Abstract
Corosolic acid (CRA) is found in various plants and has been used as a health food supplement worldwide. Although it has been reported that CRA exhibits significant anticancer activity, the effect of this compound on prostate cancer remains unknown. In this study, we investigated the effect of CRA on cellular transformation and the reactivation of nuclear factor erythroid 2-related factor 2 (Nrf2) through epigenetic regulation in TRAMP-C1 prostate cells. Specifically, we found that CRA inhibited anchorage-independent growth of prostate cancer TRAMP-C1 cells but not Nrf2 knockout prostate cancer TRAMP-C1 cells. Moreover, CRA induced mRNA and protein expression of Nrf2, heme oxygenase-1 (HO-1) and quinone oxidoreductase-1 (NQO1). Bisulfate genomic sequencing and methylated DNA immunoprecipitation results revealed that CRA treatment decreased the level of methylation of the first five CpG sites of the Nrf2 promoter. Histone modification was analyzed using a chromatin immunoprecipitation (ChIP) assay, which revealed that CRA treatment increased the acetylation of histone H3 lysine 27 (H3K27ac) while decreasing the trimethylation of histone H3 lysine 27 (H3K27me3) in the promoter region of Nrf2. Furthermore, CRA treatment attenuated the protein expression of DNA methyltransferases (DNMTs) and histone deacetylases (HDACs). These findings indicate that CRA has a significant anticancer effect in TRAMP-C1 cells, which could be partly attributed to epigenetics including its ability to epigenetically restore the expression of Nrf2.
Keywords: Corosolic acid, Nrf2, Methylation, HDAC, DNMT
Introduction
In the United States, prostate cancer (PCa) is the most prevalent subtype of non-cutaneous male cancers and one of the leading causes of cancer-related death in men. In 2016, approximately 180,890 new PCa cases were diagnosed and 26,120 PCa deaths were estimated to have occurred in the USA [1]. Similar to most cancers, PCa is a genetic disease in which genetic events, including germline and somatic mutations, occur during cancer progression [2,3]. Moreover, accumulating data have revealed that diverse epigenetic changes significantly contribute to PCa development [2,3]. The major epigenetic processes that regulate gene expression are DNA methylation, alterations in the chromatin structure due to the post-translational modification of histones and certain non-histone proteins, and microRNAs. Epigenetics has attracted much attention because epigenetic interventions may reverse aberrant epigenetic modifications, and epigenetic therapy has been hypothesized to be a viable strategy for treating cancer. Many studies have attempted to develop inhibitors of epigenetic mediators, such as DNA methyltransferase (DNMT), histone deacetylase (HDAC), enhancer of zeste homolog 2, DOT1-like histone H3K79 methyltransferase, and bromodomain and extraterminal domain proteins.
Oxidative stress is the overproduction or imbalanced production of reactive oxygen species (ROS), which significantly influence cancer cell survival, proliferation, invasion, angiogenesis, apoptosis and chemo- and radio-therapy resistance [4]. To prevent the DNA damage and mutagenic events caused by oxidative stress, cells have evolved an elaborate and powerful cellular defense machinery against ROS, and the transcription factor nuclear factor erythroid 2-related factor 2 (Nrf2) is central to this cellular defensive machinery. Specifically, the activation of Nrf2 signaling induces the antioxidant response element (ARE)-dependent expression of antioxidative stress proteins, such as quinone oxidoreductase-1 (NQO1), the antioxidant protein heme oxygenase-1 (HO-1), and glutathione peroxidases (Gpx), which protects cells from oxidative stress and prevents carcinogenesis. Nrf2-deficient mice have proven to be more susceptible to carcinogen-induced tumorigenesis [5,6,7], and our previous works have shown that the expression levels of Nrf2 and its downstream genes are downregulated during the development of prostate tumors in TRAMP mice [8,9], highlighting the crucial role of Nrf2 in carcinogenesis. The primary mechanism of Nrf2 activation is based on the interaction between Keap1 and Nrf2. However, the transcriptional activity of Nrf2 is regulated by a complex signaling network, and other molecular mechanisms are also involved. Among these mechanisms, the epigenetic regulation of Nrf2 is frequently observed in cancer. We have found that the expression of Nrf2 is suppressed by promoter CpG methylation in TRAMP prostate tumors, TRAMP-C1 cells [10, 11] and JB6 P+ cells [12]. Moreover, treatment with chemopreventive compounds such as Z-ligustilide [13], sulforaphane [14], and curcumin [15] restored the expression of Nrf2 by inducing DNA demethylation, which demonstrates that targeting the epigenetic regulation of Nrf2 may be a viable strategy in the development of therapeutic agents for cancer.
Corosolic acid (CRA, Figure 1A), a triterpenoid also known as 2α-hydroxyursolic acid, is found in various plants, including Schisandra chinensis, Eriobotrya japonica, Lagerstroemia speciosa L., Orthosiphon stamineus and Weigela subsessilis [16]. It has been demonstrated that CRA has significant anti-diabetes [17], anti-obesity [18] and anti-atherosclerosis activities [19], and CRA also exhibits anticancer activities against various cancer cell lines. Specifically, CRA induces apoptosis in HCT 116 cells by activating caspases [16]. This compound also enhances the anticancer activity of 5-fluorouracil via mTOR inhibition in SNU-620 human gastric carcinoma cells [20] and inhibits hepatocellular carcinoma cell migration by targeting the VEGFR2/Src/FAK pathway [21]. Although many studies have focused on the potential anticancer activity of CRA [16,20,21], the mechanisms of CRA action are not yet fully understood. To date, the effect of CRA on epigenetic modification in prostate cancer has not yet been explored. In this study, we investigated the ability of CRA to restore the expression of Nrf2 via epigenetic modification in TRAMP-C1 cells.
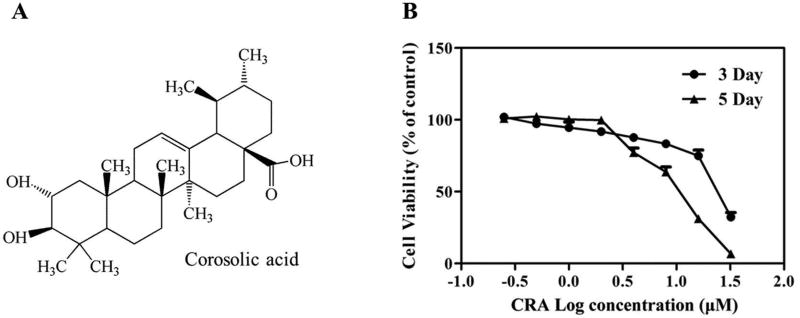
Figure 1. Chemical structure of corosolic acid (CRA, A) and the cytotoxicity of CRA to TRAMP-C1 cells (B).
