Abstract
Dental caries is a chronic infectious disease that affects billions of people with large individual differences in activity. We investigated whether PRH1 and PRH2 polymorphisms in saliva acidic proline-rich protein (PRP) receptors for indigenous bacteria match and predict individual differences in the development of caries. PRH1 and PRH2 variation and adhesion of indigenous and cariogenic (Streptococcus mutans) model bacteria were measured in 452 12-year-old Swedish children along with traditional risk factors and related to caries at baseline and after 5-years. The children grouped into low-to-moderate and high susceptibility phenotypes for caries based on allelic PRH1, PRH2 variation. The low-to-moderate susceptibility children (P1 and P4a−) experienced caries from eating sugar or bad oral hygiene or infection by S. mutans. The high susceptibility P4a (Db, PIF, PRP12) children had more caries despite receiving extra prevention and irrespective of eating sugar or bad oral hygiene or S. mutans-infection. They instead developed 3.9-fold more caries than P1 children from plaque accumulation in general when treated with orthodontic multibrackets; and had basic PRP polymorphisms and low DMBT1-mediated S. mutans adhesion as additional susceptibility traits. The present findings thus suggest genetic autoimmune-like (P4a) and traditional life style (P1) caries, providing a rationale for individualized oral care.
Keywords: Dental caries, Chronic infections, PRH1, PRH2, Host susceptibility, Acidic proline-rich proteins
Highlights
-
•
Allelic PRH1, PRH2 variation group children into high, moderate and low susceptibility phenotypes for caries.
-
•
Low susceptibility phenotypes experience caries from eating sugar and bad oral hygiene and infection by cariogenic S. mutans.
-
•
High susceptibility phenotypes may represent an autoimmune condition susceptible to bacteria in general.
Dental caries is a chronic infectious disease affecting billions of people with large individual differences in activity. The present study provides the first evidence of variation in specific human genes, PRH1 and PRH2, that matches and predicts individual experiences with caries in children. The high susceptibility phenotype (Db, PIF, PRP12) suggests an autoimmune-like condition, whereas those with low susceptibility (PIF2, PRP12) experiences caries from eating sugar and bad oral hygiene. These genetic and traditional life style sub types of caries suggest novel approaches for their diagnosis, prevention and treatment.
1. Introduction
Dental caries, a chronic infectious disease, affects billions of people with large individual differences in disease activity (Kassebaum et al., 2015, Selwitz et al., 2007). The global economic burden of dental diseases amounted to 442 billion USD in 2010 and was 4.6% of global health expenditure (Listl et al., 2015). The prevalence of caries has declined dramatically in Western countries, with ~ 80% of children being healthy or nearly caries-free and ~ 20% children having a high caries burden (Källestål, 2005). The high caries burden is poorly explained by eating and oral hygiene habits (relative risk 0.9–1.2) and is almost unaffected by prevention based on fluorides and life style (Källestål, 2005). Dental caries is detected when clinical symptoms arise but not by saliva, bacteria, or life style biomarkers, which do predict cross-sectional caries (Nordlund et al., 2009, Selwitz et al., 2007). Thus, new models to diagnose, prevent, and treat caries based on etiology are needed.
The traditional concept of dental caries is an imbalance in saliva defense, microbial load and life style habits (Nordlund et al., 2009, Selwitz et al., 2007). Saliva and salivary protein pellicles on teeth provide innate immunity clearance of bacteria, tooth homeostasis and adhesion-mediated colonization of both indigenous biofilms of Actinomyces and Streptococcus species and the caries pathogen Streptococcus mutans (Dawes, 2012, Gibbons, 1989, Esberg et al., 2017). Acidification of plaque from eating sugar emanates from S. mutans and other acid tolerant species, such as the abundant but moderate acid-producing non-mutans streptococci, and increases caries risk and shift ecology toward acid-producing species (Bradshaw et al., 1989, de Soet et al., 2000, Esberg et al., 2017). Bad oral hygiene, which increases the number of plaque bacteria, and eating sugar may act synergistically in plaque acidification. However, the presence of highly cariogenic adhesin biotypes of S. mutans, some with stroke and endocarditis risk (Nakano et al., 2011), points to specific disease mechanisms (Esberg et al., 2017). Despite such advances, dental caries is still grouped into different types based on affected surfaces (e.g. root, occlusal, approximal, coronal), dentition (i.e. primary, permanent) and age group (e.g. early childhood, adolescents, elderly) (Selwitz et al., 2007).
Twin (Boraas et al., 1988), human experimental (Krasse, 2001) and association studies, including genome wide association studies (GWAS) (Werneck et al., 2010), have suggested that genetic factors play a role in caries. Salivary acidic and basic proline-rich proteins (PRPs) (Ayad et al., 2000, Stenudd et al., 2001), agglutinin/DMBT1 (Jonasson et al., 2007) and HLA (Lehner et al., 1981) involved in innate and adaptive immunity, and proteins involved in enamel formation (Vieira et al., 2008), have been associated with susceptibility to caries and may be used to resolve the etiological types of disease.
Acidic and basic PRPs, encoded by PRH1-2 and PRB1-4 respectively, are major salivary innate immunity polypeptides (Bennick, 1987, Hay et al., 1994). Whereas basic PRPs survey and neutralize microbial pathogens in saliva (Burgener et al., 2012), acidic PRPs are pellicle receptors for adhesion of indigenous Actinomyces (e.g. A. oris). and Streptococcus (e.g. S. gordonii) species (Gibbons, 1989), which co-operate (mutualism) in biofilm formation (Kolenbrander et al., 2002). Acidic PRPs also modulate adhesion of S. mutans (Esberg et al., 2012), provide a barrier against bacterial acids (Bennick, 1987), and are degraded into bioactive peptides that modulate biofilm properties (Drobni et al., 2006). Allelic acidic PRP variants PRP1, PRP2 (PRH2) and Pa, Db, PIF (PRH1) and their post-translational variants generate many mixed PRP phenotypes in saliva (Azen, 1993, Hay et al., 1994). Db-positive phenotypes have been implicated in the susceptibility to caries and S. mutans adhesion and Db-negative phenotypes in resistance to caries and indigenous adhesion (Stenudd et al., 2001, Jonasson et al., 2007).
Salivary agglutinin, which aggregates S. mutans, is also known as DMBT1 or gp340 (Prakobphol et al., 2000). The DMBT1 pattern-recognition molecule binds to S. mutans and a wide array of microbes, as well as innate and adaptive immunity factors, through its multiple domains (Madsen et al., 2010, Loimaranta et al., 2005). Thus, DMBT1 modulates innate and adaptive immunity, including complement activation, NF-κb signaling via Toll receptors and cellular proliferation (Madsen et al., 2010). A 6.2 kb dmbt1 deletion variant has been associated with cancer (Madsen et al., 2010) and inflammatory bowel disease via increased NF-κB mediated inflammation in humans (Renner et al., 2007). The corresponding salivary size variant (designated gp340 or DMBT1 size variant I) coincides with increased caries and S. mutans adhesion in children (Jonasson et al., 2007).
The present study was performed to further explore the etiology of caries. The primary aim was to replicate and further explore the role of acid PRP polymorphisms and adhesion of indigenous (A. oris LY7) and cariogenic (S. mutans Ingbritt) model bacteria in caries development. We therefore matched PRH1, PRH2 genetic variation with individual differences in baseline caries and 5-year development of caries in Swedish children measured also for traditional variables and bacterial adhesion.
2. Materials and Methods
2.1. Study Participants
A total of 452 12-year-old children were enrolled as two independent samples (n = 218, n = 234) from 13 Public Dental Service Clinics in the county of Västerbotten, Sweden. The first sample collected in 2008 included children born in 1996 with caries cases (≥ 1 decayed, filled surfaces [DFS] in the permanent dentition) and controls (DFS = 0 in 2007) in a 1:1 ratio. The second sample collected in 2010 included children born in 1998 and caries cases (≥ 2 DFS lesions) and controls (DFS = 0 in 2009) in a 2:1 ratio to increase the portion of caries cases in the total sample of 452 children (Table S1). All children in the study received ordinary dental care at the clinics. The exclusion criterion was unwillingness to participate in the study. Both samples were re-examined after 5 years (n = 390 children, 14% drop-out). Twenty children had moved out of the area and 42 had repeatedly missed the examination so they were not included in the follow-up. The children received operative treatment and caries prevention between 12 and 17 years of age, and 15% of the children underwent orthodontic treatment with multibrackets after 12 years of age (as established from dental records) according to ordinary routines and policies at the clinics. The Ethics Committee for Human Experiments at Umeå University, Sweden, approved the study, and informed consent was obtained from the children and their parents prior to participation. All parents signed consent to participate.
