. Microarrayed allergen molecules: diagnostic gatekeepers for allergy treatment. FASEB J. 2002;16(3):414–416. doi: 10.1096/fj.01-0711fje. [DOI] [PubMed] [Google Scholar]
- Illi S., von Mutius E., Lau S. The pattern of atopic sensitization is associated with the development of asthma in childhood. J. Allergy Clin. Immunol. 2001;108(5):709–714. doi: 10.1067/mai.2001.118786. [DOI] [PubMed] [Google Scholar]
- Illi S., von Mutius E., Lau S. The natural course of atopic dermatitis from birth to age 7 years and the association with asthma. J. Allergy Clin. Immunol. 2004;113(5):925–931. doi: 10.1016/j.jaci.2004.01.778. [DOI] [PubMed] [Google Scholar]
- Lupinek C., Wollmann E., Baar A. Advances in allergen-microarray technology for diagnosis and monitoring of allergy: the MeDALL allergen-chip. Methods. 2014;66(1):106–119. doi: 10.1016/j.ymeth.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupinek C., Wollmann E., Valenta R. Monitoring allergen immunotherapy effects by microarray. Curr. Treat Options Allergy. 2016;3:189–203. doi: 10.1007/s40521-016-0084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melioli G., Passalacqua G., Canonica G.W., Baena-Cagnani C.E., Matricardi P. Component-resolved diagnosis in pediatric allergic rhinoconjunctivitis and asthma. Curr. Opin. Allergy Clin. Immunol. 2013;13(4):446–451. doi: 10.1097/ACI.0b013e32836274d8. [DOI] [PubMed] [Google Scholar]
- Patelis A., Gunnbjornsdottir M., Malinovschi A. Population-based study of multiplexed IgE sensitization in relation to asthma, exhaled nitric oxide, and bronchial responsiveness. J. Allergy Clin. Immunol. 2012;130(2):397–402. doi: 10.1016/j.jaci.2012.03.046. (e2) [DOI] [PubMed] [Google Scholar]
- Posa D., Perna S., Resch Y. Evolution and predictive value of IgE responses toward a comprehensive panel of house dust mite allergens during the first 2 decades of life. J. Allergy Clin. Immunol. 2017;139(2):541–549. doi: 10.1016/j.jaci.2016.08.014. [DOI] [PubMed] [Google Scholar]
- Prosperi M.C., Belgrave D., Buchan I., Simpson A., Custovic A. Challenges in interpreting allergen microarrays in relation to clinical symptoms: a machine learning approach. Pediatr. Allergy Immunol. 2014;25(1):71–79. doi: 10.1111/pai.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranciere F., Nikasinovic L., Bousquet J., Momas I. Onset and persistence of respiratory/allergic symptoms in preschoolers: new insights from the PARIS birth cohort. Allergy. 2013;68(9):1158–1167. doi: 10.1111/all.12208. [DOI] [PubMed] [Google Scholar]
- Rhodes H.L., Sporik R., Thomas P., Holgate S.T., Cogswell J.J. Early life risk factors for adult asthma: a birth cohort study of subjects at risk. J. Allergy Clin. Immunol. 2001;108(5):720–725. doi: 10.1067/mai.2001.119151. [DOI] [PubMed] [Google Scholar]
- Roberts G., Lack G. Food allergy and asthma--what is the link? Paediatr. Respir. Rev. 2003;4(3):205–212. doi: 10.1016/s1526-0542(03)00058-7. [DOI] [PubMed] [Google Scholar]
- Rönmark E., Perzanowski M., Platts-Mills T., Lundbäck B. Obstructive lung disease in northern Sweden study G. Four-year incidence of allergic sensitization among schoolchildren in a community where allergy to cat and dog dominates sensitization: report from the obstructive lung disease in northern Sweden study group. J. Allergy Clin. Immunol. 2003;112(4):747–754. doi: 10.1016/S0091. [DOI] [PubMed] [Google Scholar]
- Skrindo I., Lupinek C., Valenta R. The use of the MeDALL-chip to assess IgE sensitization: a new diagnostic tool for allergic disease? Pediatr. Allergy Immunol. 2015;26(3):239–246. doi: 10.1111/pai.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit H.A., Pinart M., Anto J.M. Childhood asthma prediction models: a systematic review. Lancet Respir. Med. 2015;3(12):973–984. doi: 10.1016/S2213-2600(15)00428-2. [DOI] [PubMed] [Google Scholar]
- Valenta R., Campana R., Marth K., van Hage M. Allergen-specific immunotherapy: from therapeutic vaccines to prophylactic approaches. J. Intern. Med. 2012;272(2):144–157. doi: 10.1111/j.1365-2796.2012.02556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenta R., Campana R., Niederberger V. Recombinant allergy vaccines based on allergen-derived B cell epitopes. Immunol Lett. 2017;189:19–26. doi: 10.1016/j.imlet.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hage M., Hamsten C., Valenta R. ImmunoCAP assays: Pros and cons in allergology. J. Allergy Clin. Immunol. 2017;140(4):974–977. doi: 10.1016/j.jaci.2017.05.008. [DOI] [PubMed] [Google Scholar]
- Westman M., Lupinek C., Bousquet J. Early childhood IgE reactivity to pathogenesis-related class 10 proteins predicts allergic rhinitis in adolescence. J. Allergy Clin. Immunol. 2015;135(5):1199–1206. doi: 10.1016/j.jaci.2014.10.042. e1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westman M., Asarnoj A., Hamsten C., Wickman M., van Hage M. Windows of opportunity for tolerance induction for allergy by studying the evolution of allergic sensitization in birth cohorts. Semin. Immunol. 2017;30:61–66. doi: 10.1016/j.smim.2017.07.005. [DOI] [PubMed] [Google Scholar]
- Wickman M., Kull I., Pershagen G., Nordvall S.L. The BAMSE project: presentation of a prospective longitudinal birth cohort study. Pediatr. Allergy Immunol. 2002;13(Suppl. 15):11–13. doi: 10.1034/j.1399-3038.13.s.15.10.x. [DOI] [PubMed] [Google Scholar]
- Wickman M., Melen E., Berglind N. Strategies for preventing wheezing and asthma in small children. Allergy. 2003;58(8):742–747. doi: 10.1034/j.1398-9995.2003.00078.x. [DOI] [PubMed] [Google Scholar]
- Wickman M., Asarnoj A., Tillander H. Childhood-to-adolescence evolution of IgE antibodies to pollens and plant foods in the BAMSE cohort. J. Allergy Clin. Immunol. 2014;133(2):580–582. doi: 10.1016/j.jaci.2013.09.009. [DOI] [PubMed] [Google Scholar]
- Zieglmayer P., Focke-Tejkl M., Schmutz R. Mechanisms, safety and efficacy of a B cell epitope-based vaccine for immunotherapy of grass pollen allergy. EBioMedicine. 2016;11:43–57. doi: 10.1016/j.ebiom.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table 1 A-G and supplementary figure 1.