Cells were treated with various concentrations of CRA for 3 or 5 days. The MTS assay was performed to assess cell viability. The data are presented as the mean ± SEM.
Materials and Methods
Materials
CRA (≥ 98% in purity) was purchased from Chendu Biopurify Medical Technology Co., Ltd. (Chendu, China; 13120404). 5-Aza-2′-deoxycytidine (5-AZA), trichostatin A (TSA), ampicillin, bovine serum albumin (BSA), and a protein inhibitor cocktail were purchased from Sigma-Aldrich (St. Louis, MO, USA). Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), and trypsin-EDTA (0.25%) were purchased from Gibco (Carlsbad, CA, USA). The anti-Nrf2, anti-HO-1, and anti-NQO-1 antibodies were purchased from Abcam (Cambridge, MA, USA). Anti-DNMT1, anti-DNMT3a and anti-DNMT3b antibodies were supplied by Novus Biologicals (Littleton, CO, USA). The antibodies against HDACs (HDAC1, HDAC2, HDAC4, HDAC6 and HDAC7) were purchased from Cell Signaling Technology (Boston, MA, USA). The anti-HDAC8 antibody was obtained from Proteintech Group (Chicago, IL, USA). The anti-β-actin primary antibody and secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Cell culture, treatment and lentiviral transduction
TRAMP-C1 cells were maintained in DMEM containing 10% FBS at 37°C in a humidified atmosphere of 5% CO2, as described in our previous publications [13, 14, 15]. Cells were seeded in 10-cm dishes at a density of 2 × 105 cells/dish, incubated for 24 h and then treated with either 0.1% DMSO, different concentrations of CRA, or 5-AZA (250 nM) in DMEM containing 1% FBS. The medium was exchanged every 2 days. For the 5-AZA and TSA combination treatment, TSA (100 nM) was added to the 5-AZA-containing medium on day 4. The cells were harvested on day 5 for DNA, protein or total RNA extraction.
Lentivirus mediated short hairpin RNAs were used to establish stable mock (scramble control, sh-Mock) and Nrf2 knockdown (sh-Nrf2) TRAMP-C1 cells. The shRNA clone sets were obtained from Genecopoeia (Rockville, MD, USA), and lentiviral transduction was performed according to the manufacturer’s manual. After selection in DMEM medium supplemented with 10% FBS and 2 µg/mL puromycin for 3 weeks, the sh-Mock and sh-Nrf2 cells were further used to evaluate the functional roles of Nrf2.
Cell viability assay
TRAMP-C1 cells were seeded in 96-well plates (1,000 cells/well). After 24 h, the cells were treated with various concentrations of CRA ranging from 0.25 µM to 32 µM. On day 3 or 5, the CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS) (Promega, Madison, WI, USA) was used to assess cell viability.
Colony formation assay
The colony formation assay was performed as previously described [22]. Briefly, sh-Mock and sh-Nrf2 cells were treated with 0.1% DMSO, either 2, 4, or 8 µM CRA for 5 days. On day 5, the pretreated cells were transferred to 1 mL of BME containing 0.33% agar, which was layered over 3 mL of BME containing 0.5% agar and 10% FBS in 6-well plates. The cells were maintained in soft agar at 37°C in a humidified atmosphere with 5% CO2 for an additional 14 days. Colonies were photographed using a computerized microscope system with the Nikon ACT-1 program (Version 2.20) and counted using ImageJ software (Version 1.48d; NIH).
RNA extraction and quantitative real-time PCR
Total RNA was extracted from cells using the GeneJET RNA Purification Kit (ThermoFisher Scientific, Waltham, MA, USA), and cDNA was synthesized from the total RNA using the SuperScript III First-Strand Synthesis System (Invitrogen, Grand Island, NY, USA) according to the manufacturer’s instructions. To assess the RNA expression of specific genes, the cDNA was used as the template for real-time PCR using Power SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA) with β-actin serving as an internal control.
The following primers were used for real-time PCR: Nrf2, 5’-AGCAGGACTGGAGAAGTT-3’ (sense) and 5’-TTCTTTTTCCAGCGAGGAGA-3’ (antisense); HO-1, 5’-CCTCACTGGCAGGAAATCATC-3’ (sense) and 5’-CCTCG TGGAGACGCTTTACATA-3’ (antisense); and NQO1, 5’-AGCCCAGATATTGTGGCC G-3’ (sense) and 5’-CTTTCAGAATGGCTGGCAC-3’ (antisense); β-actin was used as an internal control with sense (5’-CGTTCAATACCCCAGCCATG-3’) and antisense (5’-ACCCCGTCACCAGAGTCC-3’) primers.
DNA extraction and bisulfite genomic sequencing (BGS)
Total DNA was extracted from TRAMP-C1 cells using the GeneJET DNA Purification Kit (ThermoFisher Scientific). The bisulfite conversion of genomic DNA was performed using the EZ DNA Methylation-Gold Kit following the manufacturer's instructions (Zymo Research, Orange, CA, USA). The bisulfite-modified DNA was amplified by PCR using Platinum Taq DNA polymerase (Invitrogen, Grand Island, NY) and primers that amplify the first 5 CpGs located between −1085 bp and −1226 bp of the murine Nrf2 gene, with the translation start site defined as +1. The primer sequences were 5’-AGTTATGAAGTAGTAGTAAAAA-3’ (sense) and 5’-AATATAATCTCATAAAACCCCAC-3’ (antisense). The PCR product was cloned into the pCR4 TOPO vector using a TOPO™TA Cloning Kit (Invitrogen, Carlsbad, CA). Plasmids from at least ten colonies of each group were prepared using the QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA) and sequenced (Genwiz, Piscataway, NJ).