2.2. Measurement of Caries
Caries were recorded by three dentists (intra- and inter-examiner kappa ≥ 0.98) using a mirror, probe and two bitewing radiographs. The mean number of decayed (enamel caries included), filled surfaces in the permanent dentition (DeFS) were the primary caries outcome measure (Källestål, 2005, Nordlund et al., 2009). The 5-year change in caries (ΔDeFS, increment), a good estimate of incidence rate in this age group and population (Källestål and Stenlund, 2003), was calculated by subtracting latest DeFS measure from earliest DeFS measure, dividing it by the number of observed years, and then multiplying by 5. The 1:1 ratio in the first sample and increased 2:1 ratio in the second sample and DeFS index generated a continuous gradient of discriminatory caries DeFS scores in the entire sample at baseline (Table S1) (Esberg et al., 2017).
2.3. Collection of Biological Samples
Parotid and whole saliva (PS and WS, respectively) secretions were measured (mL/min), collected and stored frozen at − 80 °C (Nordlund et al., 2009, Stenudd et al., 2001). All saliva analyses used baseline samples. Plaque was collected by pelleting from the buccal surfaces of premolars and the first molar of the left lower jaw (teeth 34–36). Fresh WS and plaque (pl) sampled at baseline were cultured on selective substrates to establish whether children infected with S. mutans (ms) or lactobacilli (lbc), as well as the proportion of ms out of oral streptococci (strept) in saliva (% ms) and plaque (% ms pl) (Nordlund et al., 2009).
2.4. Sociodemographic and Life Style Data
Sociodemographic data (sex and ethnicity) were collected from parents via pre-posted questionnaires, and oral behavior data, such as oral hygiene, intake frequency of sweets (e.g., cookies, biscuits, ice cream, or dried fruit) and sugary drinks (never, once per month, once per week, several times per week, once per day, several times per day), and the use of extra fluoride were collected from the children at the clinic using a questionnaire that was previously utilized in a cohort of 3400 12-year-old children (Källestål, 2005). Strict Swedish ethnicity was defined when both parents reported Swedish ethnicity. Oral hygiene was measured as how often the child brushes his/her teeth: irregularly, once per day, twice per day, more than twice per day. Extra fluoride treatment in addition to fluoride in toothpaste was recorded as: none, fluoride mouth rinse more than once per month, fluoride-containing chewing gum 1–3 pieces per day, or 1–3 fluoride tablets per day.
2.5. Typing Acidic PRP Phenotypes in Saliva
Acidic PRPs in PS were typed using native alkaline electrophoresis as described (Azen and Yu, 1984, Esberg et al., 2012). Saliva was lyophilized and re-dissolved to the same volume using distilled water containing 1% glycine, 10% glycerol and 0,025% bromphenol blue. Roughly 40 test and reference saliva samples (each 25 μL) were separated in a single run under native conditions on 7.0% (w/w) polyacrylamide (27:1 bisacrylamide w/w) gels using a 1.5 mM TRIS, 38 mM glycine electrophoresis buffer (pH 8.4) and a PROTEAN IIxi apparatus (BioRad). The gels were stained with 0.1% Commassie Brilliant Blue (CBB-R250) in 20% trichloroacetic acid for 1 h and destained over night in 2% acetic acid. A total of 218 samples were analyzed and typed in a double-blind fashion, with the vast majority typable (n = 213), and quantified via densitometry (optical density x mm2) using Molecular Analyst 1.5 software (BioRad). Single representatives of each PRP phenotype were validated using high-performance liquid chromatography on a Bischoff LC-caDI22-14 HPLC system (Bischoff, Germany) and a Gen-Pak™ FAX anionic exchange column (Waters, MA) as described previously (Hay et al., 1994).
2.6. Slot-blot Measurements of DMBT1 in Saliva
DMBT1 in PS was quantified using a slot-blot assay and densitometry (Stenudd et al., 2001, Esberg et al., 2012). Briefly, saliva diluted 1:50 was transferred to a 0.45 μm nitrocellulose membrane (Protran BA85-SB, Whatman) followed by blocking with 5% dry milk in buffer (50 mM Tris, 150 mM NaCl, pH 7.4) containing 0.05% Tween-20. The membrane was then incubated with anti-gp340 antibody (mAb-143, 1:60,000, kindly provided by Dr. Malamud, University of Pennsylvania, Philadelphia), for 1 h, washed twice for 10 min and incubated with secondary antibody SAB-100 (1:60,000, Nordic Biosite AB, Sweden). After two washes, binding was visualized using the Super Signal West Dura Detection kit (Pierce Protein Research Products, IL) and quantified by densitometry (INTxmm2) using Chemidoc (Bio-Rad).
2.7. Adhesion of Indigenous and Pathogenic Model Bacteria by Saliva
Adhesion of 35S-Met metabolically labeled reference strains were done with individual saliva's coated on hydroxyapatite beads (Bio-Rad Laboratories) in 96-well plates (Nunclon™ surface, Thermo Scientific Fischer, Denmark) (Loimaranta et al., 2005, Esberg et al., 2012). Briefly, after hydration of the hydroxyapatite beads in adhesion buffer at 4 °C overnight (5 mg beads in 125 μL per well), the beads were coated with 125 μL of PS diluted in adhesion buffer (1:64 for A. naeslundii or S. gordonii and 1:1 for S. mutans or S. pyogenes) for 1 h, washed, and incubated with 35S-labeled bacteria (125 μL, 108 cells/mL; 1000 cells/cpm) for 1 h under agitation. After three washes with 200 μL adhesion buffer, the beads were counted in a scintillation counter (MicroBeta2, PerkinElmer) and attached bacteria expressed as percent bound out of added bacteria (percent adhesion, adh%) or total radioactivity count (adhtot). Reference strains used were: Streptococcus gordonii strain SK12 and Actinomyces oris strains LY7 and T14 V with types 1 and 2 pili (and its mutant T14Vm without type-2 pili) as representative for clinical isolates of indigenous actinomycetes and streptococci (Loimaranta et al., 2005); strain Ingbritt as representative for clinical isolates of S. mutans (Esberg et al., 2012) and strain A8173 for isolates of Streptococcus pyogenes involved in GAS infections (Loimaranta et al., 2005).
2.8. PRH1, PRH2 and PRB1-4 Genotyping
Buccal epithelia were collected using swabs (Catch-All™ Sample Collection Swab, Epicentre Biotechnologies) and genomic DNA isolated using the QIAamp DNA Micro Kit (Qiagen). The genomic DNA was whole genome amplified (Illustra Ready-To-Go™ GenomiPhi V3 DNA kit, GE Healthcare) and genotyped using 6 tag and 39 additional single nucleotide polymorphisms (SNPs) with an allele frequency generally > 5% of PRH1 and PRH2 and 16 tag SNPs of PRB1–4 in an Illumina Golden Gate array at the SNP&SEQ Technology Platform in Uppsala (Table S2). Five PRH1, PRH2 SNPs and one PRB3 SNP with no call frequencies were excluded. Mixed PRH1, PRH2 genotypes were resolved in the vast majority of subjects (n = 441) by principal component analysis of the remaining PRH1, PRH2 SNPs (n = 40). The 40 SNPs used to resolve the mixed PRH1, PRH2 genotypes were highly conserved in the Db, PIF, Pa (PRH1) and PRP1, and PRP2 (PRH2) alleles. Illumina and protein typing as well as TaqMan typing using protein coding SNPs rs2923234, rs1049117 and rs1049112 (Table S2) revealed 98–100% typing congruence and generated essentially identical caries-related outcome results.
2.9. Measurement of S. mutans in Whole Saliva by qPCR
Bacterial DNA was prepared from WS using the GenElute Bacterial Genomic DNA Kit (Sigma). S. mutans was measured by qPCR (Corbett Rotor Gene 6000 apparatus) from the purified DNA using the KAPA Sybr Fast Universal qPCR kit (Kapa Biosystems) and S. mutans-specific primers as described (Yano et al., 2002, Esberg et al., 2017).
2.10. Statistical Analysis
Multivariate partial least squares (PLS) models, which relates two data matrices (X and Y) to each other, were generated using Simca+ P12.01 (Eriksson et al., 2004). The PLS models show the ability of the X variables to explain (R2) or predict (Q2) the variation in Y and the relative variable importance in the projection is given by VIP-values (VIP ≥ 1.0 marks influential x variables). The predictive ability (Q2) is estimated by cross-validation via step-wise exclusion of 1/7th of the data in the model which is used to predict the left out data. Data were log transformed and auto-scaled to unit variance before PLS models were generated.
Univariate statistical analyses were performed using the chi-squared test, Fischer's exact test, Mann-Whitney U test, or Spearman's rank correlation (SPSS version 23 or GraphPad software 5) with p < 0.05 considered significant.
3. Results
3.1. Role of Allelic PRH1, PRH2 Variation in Caries
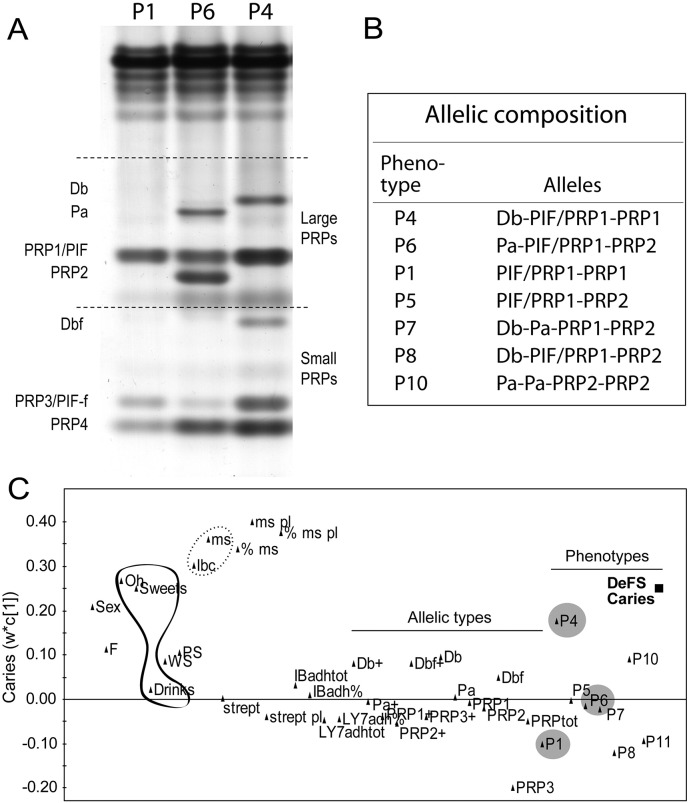
We previously showed that allelic PRH1, PRH2 acidic PRP variation coincides with caries and adhesion of indigenous (A. oris LY7) and cariogenic (S. mutans Ingbritt) model bacteria (Stenudd et al., 2001, Jonasson et al., 2007, Esberg et al., 2012). To replicate and further explore allelic PRH1, PRH2 variation in caries and bacterial adhesion, we measured the parotid saliva (PS) of 218 12-year-old children for acidic PRP allelic variants and adhesion of S. mutans Ingbritt and indigenous A. oris LY7 model bacteria with specificity for acidic PRP receptors (Fig. 1).
Fig. 1.
Acidic PRP phenotypes, their allelic composition and association with baseline caries. (AB) Allelic types and amounts of mixed acidic PRP phenotypes P1-P10 from gel electrophoresis of parotid saliva in 218 12-year-old children. (C) PLS loading plot showing the association of P4, P6 and P1 children (grey circles) and traditional risk factors (S. mutans, lactobacilli > oral hygiene > sweets >> drinks) with baseline caries in children with Swedish ethnicity (n = 185); Sex = 0 boys, 1 girls; F = extra fluoride; Oh = oral hygiene; WS = whole saliva flow; PS = parotid saliva flow; lbc = lactobacilli in saliva; ms, ms pl, % ms and % ms pl = S. mutans and % of total streptococci in saliva and plaque; strept, strept pl = total streptococcal in saliva, plaque; IBadh = S. mutans adhesion, % and tot; LY7adh = A. oris adhesion, % and tot); Db + etc. and Db etc. = qualitative, +, and quantitative acidic PRP variants from electrophoresis; PRPtot = total amounts of PRPs; P1, P4 etc. = mixed acidic PRP phenotypes from electrophoresis.
The children grouped into three major P4 [Db, PIF/PRP12], P6 [Pa, PIF/PRP1, PRP2] and P1 [PIF2, PRP12] and minor allelic acidic PRP phenotypes based on the composition of allelic variants Db, Pa, PRP2, PIF or PIF/PRP1 (which was unresolved by electrophoresis) (Fig. 1A,B).
We explored the association of PRP phenotype and bacterial adhesion with caries by modeling of these variables among traditional risk factors against baseline caries in the 185 children of strict Swedish ethnicity (i.e. omitting the 33 children of other ethnicities) using PLS prediction models (Fig. 1C). Traditional risk factors (e.g. oral hygiene, sweet consumption, cariogenic S. mutans and lactobacilli) and P4 children (VIP = 1.11) correlated positively with caries, whereas P6 was non-influential and P1 negatively correlated with caries. The P4, P6 and P1 children accordingly had high (DeFS ± SD 2.75 ± 1.91), moderate (1.66 ± 3.07) and low (2.41 ± 2.26) number of caries, respectively, with P4 children presenting 1.6-fold more caries than P1 children (p = 0.049). However, the saliva adhesion of indigenous or cariogenic model bacteria did not correlate with caries (Fig. 1C).
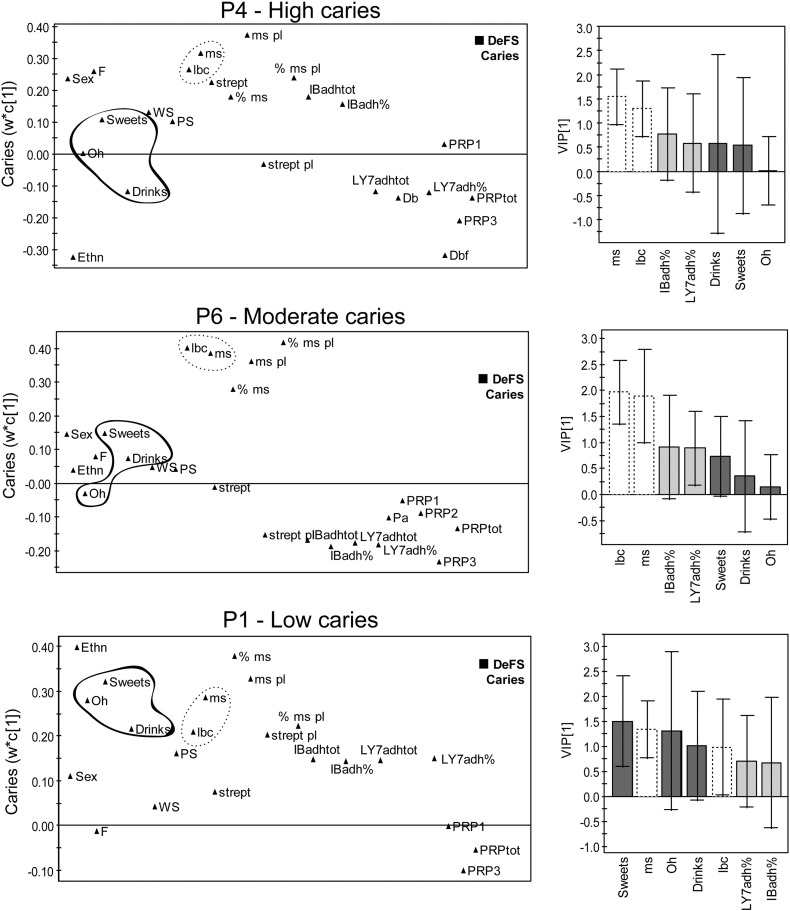
3.2. Risk Factor Profiling in P4a, P6 and P1 Children
Having identified acidic PRP phenotypes with high (P4), moderate (P6) and low (P1) caries, we explored their intra type risk factor patterns by modeling against baseline caries (Fig. 2). The P4, P6 and P1 prediction models, which were stronger for P4 (R2 = 42% Q2 = 23%, n = 40) and equally strong for P6 (R2 = 33% Q2 = 20%, n = 62) and P1 (R2 = 36% Q2 = 16%, n = 72) compared to the model for the whole sample (R2 = 25% Q2 = 18%, n = 218), had influential S. mutans and lactobacilli risk factors (VIP ~ 1.5) (Fig. 2). However, oral hygiene and sweet consumption were risk factors only in P1 children (VIP = 1.3–1.5), and extra fluoride treatment in P4 children (VIP > 1.0). Moreover, bacterial adhesion risk profiles were different in P4, P6 and P1 children for baseline caries, but these differences were not significant. Thus, P1 children experience fewer caries lesions and are dependent on high loads of negative life style factors (life style type), P4 children experience more caries though they are diagnosed and treated as high risk subjects at our clinics (genetic type).