Methylated DNA immunoprecipitation (MeDIP) analysis
To confirm the results of the BGS, MeDIP was performed using the Methylamp Methylated DNA Capture (MeDIP) Kit (Epigentek, Farmingdale, NY, USA), as described in our previous work [14]. Briefly, DNA extracted from treated cells was sonicated using a Bioruptor sonicator (Diagenode Inc., Sparta, NJ, USA) to shear the DNA into lengths of 200–1000 bp. The sonicated DNA was incubated at 95°C for 2 min, followed by immunoprecipitation with an anti-5-methylcytosine antibody at room temperature for 2 h. After washing and purification, the methylation status was quantified via a qPCR assay using the primer set 5’-GAGGTCACCACAACACGAAC-3’ (forward) and 5’-ATCTCATAAGGCCCCACCTC-3’ (reverse) to cover the DNA sequence of the first five CpGs of murine Nrf2. The enrichment of methylated DNA in each treatment was calculated according to the standard curve of the serial diluted input DNA. The relative ratios of methylated DNA were then calculated based on the control, which was defined as 100% DNA methylation.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed using the MAGnify™ Chromatin Immunoprecipitation system according to manufacturer’s protocol (ThermoFisher Scientific, Waltham, MA, USA). Briefly, the chromatin extracted from cells was fixed with 1% formaldehyde and sonicated to generate fragments of 200–500 bp using a Bioruptor sonicator (Diagenode Inc., Sparta, NJ, USA). The sonicated solutions were incubated with protein A/G Dynabeads and a specific antibody against H3K27AC or H3K27me3 (Abcam, Cambridge, MA, USA) or rabbit IgG overnight at 4°C. After reverse cross-linking, the DNA was purified using the ChIP DNA Clean Concentrator (Zymo Research) according to the manufacturer’s instructions. The enrichment of the diluted DNA was quantified via a qPCR assay using the following primers: 5’-ACCCTGGGTCAACAAGTC-3’ (forward) and 5’-TGCCTGTGCTTTTCATAACG-3’ (reverse).
Western blot analysis
After washing with PBS, treated cells were harvested with trypsin and lysed using radio-immunoprecipitation assay buffer (Cell Signaling Technology, Boston, MA, USA) containing a protein inhibitor cocktail. The concentrations of the protein extracts were measured using a bicinchoninic acid assay (Pierce, Rockford, IL, USA). Equal amounts of protein were resolved using SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad), and then blocked at room temperature for 2 h. For immunoblotting, the membranes were incubated with the respective primary antibodies at 4°C overnight, followed by a 2-h incubation with the secondary antibody at room temperature. The bands were visualized using SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher Scientific) and were analyzed using ImageJ version 1.48d.
HDAC and DNMT activity
DNMT activity and HDAC activity assays were conducted using a DNMT activity/inhibition assay kit and an HDAC activity/inhibition assay kit (Epigentek, Farmingdale, NY). The nuclear protein was isolated using the NEPER Nuclear and Cytoplasmic Protein Extraction Kit (Thermo Scientific, Pittsburgh, PA). The relative DNMT and HDAC activities were calculated based on the ratio of the CRA treatment group to the control group after normalization to the protein amount.
Statistical analysis
All analyses were performed using SPSS software, version SPSS19.0 (Chicago, IL, USA). The data are presented as mean ± standard error (SEM). The statistical analyses were conducted using a one-way analysis of variance (ANOVA) or Student’s t-test. P values less than 0.05 were considered statistically significant.
Results
Cytotoxicity of CRA against TRAMP-C1 cells
To investigate the effect of CRA on the viability of TRAMP-C1 cells, cells were treated with CRA for 3 and 5 days, and the growth of the cells was then measured using an MTS assay. The results (Figure 1B) showed that CRA treatment decreased cell viability in a time- and dose-dependent manner after 3 and 5 days of treatment. Similar results were also observed in LNCaP cells (Figure S1A).
Knockdown of Nrf2 attenuated the inhibitory effect of CRA on anchorage independent growth of TRAMP-C1 cells
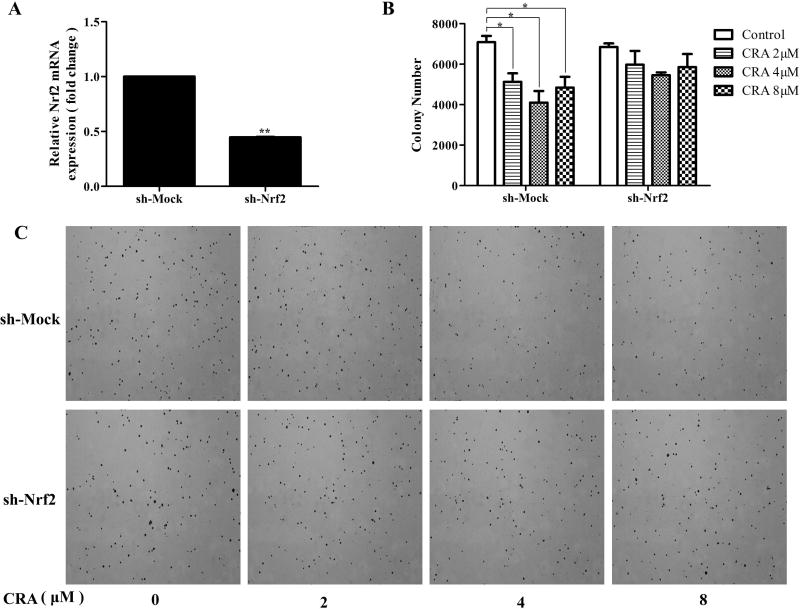
The anchorage-independent growth capacity of cells reflects their tumorigenicity. Nrf2 acts as a sensor of oxidative or electrophilic stress and prevents genome instability. To test the effect of CRA on the anchorage-independent growth of TRAMP-C1 cells and investigate the role of Nrf2 in this process, sh-Mock (wild-type) and sh-Nrf2 (knockdown) cell lines were established using lentivirus shRNA vectors. qPCR confirmed Nrf2 knockdown in stably transduced cells as compared with control cells (Figure 2A). CRA at the concentrations of 2, 4 and 8 µM significantly suppressed colony formation of sh-Mock cells by 27.6%, 42.1% and 31.7%, respectively (Figure 2B and 2C), indicating that this compound has great potential of decreasing tumorigenicity. By contrast, the CRA-mediated inhibitory effect on colony formation of sh-Nrf2 cells was reduced to around 12.7% to 20.3% (Figure 2B–C). These results suggest that Nrf2 played an important role in the CRA-mediated suppression of anchorage-independent growth of TRAMP-C1 cells.
Figure 2. Nrf2 knockdown attenuated the inhibitory effects of CRA on the anchorage-independent growth of TRAMP-C1 cells.
Stable mock (scramble-sequence control, sh-Mock) and Nrf2 knockdown (sh-Nrf2) TRAMP-C1 cells were established using lentivirus mediated short hairpin RNAs and were selected with puromycin for 3 weeks. (A) Reduced mRNA expression of Nrf2 in knockdown cells was confirmed by qPCR. (B) Anchorage-independent growth of sh-Mock and sh-Nrf2 with or without the presence of CRA for 14 days. (C) Representative images of each group under a microscope. All the data are presented as the mean ± SEM. *P < 0.05 compared to the control.