Fig. 2.
Differential life style risk factor profiles for the high (P4), low (P1) and moderate (P6) phenotypes. PLS loading plots of various factors (left) and influential (VIP) values for selected factors (right) showing their association with caries (DeFS) in P4, P6, and P1 phenotypes (P4 = 40, P6 = 62, P1 = 72 children). Sex = 0 boys, 1 girls; Ethn = ethnicity; F = extra fluoride; Oh = oral hygiene; WS = whole saliva flow; PS = parotid saliva flow; lbc = lactobacilli counts in saliva; ms, ms pl, % ms and % ms pl = S. mutans counts and % of total streptococci in saliva and plaque; strept, strept pl = total streptococcal counts in saliva, plaque; IBadh = S. mutans adhesion, % and tot; LY7adh = A. oris adhesion, % and tot; Db, Pa etc. = quantitative acidic PRP variants from electrophoresis; PRPtot = total amounts of PRPs.
3.3. Verification of Genetic (P4a) and Life Style (P1) Caries by Genotyping
To substantiate our findings of genetic- and life style-dependent caries based on PRH1 and PRH2 variation, we recruited a second sample of 234 children and analyzed the pooled sample of 452 children by PRH1 and PRH2 genotyping in relation to caries at baseline and 5-year prospectively (Table 1).
Table 1.
Association of mixed PRH1, PRH2 genotypes with caries in Swedish children.
| Genotypea | DeFS-12yb |
DeFS-17yb |
ΔDeFSc |
||||||
|---|---|---|---|---|---|---|---|---|---|
| n | mean ± SD | pd | n | mean ± SD | pd | n | mean ± SD | pd | |
| Whole sample | |||||||||
| P4a | 75 | 3.1 ± 3.1 | 0.008 | 65 | 8.1 ± 8.3 | 0.004 | 65 | 4.7 ± 6.6 | 0.102 |
| P6 | 102 | 2.3 ± 2.5 | 0.285 | 93 | 6.1 ± 6.7 | 0.209 | 93 | 3.6 ± 5.8 | 0.477 |
| P1 | 115 | 1.9 ± 2.3 | Ref. | 96 | 4.9 ± 5.7 | Ref. | 96 | 3.0 ± 4.7 | Ref. |
| P4a− | 279 | 2.2 ± 2.4 | 0.022 | 245 | 5.8 ± 6.8 | 0.024 | 245 | 3.6 ± 5.6 | 0.214 |
| Orthodontic treatmente | |||||||||
| P4a | 11 | 4.6 ± 3.9 | 0.032 | 11 | 10.6 ± 6.0 | 0.002 | 11 | 5.5 ± 5.0 | 0.021 |
| P6 | 14 | 2.4 ± 2.4 | 0.382 | 14 | 5.6 ± 8.2 | 0.631 | 14 | 2.7 ± 5.4 | 0.923 |
| P1 | 20 | 1.9 ± 2.4 | Ref. | 18 | 3.3 ± 3.1 | Ref. | 18 | 1.4 ± 2.2 | Ref. |
| P4a− | 47 | 2.2 ± 2.3 | 0.040 | 43 | 5.2 ± 6.0 | 0.005 | 43 | 2.7 ± 4.4 | 0.044 |
Virtually identical results were obtained for P4a (13 children) vs. P1 (23 children) in the whole sample; ΔDeFS ± SD 5.34 ± 4.82 vs. 1.33 ± 2.16, p = 0.016.
Swedish ethnicity (n = 369). Mixed acidic PRP phenotypes from PRH1, PRH2 genotyping.
DeFS, decayed, enamel included, filled surfaces.
ΔDeFS = 5 year caries increment.
p-Values were obtained using the Mann-Whitney U test compared to the reference (Ref.).
Children treated with orthodontic multibrackets between 12 and 17 years of age.
Illumina array and TaqMan genotyping of 45 and 3 diagnostic SNPs, respectively, in the 452 children resolved P4 (Db-PIF/PRP12, n = 84) into P4a (Db-PIF-PRP12, n = 75) and P4b (Db2-PIF-PRP1, n = 9) with low and high caries experiences, respectively (3.08 vs. 1.44 DeFS; Table 1 and Table S2 and S3). The P4a children had 1.6- to 1.7-fold more caries at 12 and 17 years than P1 children (p = 0.008, p = 0.004), and 1.4-fold more caries at 12 and 17 years than P4a-negative children (p = 0.022, p = 0.024) (Table 1 and Table S3). Thus, P4a children developed more baseline and prospective caries, although they experienced adolescence, puberty and treatment for caries by operative and preventive means (Table S1), during the study period.
In addition, a dependence of baseline caries on oral hygiene in P1 and P4a− (both p < 0.007), but not in P1−, P4a or P6 (all p > 0.12), children was verified (Table S4).
3.4. Role of Plaque and S. mutans Infection in 5-year Caries Increment in P4a, P6 and P1 Children
As many factors may coincide or predict cross-sectional caries but only true etiological or prognostic factors predict the development of caries (Nordlund et al., 2009), we further evaluated how presence of S. mutans infection ± or plaque-accumulating orthodontic multibrackets, a well known risk factor for caries (Ren et al., 2014), affected 5-year caries development, ΔDeFS, in P4a, P6 and P1 children (Table 1, Table 2 and Fig. 3A).
Table 2.
Development of caries in P4a, P6 and P1 children related to S. mutans or lactobacilli infection and S. mutans-binding.