CRA increased the mRNA and protein expression levels of Nrf2 and its downstream genes
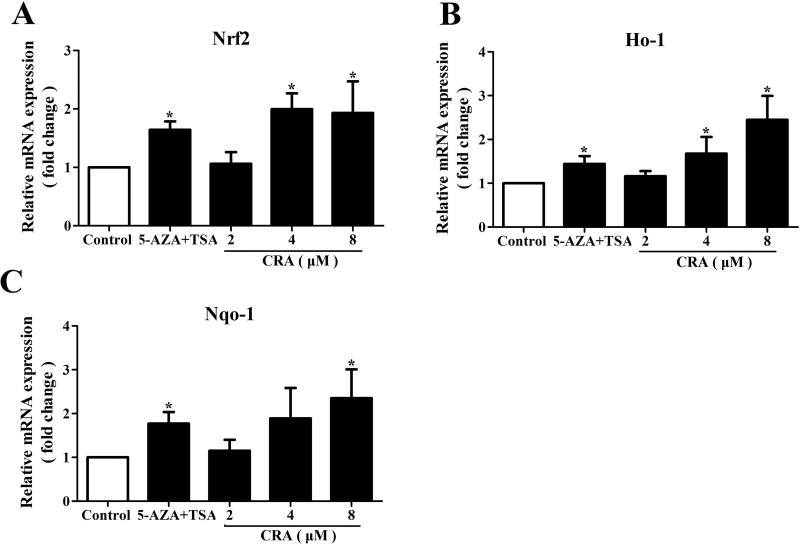
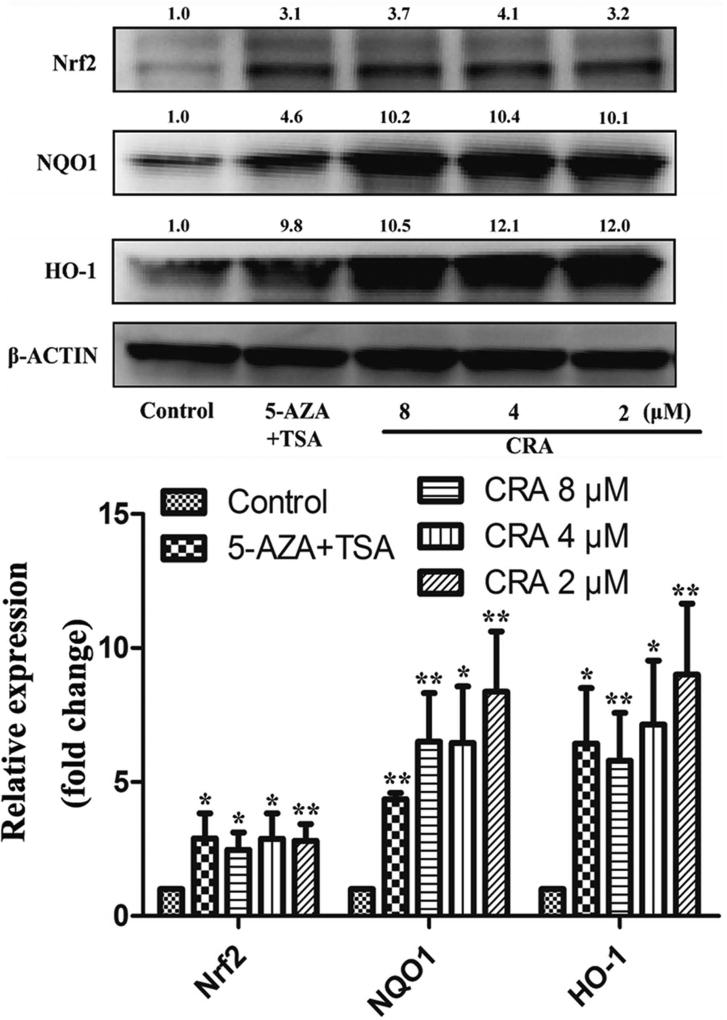
Nrf2 plays a crucial role during oxidative stress in cells. Previous studies have proven that the expression levels of Nrf2 and its downstream genes are decreased in TRAMP prostate cancer [8,9]. In this study, qPCR assays were applied to investigate the effect of CRA on the transcriptional levels of Nrf2 and its downstream genes. The results indicated that CRA significantly increased the level of Nrf2 mRNA expression (P < 0.05) and induced HO-1 and NQO1 mRNA expression (P < 0.05, Figure 3). Similar results were also observed in LNCaP cells (Figure S1B–D). The protein levels of Nrf2, HO-1 and NQO1 in TRAMP-C1 cells treated with CRA were evaluated via western blot analysis. In accordance with the qPCR results, CRA increased the protein expression of Nrf2, HO-1 and NQO1 in a dose-dependent manner at concentrations ranging from 2 µM to 8 µM (Figure 4).
Figure 3. CRA increased the mRNA expression levels of Nrf2 and Nrf2 target genes in TRAMP-C1 cells.
TRAMP-C1 cells were treated with CRA (2, 4 and 8 µM) for 5 days. After treatment, mRNA was isolated from the TRAMP-C1 cells, and the expression levels of target genes were determined using real-time qPCR assays with β-actin as an internal control. The graphical data are expressed as the mean ± SEM from three independent experiments. *P < 0.05 compared to the control.
Figure 4. CRA increased the expression levels of Nrf2, HO-1, and NQO1 in TRAMP-C1 cells.
TRAMP-C1 cells were treated with CRA (2, 4 and 8 µM) for 5 days. After treatment, the cells were lysed, and the expression levels of proteins were determined by western blot analysis. The relative protein levels in the treatment groups were quantified and compared to that of the control group. β-actin was used as an internal control to normalize loading. The data are presented as the mean ± SEM from 3 independent experiments. *P < 0.05 compared to the control.