| Group and genotypea | Group | DeFS-12yb |
DeFS-17yb |
ΔDeFSb |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Mean ± SD | pc | n | Mean ± SD | pc | n | Mean ± SD | pc | |||
| S. mutans infectiond | P4a | + | 34 | 4.3 ± 3.7 | 0.004 | 28 | 9.3 ± 8.5 | 0.114 | 28 | 4.5 ± 5.7 | 0.871 |
| – | 40 | 2.1 ± 2.1 | 36 | 7.1 ± 8.2 | 36 | 4.9 ± 7.4 | |||||
| P6 | + | 47 | 3.5 ± 2.9 | < 0.001 | 42 | 8.9 ± 10.1 | < 0.001 | 42 | 5.2 ± 7.8 | 0.025 | |
| – | 55 | 1.4 ± 1.6 | 51 | 3.8 ± 3.7 | 51 | 2.3 ± 2.7 | |||||
| P1 | + | 48 | 2.5 ± 2.4 | 0.012 | 43 | 6.8 ± 6.9 | 0.004 | 43 | 4.4 ± 6.1 | 0.013 | |
| – | 67 | 1.5 ± 2.1 | 53 | 3.4 ± 3.8 | 53 | 1.9 ± 2.7 | |||||
| Lactobacilli infectiond | P4a | + | 39 | 3.2 ± 2.6 | 0.275 | 33 | 10.0 ± 8.8 | 0.026 | 33 | 6.3 ± 8.2 | 0.101 |
| – | 36 | 3.0 ± 3.7 | 32 | 6.2 ± 7.4 | 32 | 3.2 ± 4.2 | |||||
| P6 | + | 48 | 2.9 ± 2.4 | 0.013 | 44 | 8.5 ± 9.8 | 0.002 | 44 | 5.2 ± 7.5 | 0.007 | |
| – | 54 | 1.9 ± 2.6 | 49 | 4.0 ± 4.0 | 49 | 2.2 ± 2.9 | |||||
| P1 | + | 47 | 2.5 ± 2.6 | 0.030 | 55 | 5.7 ± 6.5 | 0.332 | 55 | 3.4 ± 5.3 | 0.788 | |
| – | 68 | 1.6 ± 2.0 | 41 | 4.4 ± 4.9 | 41 | 2.8 ± 4.1 | |||||
| S. mutans bindingd | P4a | High | 14 | 2.6 ± 2.1 | 0.984 | 13 | 4.8 ± 2.9 | 0.028 | 13 | 1.9 ± 1.8 | 0.005 |
| Low | 16 | 3.3 ± 3.9 | 15 | 12.7 ± 10.8 | 15 | 7.5 ± 6.2 | |||||
| P6 | High | 31 | 1.4 ± 2.1 | 0.141 | 29 | 5.6 ± 6.9 | 0.542 | 29 | 3.5 ± 4.5 | 0.834 | |
| Low | 26 | 2.4 ± 2.8 | 25 | 7.2 ± 11.4 | 25 | 4.4 ± 8.9 | |||||
| P1 | High | 28 | 2.1 ± 2.7 | 0.547 | 23 | 5.6 ± 5.6 | 0.231 | 23 | 3.3 ± 3.9 | 0.241 | |
| Low | 31 | 1.4 ± 1.7 | 27 | 4.3 ± 4.7 | 27 | 2.5 ± 3.4 | |||||
| Correlatione | n | r-value | p | n | r-value | p | n | r-value | p | ||
| P4a | Binding | 30 | 0.257 | 0.156 | 28 | − 0.212 | 0.252 | 28 | − 0.490 | 0.007 | |
| P4a | DMBT1 | 30 | 0.502 | 0.003 | 28 | 0.124 | 0.522 | 28 | − 0.076 | 0.694 | |
Children of Swedish ethnicity (n = 369). Allelic acidic PRP saliva phenotypes from PRH1, PRH2 genotyping.
DeFS, decayed, enamel included, filled surfaces, ΔDeFS, 5-year increment of between 12 and 17 years of age.
p-Values obrained using the Mann-Whitney U test.
Infection by S. mutans (+ > 10,000 cfu), lactobacilli (+ > 20,000 cfu) and S. mutans binding to individual parotid salivas.
Spearman correlation of S. mutans saliva binding and DMBT1 (quantified by mAb143) with caries.
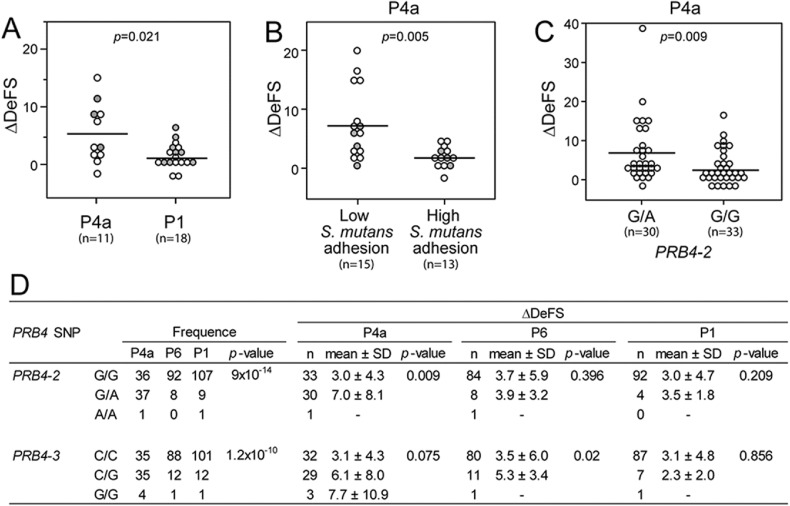
Fig. 3.
P4a children had more caries from orthodontic treatment, low S. mutans-binding and basic PRB4 allelic variation. (A) Treatment with orthodontic multibrackets between 12 and 17 years of age compared to P1 children, (B) Low versus high DMBT1-mediated binding of S. mutans to individual salivas coated onto hydroxyapatite beads; (CD) Heterozygocity at PRB4 SNP 4-2 (rs7138858) and SNP 4-3 (rs1863843) coincided with increased caries in P4a and P6 children respectively. The p-values were obtained using chi2 test (frequencies) or Mann-Whitney U test (ΔDeFS).
Treatment with multibrackets, but not infection with S. mutans or lactobacilli, increased ΔDeFS 3.9-fold in P4a children compared to P1 children (5.5 vs. 1.4, p = 0.021; Table 1 and Fig. 3A). In contrast, ΔDeFS increased in P1 and P6 children due to infection with S. mutans alone and S. mutans or lactobacilli, respectively, but not treatment with multibrackets (Table 1, Table 2). Moreover, P1 children with multibrackets developed markedly fewer caries lesions than those without, indicating that P1 children respond to the prevention given as part of the orthodontic treatment. Thus, high susceptible P4a children seemed sensitive to plaque accumulation in general, whereas the low P1 and moderate P6 susceptible phenotypes required infection by specific cariogenic species for the development of caries over 5 years.
3.5. Role of S. mutans and Indigenous Bacterial Adhesion in P4, P6 and P1 Children
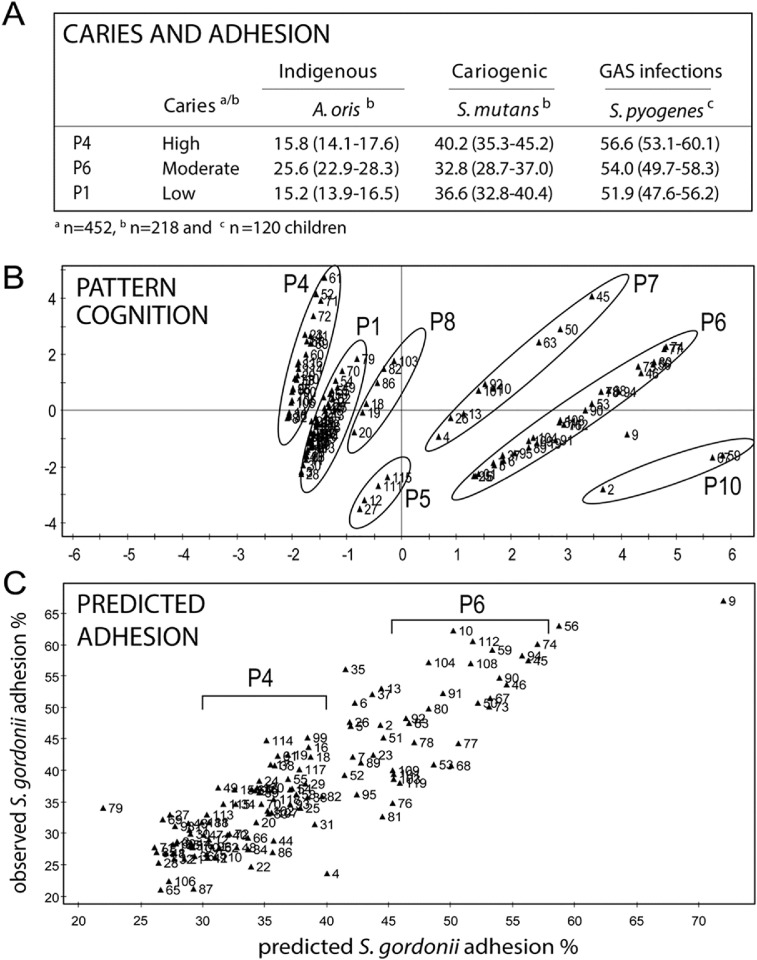
Because P4, P6 and P1 children seemed to exhibit different adhesion risk factor patterns at baseline (Fig. 2), we investigated whether saliva binding of S. mutans Ingbritt (with DMBT1 and acidic PRP receptors and coreceptors, respectively) and indigenous A. oris LY7 (with acidic PRP receptors) affected ΔDeFS in P4a, P6 and P1 children (Table 2 and Fig. 3B).