CRA decreased CpG methylation in the promoter region of Nrf2
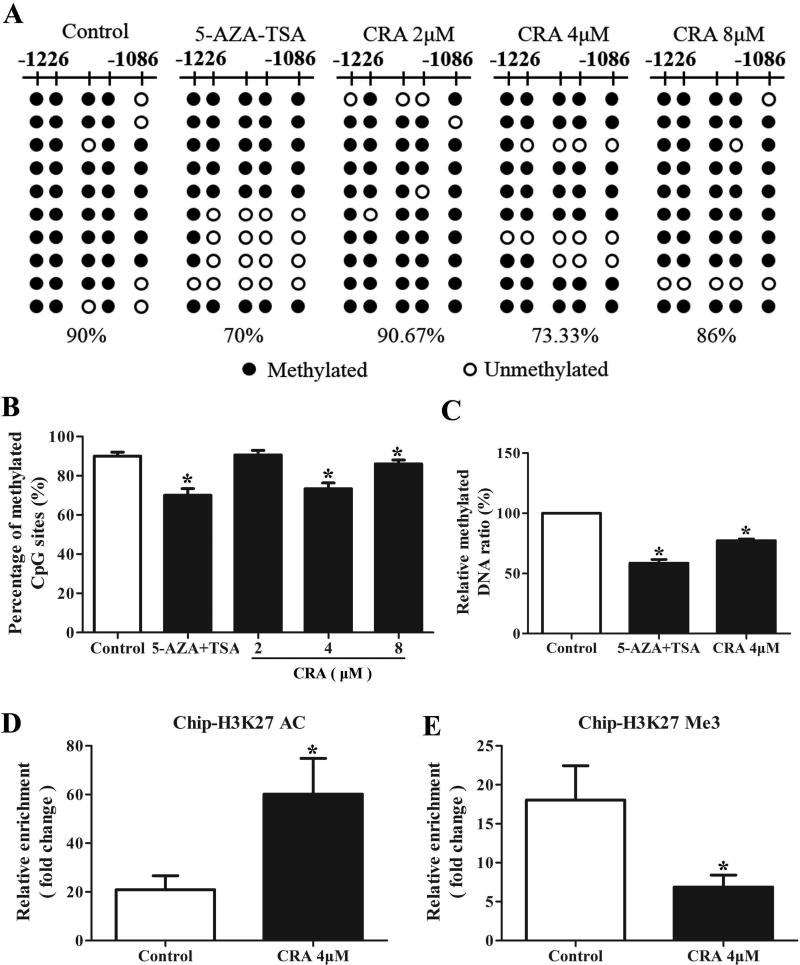
Our previous report identified the first five CpG sites in the promoter of Nrf2 as being hypermethylated in the TRAMP-C1 cell line [14, 15, 23]. In this study, we examined whether CRA could reverse the methylation in the promoter region of Nrf2 using BGS and MeDIP assays. As shown in Figure 5A and B, the BGS results revealed a hypermethylated region in the promoter of Nrf2 in TRAMP-C1 cells, and 90% of the CpG sites were methylated. Notably, after treatment with 4 µM CRA or AZA plus TSA (positive control) for 5 days, the percentage of methylated CpG sites significantly decreased to 75% or 70%, respectively (P < 0.05, one-way ANOVA). However, there was no significant change in the methylation of CpG sites when the cells were treated with 2 and 8 µM CRA. These results indicate that CRA can reverse the CpG methylation status of the Nrf2 promoter, which may contribute to the ability of this drug to increase Nrf2 expression.
Figure 5. CRA altered the DNA methylation of the Nrf2 promoter and regulated its histone modification in TRAMP-C1 cells.
TRAMP-C1 cells were treated with various concentrations of CRA or 5-AZA/TSA for 5 days. The analyzed CpG sites were located at positions −1085 bp to −1226 bp in the promoter region of Nrf2. (A and B) BGS was used to analyze DNA methylation. Black spots denote methylated CpGs, and hollow dots denote unmethylated CpGs. (C) An MeDIP assay was used to estimate the enrichment of methylated DNA fragments captured by the anti-5-methylcytosine antibody. A ChIP assay was used to assess the enrichment of H3K27AC (D) and H3K27Me3 (E) at the promoter of Nrf2. The graphical data are expressed as the mean ± SEM. *P < 0.05 compared to the control.
To verify the BGS results, an MeDIP assay was performed, which uses an anti-5-methylcytosine antibody to capture methylated genomic DNA without bisulfite conversion. The enriched methylated fractions were then measured using a qPCR assay. As shown in Figure 5C, the ratio of methylated DNA relative to the control was significantly reduced in the 4 µM CRA or the combination of 5-AZA/TSA treatment groups (P < 0.05, one-way ANOVA). Taken together, these data indicate that 4 µM CRA can decrease CpG methylation in the promoter region of Nrf2 in TRAMP-C1 cells.
CRA increased the acetylation of histone H3 lysine 27 (H3K27ac) and decreased the trimethylation of histone H3 lysine 27 (H3K27Me3) in the promoter region of Nrf2
In addition to DNA methylation, histone modification is also a critical process in the regulation of gene expression. In this study, a ChIP analysis was performed to investigate the enrichment of H3K27ac and H3K27Me3 at the promoter of Nrf2 in TRAMP-C1 cells. Compared to the control, H3K27ac enrichment was increased after treatment with 4 µM CRA (Figure 5D), while the association of H3K27Me3 with the promoter of Nrf2 was decreased, as shown in Figure 5E (P < 0.05, Student's t-test).
CRA downregulated the expressions and activities of epigenetically modifying enzymes in TRAMP-C1 cells
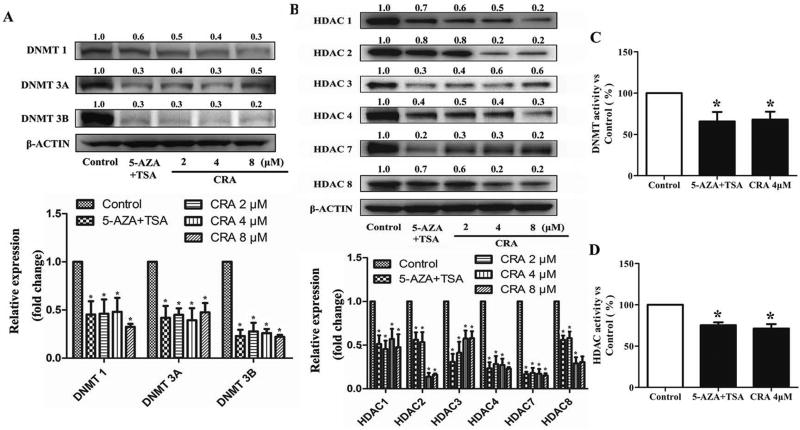
To explore the potential epigenetic mechanism by which CRA decreases Nrf2 promoter methylation and increases the Nrf2 level, we examined the effect of CRA on the expression of epigenetically modifying enzymes. DNA methylation is catalyzed by enzymes known as DNMTs, including DNMT1, DNMT3a, and DNMT3b. As shown in Figure 6A, CRA treatment decreased the protein levels of DNMT1, DNMT3a and DNMT3b in a dose-dependent manner in TRAMP-C1 cells (P < 0.05, one-way ANOVA).
Figure 6. CRA decreased the protein levels and activities of DNMTs (A and C) and HDACs (B and D).
DNMT and HDAC protein levels were determined by a western blot analysis. The relative expression was calculated according to the intensity of each band normalized to β-actin. *P < 0.05 compared to the control.
Moreover, HDACs are well-known enzymes that dynamically catalyze histone deacetylation. Treatment with CRA significantly reduced the protein levels of HDAC1, HDAC2, HDAC3, HDAC4, HDAC7 and HDAC8 (Figure 6B, P < 0.05, one-way ANOVA) in TRAMP-C1 cells. With respect to DNMT and HDAC activities, CRA at the dose of 4 µM significantly decreased the DNMT and HDAC activities (Figures 6C–D, *P < 0.05).
Discussion
PCa is one of the most common non-cutaneous malignancies among men worldwide [1]. The development and formation of a prostate carcinoma from a preneoplastic lesion to androgen-independent invasive carcinoma usually occurs over several decades [2], allowing the possibility of using chemopreventive agents to delay this process and reduce the morbidity of PCa. Many natural phytochemicals, such as curcumin [15], sulforaphane [14] and Z-ligustilide [13], have been shown to have cancer chemopreventive activity regarding PCa. CRA, a triterpenoid, has been identified in many Chinese medicinal herbs and has been used as an anti-diabetes agent in many food supplementary products [24]. In addition to the excellent anti-diabetic activity of this compound, cytotoxic activity against several human cancer cell lines has also been reported for CRA [25]. Specifically, CRA induced mitochondria-mediated and caspase-dependent apoptosis in A549 cells [26] and also showed anticancer activity against colon cancer cells by promoting the degradation of β-catenin [27]. In our study, CRA significantly inhibited the proliferation of TRAMP-C1 cells. Moreover, this compound also significantly inhibited sh-mock TRAMP-C1 cell colony formation, demonstrating the ability of CRA to prevent prostate carcinogenesis in vitro.
Oxidative stress, which results from a perturbation in the production and removal of ROS, may damage DNA and proteins, thereby disturbing cellular processes [28]. To respond to oxidative stress, organisms create a highly reducing intracellular environment by regulating phase II antioxidant and detoxification enzymes, such as HO-1 (antioxidant), NQO1, and UGT1A1 (detoxification) [29]. It is widely accepted that the transcription of these enzymes is regulated by the ARE [29, 30]. Nrf2 is a transcription factor that binds to the ARE and mediates ARE-dependent gene activation. Therefore, Nrf2 plays a crucial role in the oxidative stress response and its related diseases [3, 4, 30, 31, 33]. The expression of Nrf2 is decreased in human prostate cancer [32] and in TRAMP mice [10]. Many natural compounds, such as Z-ligustilide [13], sulforaphane [14], and curcumin [15], may inhibit prostate tumorigenesis by enhancing the expression of Nrf2, suggesting that Nrf2 could be a target in the development of cancer therapeutic agents. To further investigate the role of Nrf2 in mediating CRA’s inhibitory effects on colony formation, we generated stable Nrf2 knockdown TRAMP C1 cells via shRNA expression through lentiviral transduction. Inhibition of anchorage-independent growth by CRA was considerably attenuated by the ablation of Nrf2 expression in TRAMP-C1 cells (Figure 2B–C). Hence, we conclude that Nrf2 plays an important role in CRA-mediated inhibition of colony formation in TRAMP-C1 cells. CRA also enhanced the mRNA and protein expression of Nrf2 and its downstream genes. These results suggest that the inhibitory effect of CRA on prostate cancer cells at least partly through the modulation of Nrf2 pathway.
Epigenetic aberrations, including changes in DNA methylation patterns or histone modifications, are well known as key drivers of prostate carcinogenesis [2]. Our previous studies demonstrated that Nrf2 is epigenetically silenced by the hypermethylation of the first five CpG sites in TRAMP mice and TRAMP-C1 cells [8, 9, 10, 11], which suggests that the epigenetic modulation of Nrf2 is likely a critical mechanism for Nrf2 activation. It has been reported ursolic acid can activate Nrf2 in JB6 P+ cells [34]. Because the structure of CRA is similar to that of ursolic acid, we hypothesized that epigenetic modifications may contribute to the CRA-induced activation of Nrf2. In the bisulfite sequencing test, we found that CRA treatment (4 and 8 µM) caused the demethylation of the first 5 CpGs in the Nrf2 promoter in TRAMP-C1 cells (Figure 5A). This hypomethylation was confirmed by the MeDIP assay, which showed that the ratio of methylated DNA of the first 5 CpGs in the Nrf2 promoter was lower in the TRAMP-C1 cells treated with CRA (4 µM) than in the control cells. These results suggest that CRA may modulate DNA methylation, which is implicated in carcinogenesis. Furthermore, DNA methylation is a covalent chemical modification that is catalyzed by DNMT [35]. We found that CRA decreased the protein levels of DNMT1, DNMT3a and DNMT3b, which may be an underlying mechanism for the CRA-induced demethylation of Nrf2.
In addition to DNA methylation, histone modifications have also been implicated in prostate carcinogenesis [36, 37]. The N-terminal tails of histones can undergo a variety of post-translational covalent modifications, including acetylation, methylation, phosphorylation, ubiquitination, and glycosylation [38]. In general, histone acetylation, which is regulated by HDACs, correlates with transcriptional activation. In PCa, HDACs 1, 2 and 3 are strongly expressed and may correlate with increased proliferative capacity [38]. Therefore, the discovery of selective HDAC inhibitors has had significant implications for cancer therapy [36, 37]. Notably, in our study, CRA significantly decreased the protein levels of HDAC1, HDAC3, HDAC4, HDAC5 and HDAC6 in TRAMP-C1 cells. Moreover, CRA increased the enrichment of H3K27AC at the promoter of Nrf2. In contrast to histone acetylation, histone methylation on arginine and lysine may be associated with either gene activation or repression [36, 37, 39]. Many studies have characterized H3K27me3 as an epigenetic feature of histone methylation-related prostate cancer progression [40, 41]. In our study, CRA decreased the enrichment of H3K27Me3 at the promoter of Nrf2. Taken together, these results imply that CRA prevents DNA hypermethylation by regulating DNMTs and HDACs, unlike 5-AZA and TSA, which specifically inhibit only DNA methylation and histone deacetylation.