Low saliva binding of S. mutans Ingbritt correlated with increased ΔDeFS in P4a children exclusively, whereas amount of DMBT1 did not (Table 2 and Table S5). Thus, P4a children may harbor a qualitatively different DMBT1 and acidic PRP innate immunity complex. In addition, P4 children had comparably high S. mutans but low indigenous adhesion at baseline (Fig. 4A; Esberg et al., 2012).
Fig. 4.
Relationship between acidic PRP phenotypes and adhesion of indigenous and cariogenic model bacteria. (A) Caries and saliva adhesion of indigenous and pathogenic bacteria in P4, P6 and P1 children. Data are from adhesion to individual saliva's and presented as mean adhesion (95% CI). (B) Pattern cognition of phenotypes P1-P10 and (C) nearly linear prediction of observed adhesion of S. gordonii SK12 (R2 = 78%, Q2 = 56%) upon PLS prediction modeling of acidic PRP allelic composition and amounts against indigenous adhesion of A.oris and S. gordonii model bacteria to individual saliva's (n = 218 children). Adhesion increases from left to right in each phenotype as the amount of acidic PRPs increases (marked by circles and brackets).
High versus low saliva adhesion of indigenous A. oris LY7 did not affect ΔDeFS in P4a, P1 or P6 children (Table S6, S7). However, PRH1, PRH2 variation modulated saliva adhesion of indigenous A. oris (strains LY7 and T14 V) and S. gordonii (strain SK12) with different PRP specificities similarly as shown by PLS modeling (Figs. 4C and S1). The adhesion of indigenous bacteria was nearly linearly predicted by acidic PRP composition regardless of species or strain (Fig. 4C), and positively correlated with phenotype P6, as well as Pa, PRP2 and acidic PRP amount, and negatively with phenotypes P4 and P1, but was non-influential with Db (Fig. S1).
P4a, P1 and P6 children did not differ in gross saliva or plaque numbers of indigenous streptococci or S. mutans (data not shown). Thus, saliva-mediated adhesion of indigenous model bacteria and the S. mutans Ingbritt adhesin type, as well as colonization of indigenous streptococci or S. mutans, did not explain caries experience or development.
3.6. P4a and P6 Children Differ in Basic PRB4 Polymorphisms
Because PRH1, PRH2 and PRB1-4 encoding basic PRPs are closely linked on chromosome 12, and basic PRPs are salivary innate immunity polypeptides, we screened PRB1-4 tag SNPs in P4a, P6 and P1 children using an Illumina array (Fig. 3C,D). Heterozygosity at two noncoding PRB4 SNPs, PRB4-2 (rs7138858) and PRB4-3 (rs1863843), which were more frequently found in P4a children, increased ΔDeFS in P4a and P6 children, respectively. These findings suggest a PRH-PRB haplotype in which acidic and basic PRP properties co-operate to form high and moderate susceptible P4a and P6 phenotypes.
4. Discussion
The present study shows that PRH1, PRH2 composition specifies high (P4a), moderate (P6) and low (P1) caries experience and susceptibility in children/adolescents. This study is the first demonstration of specific human genes that match and predict individual differences in prospective caries development (summarized in Table 3). The susceptible P4a children may develop caries from an autoimmune-like condition, as opposed to the low-to-moderately susceptible children (P1 and P6) that experience caries from eating sugar, bad oral hygiene, or S. mutans-infection. The P4a children, who were sensitive to plaque accumulation in general but not to eating sugar, bad oral hygiene or S. mutans infection, harbored additional DMBT1 and PRB4 polymorphic behaviors. That P4a children developed 3.9-fold more caries from orthodontic multibrackets, which accumulate plaque and disrupt saliva flow through the biofilm, emphasizes that children, adults and elderly with the genetic caries type (P4a) may be at risk at vulnerable sites introduced by fillings or prosthetic replacements. Thus, secondary caries is still the major reason for failure of operative treatment. In contrast to the P1 children, the P4a children responded poorly to traditional prevention even though they were diagnosed and treated as high risk subjects at our clinics. Accordingly, P1 children will benefit from traditional oral hygiene and diet preventive measures, whereas susceptible P4a subjects will require intensified or novel prevention regimes.
Table 3.
Summary of genetic and life-style-dependent types of caries defined by PRH1 and PRH2 variation.
|
PRH1, PRH2 children |
||||
|---|---|---|---|---|
| P4a | P6 | P1 | ||
| Caries type | Genetic | Genetic/life style | Life style | |
| Prevalence | 19% | 32% | 28% | |
| Composition | PRH1 | Db, PIF | Pa, PIF | PIF, PIF |
| PRH2 | PRP1, PRP1 | PRP1, PRP2 | PRP1, PRP1 | |
| Caries DeFS score | High | Moderate | Low | |
| Susceptibility | High | Moderate | Low | |
| Dependence on | ||||
| Life style (DeFS-12y)a | No | No | Yes | |
| Orthodontic treatmentb | Yes | No | No | |
| Infection status (ΔDeFS)c | ||||
| S. mutans | No | Yes | Yes | |
| Lactobacilli | No | Yes | No | |
| DMBT1 immunity | Yes | No | No | |
| PRB4 immunity | Yes | Yes | No | |
Data from PLS models of P4 (R2 = 42%, Q2 = 23%), P6 (R2 = 33%, Q2 = 20%), P1 (R2 = 36%, Q2 = 16%) in 218 children and from chi2 analyses of P4a, P6 and P1 in 452 children using baseline caries, DeFS-12y.
Children treated with orthodontic multibrackets between 12 and 17 years of age.
Infection status (+, −) related to 5-year caries increment, ΔDeFS.
We assume that the susceptible P4a children represent an autoimmune-like condition with impaired PRH-PRB and dmbt1 innate and adaptive immunity and bacterial clearance; and that adhesion-mediated colonization of S. mutans and indigenous bacteria per se, although only a few model bacteria were used, is less likely to explain caries development. The PRH1, PRH2 background of P4a children may misbehave in acidic PRP barrier and innate defenses against dissolution of hydroxyapatite. This capacity of acidic PRPs to modulate early events at the host-microbe interface is evident from their composition predicting the binding of indigenous bacteria to salivary pellicles in a nearly linear fashion regardless of species. These individually unique adhesion patterns and the high specificity and co-operativity between indigenous biofilm bacteria may promote diverse biofilm compositions, even though they do not drive or explain caries development in the present PRP phenotypes. Because low saliva adhesion of S. mutans correlated with caries development only in P4a children, DMBT1 binds a wide array of bacteria and DMBT1 size variant I coincides with caries, P4a children may rather misbehave in DMBT1-mediated binding and neutralization of oral bacteria in general and abundant non-mutans streptococci (de Soet et al., 2000). S. mutans cariogenicity, on the other hand, seems to involve high affinity binding to and neutralization of DMBT1 (Esberg et al., 2017), emphasizing a key etiological role of DMBT1 in caries. Moreover, unique basic PRP innate immunity properties also occur because two PRB4 SNPs coincided with caries in P4a and P6 children, and P6 children alone developed caries from infection by lactobacilli. However, an autoimmune-like condition, which could make P4a individuals sensitive to various infections and bacteria-related diseases, should be further explored for genes and functions reported to be altered in autoimmune diseases (Wang et al., 2011). It should also be explored whether the potential PRH-PRB haplotype and dmbt1 complex of P4a children select for a unique oral microbiota, non-mutans streptococci or different S. mutans adhesin biotypes (Esberg et al., 2017).
Our results extend our previous notion of a role of Db and DMBT1 in caries (Stenudd et al., 2001, Jonasson et al., 2007), and fully resolve the Db, PIF, PRP12 susceptibility phenotype P4a in a larger and more representative sample of 452 Swedish children. The high prevalence of P4a (19%) and P6 (28%) children with DMBT1 and PRB4 polymorphic properties, argues for a platform to identify susceptibility genes and etiological subtypes of caries with different treatment needs. The finding that baseline factors, such as infection status, predicted 5-year caries increment markedly different in P4a, P6 and P1 children, emphasizes that subtyping caries may be a prerequisite for multimarker risk assessment. The finding that the S. mutans counts predicted cross-sectional caries in P4a children, although infection status (+ or -) of the organism failed to predict 5 year caries increment, suggests that S. mutans and sugar consumption co-operate in gross plaque acidification and the cross-sectional risk of caries. Prospective risk assessment requires stable genetic factors acting at the tooth-bacteria interface. In this respect, cross-sectional and prospective risk assessment may use different factors, and many factors identified in cross-sectional studies will not be valid for predicting the future development of caries.
The high association between genetic group and cross-sectional and prospective caries, even though the children received oral health prevention and treatment, and experienced puberty and adolescence during the study, strengthens the group-specific differences in caries. However, it is difficult to estimate susceptibility or risk in absolute terms because of these experiences, in addition to the treatment of caries with fillings reducing the surfaces at future risk. Moreover, puberty could modulate the mucine-like glycosylation of DMBT1, explaining why S. mutans binding correlated differently with baseline and prospective caries. The P4a caries type was more pronounced in children/adolescents of Swedish ethnicity than in the entire sample with greater genetic and cultural diversity, but it was evident in both samples. Therefore, we consider the present sample representative of Swedish children/adolescents, and assume that the genetic (P4a, P6) and lifestyle (P1) types of caries are generalizable, although PRH1, PRH2 variation is ethnicity-dependent (The International HapMap Consortium, 2003), in broader populations. However, the role of acidic PRP phenotypes in caries should be replicated in longitudinal studies of early primary and permanent dentitions in different populations and ethnicities and related to initiation versus progression of caries.
Taken together with our recent report on S. mutans adhesion biotypes with high caries and systemic virulence (Esberg et al., 2017), the present findings may be generalized to indicate that many of the high-risk children who are non-responders to traditional prevention carry highly cariogenic strains or defective innate and adaptive immunity. On the other hand, the many individuals of low-to-moderate genetic risk may develop caries largely from eating sugar and bad oral hygiene and respond to traditional self-care. Our findings give intriguing insights into the etiology of caries, and a rationale for typing individuals for risk in individualized oral care. In this way, high risk individuals may be identified and treated before caries and other symptoms arise, when these conditions may be more easily cured or prevented than when fully established (The Look AHEAD Research Group, 2013). Moreover, missing teeth from chronic infection and inflammation constitutes a risk factor for cardio-vascular diseases (Vedin et al., 2015) and both S. mutans adhesin biotypes of high virulence and caries susceptibility may accentuate this risk (Nakano et al., 2011, Watanabe et al., 2016). Our findings also provide a rationale for developing novel therapeutics based on PRH-PRB and dmbt1 encoded functions, as high susceptibility individuals may require intensified traditional prevention and ultimately innate immunity supplementation or immunostimulatory therapeutics.
Conflicts of Interest
The authors have no competing interests to declare.
Author Contributions
Author contributions: N.S., C.K designed research; A.E., N. Sheng and A.L-B. performed research; N.S., C.K., N. Sheng, L.M., K.D. collected clinical data and samples; N.S., C.K., A.E., N. Sheng, L.M., analyzed data; N.S. wrote the paper.
Acknowledgement
This work was supported by the Swedish Research Council (10906), the Faculty of Medicine of Umeå University, and Västerbotten County Council. We acknowledge financial support provided through a regional agreement between Umeå University and Västerbotten County Council in the field of Medicine, Odontology, and Health (377341) and through a Fund for Cutting-Edge Medical Research grant from the County Council of Västerbotten (135041). This study was also supported by the Swedish Dental Society and local foundations at Umeå University. The work was performed in part at Umeå Centre for Chronic Infectious Disease (UCCID). Ingmarie Bernhardsson, Ewa Strömqvist-Engbo, Ulla Öhman are acknowledged for their assistance. We thank all families and public health care personnel who participated in the study.
Genotyping was performed by the SNP&SEQ Technology Platform in Uppsala, which is part of the Science for Life Laboratory at Uppsala University and supported as national infrastructure by the Swedish Research Council.
The funders had no role in study design; data collection, analysis or interpretation; or writing of the report.
A patent application has been filed; no. PCT/SE/2017/000028.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2017.11.019.
Appendix A. Supplementary data
Supplementary material
References
- Ayad M., Van Wuyckhuyse B.C., Minaguchi K., Raubertas R.F., Bedi G.S., Billings R.J., Bowen W.H., Tabak L.A. The association of basic proline-rich peptides from human parotid gland secretions with caries experience. J. Dent. Res. 2000;79:976–982. doi: 10.1177/00220345000790041401. [DOI] [PubMed] [Google Scholar]
- Azen E.A. Genetics of salivary protein polymorphisms. Crit. Rev. Oral Biol. Med. 1993;4:479–485. doi: 10.1177/10454411930040033201. [DOI] [PubMed] [Google Scholar]
- Azen E.A., Yu P.-L. Genetic polymorphisms of Pe and Po salivary proteins with probable linkage of their genes to the salivary protein gene complex (SPC) Biochem. Genet. 1984;22:1065–1079. doi: 10.1007/BF00499632. [DOI] [PubMed] [Google Scholar]
- Bennick A. Structural and genetic aspects of proline-rich proteins. J. Dent. Res. 1987;66:457–461. doi: 10.1177/00220345870660021201. [DOI] [PubMed] [Google Scholar]
- Boraas J.C., Messer L.B., Till M.J. A genetic contribution to dental caries, occlusion, and morphology as demonstrated by twins reared apart. J. Dent. Res. 1988;67:1150–1155. doi: 10.1177/00220345880670090201. [DOI] [PubMed] [Google Scholar]
- Bradshaw D.J., McKee A.S., Marsh P.D. Effects of carbohydrate pulses and pH on populations shifts within oral microbial communities in vitro. J. Dent. Res. 1989;68:263–271. doi: 10.1177/00220345890680090101. [DOI] [PubMed] [Google Scholar]
- Burgener A., Mogk K., Westmacott G., Plummer F., Ball B., Broliden K., Hasselrot K. Salivary basic proline-rich proteins are elevated in HIV-exposed seronegative men who have sex with men. AIDS. 2012;26:1857–1867. doi: 10.1097/QAD.0b013e328357f79c. [DOI] [PubMed] [Google Scholar]
- Dawes C. Salivary flow patterns and the health of hard and soft oral tissues. J. Am. Dent. Assoc. 2012;39:18–24. doi: 10.14219/jada.archive.2008.0351. [DOI] [PubMed] [Google Scholar]
- Drobni M., Olsson I.-M., Eriksson C., Almqvist F., Strömberg N. Multivariate design and evaluation of a set of RGRPQ-derived innate immunity peptides. J. Biol. Chem. 2006;281:15164–15171. doi: 10.1074/jbc.M511727200. [DOI] [PubMed] [Google Scholar]
- Eriksson L., Antti H., Gottfries J., Holmes E., Johansson E., Lindgren F., Long I., Lundstedt T., Trygg J., Wold S. Using chemometrics for navigating in the large data sets of genomics, proteomics, and metabonomics (gpm) Anal. Bioanal. Chem. 2004;380:419–429. doi: 10.1007/s00216-004-2783-y. [DOI] [PubMed] [Google Scholar]