Prostate carcinogenesis is a complicated process regulated by an interlinked network of many genes. Aberrant alterations of the methylation patterns are common features of PCa development and progression [35]. More than 100 genes such as GSTP1, RARB2, RASSF1A, APC, COX2, MDR1, TIG1, HIC1 have been shown to be inactivated by promoter hypermethylation in PCa [35]. In our study, there was a significant decrease in the protein levels of DMNTs and HDACs after CRA treatment, suggesting the possibility that CRA might also change the methylation patterns of other genes besides Nrf2. However, further studies are needed to investigate the effects of CRA on the methylation of other genes.
In conclusion, our findings revealed that CRA can inhibit the growth and colony formation of TRAMP-C1 cells and increase the mRNA and protein expression levels of Nrf2 and its downstream phase II detoxifying and antioxidant enzymes, including HO-1 and NQO1. We also demonstrated for the first time that CRA restores the silenced Nrf2 gene via the demethylation of the Nrf2 promoter in TRAMP-C1 cells, and this alteration is mediated by the inhibition of DNMTs and HDACs. Collectively, our data provide new insight into the function of CRA as an epigenetic regulator for the prevention of prostate cancer.
Supplementary Material
Acknowledgments
We thank all the members of Dr. Kong’s laboratory for their helpful discussions and the preparation of this manuscript.
Funding information
This work was supported in part by institutional funds and by R01 AT009152 from the National Center for Complementary and Integrative Health (NCCIH).
Abbreviations
- CRA
corosolic acid
- ChIP
chromatin immunoprecipitation
- H3K27ac
acetylation of histone H3 lysine 27
- H3K27me3
trimethylation of histone H3 lysine 27
- PCa
prostate cancer
- ROS
reactive oxygen species
- Nrf2
nuclear factor erythroid 2-related factor 2
- NQO1
quinone oxidoreductase-1
- HO-1
heme oxygenase-1
- Gpx
glutathione peroxidases
- DNMT
DNA methyltransferase
- HDAC
histone deacetylase
- MeDIP
methylated DNA immunoprecipitation.
Footnotes
Conflict of Interest
The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Kgatle MM, Kalla AA, Islam MM, Sathekge M, Moorad R. Prostate cancer: epigenetic alterations, risk factors, and therapy. Prostate Cancer. 2016;5653862 doi: 10.1155/2016/5653862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumgart SJ, Haendler B. Exploiting epigenetic alterations in prostate cancer. Int J Mol Sci. 2017;18:1017. doi: 10.3390/ijms18051017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gào X, Schöttker B. Reduction–oxidation pathways involved in cancer development: a systematic review of literature reviews. Oncotarget. 2017 doi: 10.18632/oncotarget.17128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khor TO, Huang MT, Kwon KH, Chan JY, et al. Nrf2-deficient mice have an increased susceptibility to dextran sulfate sodium-induced colitis. Cancer Res. 2006;66:11580–11584. doi: 10.1158/0008-5472.CAN-06-3562. [DOI] [PubMed] [Google Scholar]
- 6.Saw CL, Yang AY, Huang MT, Liu Y, et al. Nrf2 null enhances UVB-induced skin inflammation and extracellular matrix damages. Cell Biosci. 2014;4:39. doi: 10.1186/2045-3701-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khor TO, Huang MT, Prawan A, Liu Y, et al. Increased susceptibility of Nrf2 knockout mice to colitis-associated colorectal cancer. Cancer Prev Res. 2008;1:187–191. doi: 10.1158/1940-6207.CAPR-08-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barve A, Khor TO, Hao X, Keum YS, et al. Murine prostate cancer inhibition by dietary phytochemicals--curcumin and phenyethylisothiocyanate. Pharm Res-dordr. 2008;25:2181–2189. doi: 10.1007/s11095-008-9574-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barve A, Khor TO, Nair S, Reuhl K, et al. Gamma-tocopherol-enriched mixed tocopherol diet inhibits prostate carcinogenesis in TRAMP mice. Int J Cancer. 2009;124:1693–1699. doi: 10.1002/ijc.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu S, Khor TO, Cheung KL, Li W, et al. Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PloS One. 2010;5:e8579. doi: 10.1371/journal.pone.0008579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu TY, Khor TO, Su ZY, Saw CL, et al. Epigenetic modifications of Nrf2 by 3,3'-diindolylmethane in vitro in TRAMP C1 cell line and in vivo TRAMP prostate tumors. AAPS J. 2013;15:864–874. doi: 10.1208/s12248-013-9493-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Zhang C, Guo Y, Su ZY, et al. Blocking of JB6 cell transformation by tanshinone IIA: epigenetic reactivation of Nrf2 antioxidative stress pathway. AAPS J. 2014;16:1214–1225. doi: 10.1208/s12248-014-9666-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su ZY, Khor TO, Shu L, Lee JH, et al. Epigenetic reactivation of Nrf2 in murine prostate cancer TRAMP C1 cells by natural phytochemicals Z-ligustilide and radix angelica sinensis via promoter CpG demethylation. Chem Res Toxicol. 2013;26:477–485. doi: 10.1021/tx300524p. [DOI] [PubMed] [Google Scholar]
- 14.Zhang C, Su ZY, Khor TO, Shu L, Kong ANT. Sulforaphane enhances Nrf2 expression in prostate cancer TRAMP C1 cells through epigenetic regulation. Biochem Pharmacol. 2013;85:1398–1404. doi: 10.1016/j.bcp.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khor TO, Huang Y, Wu TY, Shu L, et al. Pharmacodynamics of curcumin as DNA hypomethylation agent in restoring the expression of Nrf2 via promoter CpGs demethylation. Biochem Pharmacol. 2011;82:1073–1078. doi: 10.1016/j.bcp.2011.07.065. [DOI] [PubMed] [Google Scholar]
- 16.Sung B, Kang YJ, Kim DH, Hwang SY, et al. Corosolic acid induces apoptotic cell death in HCT116 human colon cancer cells through a caspase-dependent pathway. Int J Mol Med. 2014;33:943–949. doi: 10.3892/ijmm.2014.1639. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Li JJ, Wen XD, Pan R, et al. Metabonomic analysis of the therapeutic effect of Potentilla discolor in the treatment of type 2 diabetes mellitus. Mol BioSyst. 2014;10:2898–2906. doi: 10.1039/c4mb00278d. [DOI] [PubMed] [Google Scholar]