- Esberg A., Löfgren-Burström A., Öhman U., Strömberg N. Host and bacterial phenotype variation in adhesion of Streptococcus mutans to matched human hosts. Infect. Immun. 2012;80:3869–3879. doi: 10.1128/IAI.00435-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esberg A., Mårell L., Claesson R., Persson K., Borén T., Strömberg N. Streptococcus mutans adhesin biotypes that match and predict individual caries development. EBioMedicine. 2017;24:205–215. doi: 10.1016/j.ebiom.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons R.J. Bacterial adhesion to oral tissues: a model for infectious diseases. J. Dent. Res. 1989;68:750–760. doi: 10.1177/00220345890680050101. [DOI] [PubMed] [Google Scholar]
- Hay D.I., Ahern J.M., Schluckebier S.K., Schlesinger D.H. Human salivary acidic prolin-rich protein polymorphisms and biosynthesis studied by high-performance liquid chromatography. J. Dent. Res. 1994;73:1717–1726. doi: 10.1177/00220345940730110701. [DOI] [PubMed] [Google Scholar]
- Jonasson A., Eriksson C., Jenkinson H.F., Källestål C., Johansson I., Strömberg N. Innate immunity glycoprotein gp-340 variants may modulate human susceptibility to dental caries. BMC Infect. Dis. 2007;7:57. doi: 10.1186/1471-2334-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Källestål C. The effect of five years´ implementation of caries-preventive methods in Swedish high-risk adolescents. Caries Res. 2005;39:20–26. doi: 10.1159/000081652. [DOI] [PubMed] [Google Scholar]
- Källestål C., Stenlund H. Different analytical approaches in an experimental cohort study on preventive measures for caries in adolescents. A comparison between incidence density and increment analysis. Caries Res. 2003;37:44–50. doi: 10.1159/000068219. [DOI] [PubMed] [Google Scholar]
- Kassebaum N.J., Bernabé E., Dahiya M., Bhandari B., Murray C.J., Marcenes W. Global burden of untreated caries: a systematic review and metaregression. J. Dent. Res. 2015;94:650–658. doi: 10.1177/0022034515573272. [DOI] [PubMed] [Google Scholar]
- Kolenbrander P.E., Andersen R.N., Blehert D.S., Egland P.G., Foster J.S., Palmer J.R. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 2002;66:486–505. doi: 10.1128/MMBR.66.3.486-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasse B. The Vipelolm dental caries study: recollections and reflections 50 years later. J. Dent. Res. 2001;80:1785–1788. doi: 10.1177/00220345010800090201. [DOI] [PubMed] [Google Scholar]
- Lehner T., Lamb J.R., Welsh K.L., Batchelor R.J. Association between HLA-DR antigens and helper cell activity in the control of dental caries. Nature. 1981;292:770–772. doi: 10.1038/292770a0. [DOI] [PubMed] [Google Scholar]
- Listl S., Galloway J., Mossey P.A., Marcenes W. Global economic impact of dental diseases. J. Dent. Res. 2015;94:1355–1361. doi: 10.1177/0022034515602879. [DOI] [PubMed] [Google Scholar]
- Loimaranta V., Jakubovics N.S., Hytönen J., Finne J., Jenkinson H.F., Strömberg N. Fluid- or surface-phase human salivary scavenger protein gp340 exposes different bacterial recognition properties. Infect. Immun. 2005;73:2245–2252. doi: 10.1128/IAI.73.4.2245-2252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen J., Mollenhauer J., Holmskov U. Gp-340/DMBT1 in mucosal innate immunity. Innate Immun. 2010;16:160–167. doi: 10.1177/1753425910368447. [DOI] [PubMed] [Google Scholar]
- Nakano K., Hokamura K., Taniguchi N., Wada K., Kudo C., Nomura R., Kojima A., Naka S., Muranaka Y., Thura M., Nakajima A., Masuda K., Nakagawa I., Speziale P., Shimada N., Amano A., Kamisaki K., Tanaka T., Umemura K., Ooshima T. The collagen-binding protein of Streptococcus mutans is involved in haemorrhagic stroke. Nat. Commun. 2011;2:1–10. doi: 10.1038/ncomms1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund Å., Johansson I., Källestål C., Ericson T., Sjöström M., Strömberg N. Improved ability of biological and previous caries multimarkers to predict caries disease as revealed by multivariate PLS modelling. BMC Oral Health. 2009;9:1–12. doi: 10.1186/1472-6831-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakobphol A., Xu F., Hoang V.M., Larsson T., Bergstrom J., Johansson I., Frängsmyr L., Holmskov U., Leffler H., Nilsson C., Borén T., Wright J.R., Strömberg N., Fisher S.J. Salivary agglutinin, which binds Streptococcus mutans and Helicobacter pylori, is the lung scavenger receptor cysteine-rich protein gp-340. J. Biol. Chem. 2000;275:39860–39866. doi: 10.1074/jbc.M006928200. [DOI] [PubMed] [Google Scholar]
- Ren Y., Jongsma M.A., Mei L., van der Mei H.C., Busscher H.J. Orthodontic treatment with fixed appliances and biofilm formation - a potential public health threat? Clin. Oral Invest. 2014;18:1711–1718. doi: 10.1007/s00784-014-1240-3. [DOI] [PubMed] [Google Scholar]
- Renner M., Bergmann G., Krebs I., End C., Lyer S., Hilberg F., Helmke B., Gassler N., Autschbach F., Bikker F., Strobel-Friedekind O., Gronert-Sum S., Benner A., Blaich S., Wittig R., Hudler M., Lightenberg A.J., Madsen J., Holmskov U., Annese V., Latiano A., Schirmacher P., Nieuw Amerongen A.V., D'amato M., Kioschis P., Hafner M., Poutska A., Mollenhauer J. DMBT1 confers mucosal protection in vivo and a deletion variant is associated with Crohn's disease. Gastroenterology. 2007;133:1499–1509. doi: 10.1053/j.gastro.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Selwitz R.H., Ismail A., Pitts N.B. Dental caries. Lancet. 2007;369:51–59. doi: 10.1016/S0140-6736(07)60031-2. [DOI] [PubMed] [Google Scholar]
- de Soet J.J., Nyvad B., Kilian M. Strain-related acid production by oral streptococci. Caries Res. 2000;34:486–490. doi: 10.1159/000016628. [DOI] [PubMed] [Google Scholar]
- Stenudd C., Nordlund Å., Ryberg M., Johansson I., Källestål C., Strömberg N. The association of bacterial adhesion with dental caries. J. Dent. Res. 2001;80:2005–2010. doi: 10.1177/00220345010800111101. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium The international HapMap project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- The Look AHEAD Research Group Cardiovascular effects of intensive lifestyle intervention in type 2 diabetes. N. Engl. J. Med. 2013;2013(369):145–154. doi: 10.1056/NEJMoa1212914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedin O., Hagström E., Gallup D., Neely M.L., Stewart R., Koenig W., Budaj A., Sritara P., Wallentin L., White H.D., Held C. Periodontal disease in patients with chronic coronary heart disease: prevalence and association with cardiovascular risk factors. Eur. J. Prev. Cardiol. 2015;22:771–778. doi: 10.1177/2047487314530660. [DOI] [PubMed] [Google Scholar]
- Vieira A.R., Maritza M.L., Goldstein-McHenry T. Genome wide scan finds suggestive caries loci. J. Dent. Res. 2008;87:435–439. doi: 10.1177/154405910808700506. [DOI] [PubMed] [Google Scholar]
- Wang N., Shen N., Vyse T.J., Anand V., Gunnarson I., Sturfelt G., Rantapää-Dahlqvist S., Elvin K., Truedsson L., Andersson B.A., Dahle C., Örtqvist E., Gregersen P.K., Behrens T.W., Hammarström L. Mol. Med. 2011;17:1383–1396. doi: 10.2119/molmed.2011.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe I., Kuriyama N., Miyatani F., Nomura R., Naka S., Nakano K., Ihara M., Iwai K., Matsui D., Ozaki E., Koyama T., Nishigaki M., Yamamoto T., Tamura A., Mizuno T., Akazawa K., Takada A., Takeda K., Yamada K., Nakagawa M., Tanaka T., Kanamura N., Friedland R.P., Watanabe Y. Oral Cnm-positive Streptococcus Mutans expressing collagen binding activity is a risk factor for cerebral microbleeds and cognitive impairment. Sci. Rep. 2016;6:38561. doi: 10.1038/srep38561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werneck R.I., Mira M.T., Trevilatto P.C. A critical review: an overview of genetic influence on dental caries. Oral Dis. 2010;16:613–623. doi: 10.1111/j.1601-0825.2010.01675.x. [DOI] [PubMed] [Google Scholar]
- Yano A., Kaneko N., Ida H., Yamaguchi T., Hanada N. Real-time PCR for quantification of Streptococcus mutans. FEMS Microbiol. Lett. 2002;19:23–30. doi: 10.1111/j.1574-6968.2002.tb11451.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material