- 18.Yang J, Leng J, Li JJ, Tang JF, et al. Corosolic acid inhibits adipose tissue inflammation and ameliorates insulin resistance via AMPK activation in high-fat fed mice. Phytomedicine. 2016;23:181–190. doi: 10.1016/j.phymed.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 19.Chen H, Yang J, Zhang Q, Chen LH, Wang Q. Corosolic acid ameliorates atherosclerosis in apolipoprotein E-deficient mice by regulating the nuclear factor-κB signaling pathway and inhibiting monocyte chemoattractant protein-1 expression. Circ J. 2012;76:995–1003. doi: 10.1253/circj.cj-11-0344. [DOI] [PubMed] [Google Scholar]
- 20.Lee HS, Park JB, Lee MS, Cha EY, et al. Corosolic acid enhances 5-fluorouracil-induced apoptosis against SNU-620 human gastric carcinoma cells by inhibition of mammalian target of rapamycin. Mol Med Rep. 2015;12:4782–4788. doi: 10.3892/mmr.2015.3982. [DOI] [PubMed] [Google Scholar]
- 21.Ku CY, Wang YR, Lin HY, Lu SC, Lin JY. Corosolic acid inhibits hepatocellular carcinoma cell migration by targeting the VEGFR2/Src/FAK pathway. PloS One. 2015;10:e0126725. doi: 10.1371/journal.pone.0126725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo Y, Shu L, Zhang C, Su ZY, Kong AN. Curcumin inhibits anchorage-independent growth of HT29 human colon cancer cells by targeting epigenetic restoration of the tumor suppressor gene DLEC1. Biochem Pharmacol. 2015;94:69–78. doi: 10.1016/j.bcp.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li W, Pung D, Su ZY, Guo Y, et al. Epigenetics reactivation of Nrf2 in prostate TRAMP C1 cells by curcumin analogue FN1. Chem Res Toxicol. 2016;29:694–703. doi: 10.1021/acs.chemrestox.6b00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stohs SJ, Miller H, Kaats GR. A review of the efficacy and safety of banaba (Lagerstroemia speciosa L.) and corosolic acid. Phytother Res PTR. 2012;26:317–324. doi: 10.1002/ptr.3664. [DOI] [PubMed] [Google Scholar]
- 25.Cheng QL, Li HL, Li YC, Liu ZW, et al. CRA(Crosolic Acid) isolated from Actinidia valvata Dunn.Radix induces apoptosis of human gastric cancer cell line BGC823 in vitro via down-regulation of the NF-kappaB pathway. Food Chem Toxicol. 2017;105:475–485. doi: 10.1016/j.fct.2017.05.021. [DOI] [PubMed] [Google Scholar]
- 26.Nho KJ, Chun JM, Kim HK. Corosolic acid induces apoptotic cell death in human lung adenocarcinoma A549 cells in vitro. Food Chem Toxicol. 2013;56:8–17. doi: 10.1016/j.fct.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 27.Kim JH, Kim YH, Song GY, Kim DE, et al. Ursolic acid and its natural derivative corosolic acid suppress the proliferation of APC-mutated colon cancer cells through promotion of beta-catenin degradation. Food Chem Toxicol. 2014;67:87–95. doi: 10.1016/j.fct.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 28.Udensi UK, Tchounwou PB. Oxidative stress in prostate hyperplasia and carcinogenesis. J Exp Clin Cancer Res CR. 2016;35:139. doi: 10.1186/s13046-016-0418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J, Keum YS. NRF2, a Key Regulator of antioxidants with two faces towards cancer. Oxid Med Cell Longe. 2016;2746457 doi: 10.1155/2016/2746457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Y, Yu S, Zhang C, Kong AN. Epigenetic regulation of Keap1-Nrf2 signaling. Free Radical Biol Med. 2015;88:337–349. doi: 10.1016/j.freeradbiomed.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taguchi K, Yamamoto M. The KEAP1-NRF2 system in cancer. Frontiers in oncology. 2017;7:85. doi: 10.3389/fonc.2017.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frohlich DA, McCabe MT, Arnold RS, Day ML. The role of Nrf2 in increased reactive oxygen species and DNA damage in prostate tumorigenesis. Oncogene. 2008;27:4353–4362. doi: 10.1038/onc.2008.79. [DOI] [PubMed] [Google Scholar]
- 33.Fan H, Paiboonrungruan C, Zhang X, Prigge JR, Schmidt EE, Sun Z, Chen X. Nrf2 regulates cellular behaviors and notch signaling in oral squamous cell carcinoma cells. Biochem Biophys Res Commum. 2017;493:833–839. doi: 10.1016/j.bbrc.2017.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim H, Ramirez CN, Su ZY, Kong AN. Epigenetic modifications of triterpenoid ursolic acid in activating Nrf2 and blocking cellular transformation of mouse epidermal cells. J Nutr Biochem. 2016;33:54–62. doi: 10.1016/j.jnutbio.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graca I, Pereira-Silva E, Henrique R, Packham G, et al. Epigenetic modulators as therapeutic targets in prostate cancer. Clin Epigenetics. 2016;8:98. doi: 10.1186/s13148-016-0264-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo Y, Su ZY, Kong AT. Current perspectives on epigenetic modifications by dietary chemopreventive and herbal phytochemicals. Current Pharmacology Reports. 2015;1:245–257. doi: 10.1007/s40495-015-0023-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Z, Wang L, Wang Q, Li W. Histone modifications and chromatin organization in prostate cancer. Epigenomics. 2010;2:551–560. doi: 10.2217/epi.10.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schulz WA, Hatina J. Epigenetics of prostate cancer: beyond DNA methylation. J Cell Mol Med. 2006;10:100–125. doi: 10.1111/j.1582-4934.2006.tb00293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Y, Wu R, Su ZY, Guo Y, Zheng X, Yang CS, Kong AN. A naturally occurring mixture of tocotrienols inhibits the growth of human prostate tumor, associated with epigenetic modifications of cyclin-dependent kinase inhibitors p21 and p27. J Nutr Biochem. 2017;40:155–163. doi: 10.1016/j.jnutbio.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 40.Ngollo M, Lebert A, Daures M, Judes G, et al. Global analysis of H3K27me3 as an epigenetic marker in prostate cancer progression. BMC Cancer. 2017;17:261. doi: 10.1186/s12885-017-3256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ngollo M, Lebert AD, Judes G, Karsli-Ceppioglu S, et al. The association between Histone 3 Lysine 27 Trimethylation (H3K27me3) and prostate cancer: relationship with clinicopathological parameters. BMC Cancer. 2014;14:994. doi: 10.1186/1471-2407-14-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.