Abstract
Arylamine N-acetyltransferase 1 (NAT1) expression is reported to affect proliferation, invasiveness, and growth of cancer cells. MDA-MB-231 breast cancer cells were engineered such that NAT1 expression was elevated or suppressed, or treated with a small molecule inhibitor of NAT1. The MDA-MB-231 human breast cancer cell lines were engineered with a scrambled shRNA, a NAT1 specific shRNA or a NAT1 overexpression cassette stably integrated into a single flippase recognition target (FRT) site facilitating incorporation of these different genetic elements into the same genomic location. NAT1-specific shRNA reduced NAT1 activity in vitro by 39%, increased endogenous acetyl coenzyme A levels by 35%, and reduced anchorage-independent growth (7-fold) without significant effects on cell morphology, growth rates, anchorage-dependent colony formation, or invasiveness compared to the scrambled shRNA cell line. Despite 12-fold overexpression of NAT1 activity in the NAT1 overexpression cassette transfected MDA-MB-231 cell line, doubling time, anchorage-dependent cell growth, anchorage-independent cell growth, and relative invasiveness were not changed significantly when compared to the scrambled shRNA cell line. A small molecule (5E)-[5-(4-hydroxy-3,5-diiodobenzylidene)-2-thioxo-1,3-thiazolidin-4-one (5-HDST) was 25-fold more selective towards the inhibition of recombinant human NAT1 than N-acetyltransferase 2. Incubation of MDA-MB-231 cell line with 5-HDST resulted in 60% reduction in NAT1 activity and significant decreases in cell growth, anchorage-dependent growth, and anchorage-independent growth. In summary, inhibition of NAT1 activity by either shRNA or 5-HDST reduced anchorage-independent growth in the MDA-MB-231 human breast cancer cell line. These findings suggest that human NAT1 could serve as a target for the prevention and/or treatment of breast cancer.
Keywords: Human arylamine N-acetyltransferase 1 (NAT1), MDA-MB-231, Breast Cancer, Acetyl coenzyme A
INTRODUCTION
Arylamine N-acetyltransferase 1 (NAT1) historically is identified as a phase II drug and carcinogen metabolizing enzyme that catalyzes the transfer of an acetyl group from acetyl-coenzyme A (AcCoA) to the amine nitrogen of arylamine and hydrazine compounds. Recently human NAT1 was shown to catalyze hydrolysis of AcCoA in the presence of folate [1,2].
NAT1 expression is elevated in invasive and lobular breast carcinomas when compared to normal breast tissue [3]. This overexpression of NAT1 was confirmed at the mRNA level and by immunohistological analysis in 108 breast cancer samples that demonstrated a strong association of NAT1 staining and estrogen receptor-positive tumors [3]. Elevated NAT1 expression in estrogen receptor-positive tumors has been observed in multiple studies [4,5]. Also, elevated NAT1 expression is associated with breast cancer bone metastasis [6,7]. Overexpression of NAT1 in a normal luminal epithelial-derived cell line conferred enhanced growth properties and etoposide resistance compared to control cells that had been transfected with an empty vector [3]. Subsequent studies demonstrated that a small molecule inhibitor of NAT1 activity caused decreased cell growth and reduced ability of MDA-MB-231 human breast cancer cells to grow in soft agar [8]. A lentiviral-based NAT1 shRNA, which reduced NAT1 activity by 74%, reduced cell invasion by 50% in MDA-MB-231 human breast cancer cells [8]. NAT1 activity is cell-cycle phase-dependent showing an association between high NAT1 activity and accelerated progression through the cell cycle in HaCaT keratinocytes [9]. RNAi-mediated knockdown of NAT1 by approximately 85% in the colon cancer cell line HT-29 is associated with increased cell-cell contact inhibition, loss of cell viability at confluence, and attenuation of anchorage-independent cell growth [10].
Human NAT1 activity is modified by polymorphism, but can also be regulated by microRNA, epigenetic, and/or translational and post-translational control [5,11–13]. NAT1 transcript levels can be regulated by extracellular stimuli acting on glucocorticoid or androgen receptors [14,15]. Exposure to NAT1 substrates can increase NAT1 degradation, which results in decreased NAT1 activity [12,13]. Oxidative stress can reversibly or irreversibly inactivate NAT1 [16,17]. NAT1 inhibitors have been reported including cisplatin [18], curcumin [19,20], disulfiram [21], and rhodanine derivatives [8,22]. The ability to control the level of NAT1 activity and expression by various means may reduce cancer growth or metastasis.
To investigate further whether modulation of NAT1 activity can affect cancer cell growth and invasion properties, MDA-MB-231 human breast cancer cells were engineered with either a scrambled shRNA, a NAT1 specific shRNA or a NAT1 overexpression cassette. These modifications were stably integrated into a single flippase recognition target (FRT) site facilitating incorporation of these different genetic elements into the same genomic location. We also investigated the effects of an NAT1 small molecule inhibitor. We used the Sure Silencing™ shRNA plasmid described previously [10], and a rhodanine derivative (5E)-[5-(4-hydroxy-3,5-diiodobenzylidene)-2-thioxo-1,3-thiazolidin-4-one] (5-HDST) similar to the small molecule NAT1 inhibitor reported previously [22].
MATERIALS AND METHODS
Cell line generation and culturing conditions
The breast adenocarcinoma cell line, MDA-MB-231, was purchased from ATCC (Manassas, Virginia). Cells were cultured in Dulbecco’s Modified Eagles Medium (DMEM) media, high glucose with the addition of fetal bovine serum to 10% and glutamine to 2 mM. SureSilencing™ Predesigned shRNA and NAT1 knockdown shRNA plasmids were purchased from SA Biosciences. Initial screening found that the NAT1 shRNA Clone 1 reduced NAT1 activity by 29% while NAT1 shRNA Clone 3 reduced NAT1 activity by 50%. Further studies were conducted with NAT1 shRNA Clone 3 because it was used in previous experiments [10] and resulted in the greatest reduction in NAT1 expression. The shRNA sequence for the scrambled shRNA and NAT1 shRNA clone 3 were transferred from the pGeneclip™ vector to the pcDNA5/FRT vector. The NAT1 overexpression plasmid NATb/NAT1*4 in pcDNA5/FRT was described previously [23].
A MDA-MB-231 breast cancer cell line with a single FRT site was constructed using the Flp-In System (Life Technologies, Grand Island, NY) as described previously [24]. Once an MDA-MB-231 cell line was constructed with a single FRT site in a transcriptionally active region, we transfected the shRNA plasmids and the NAT1 overexpression plasmid to deliver the cassettes to the same genetic location. The pcDNA5/FRT plasmids containing the scrambled shRNA, NAT1 specific shRNA plasmid or the NAT1 overexpression plasmid NATb/NAT1*4 [23] were co-transfected with pOG44, a Flp recombinase expression plasmid, using Amaxa Cell Line Nucleofector Kit V (Lonza, Basel, Switzerland) following the manufacturer’s recommendations. Since the pcDNA5/FRT vector contains a hygromycin resistance cassette, cells were passaged in complete DMEM containing 500 μg/mL hygromycin to select for cells with the pcDNA5/FRT plasmid stably integrated into their genomic DNA. Hygromycin-resistant colonies were selected approximately 2 weeks after transfection and isolated using cloning cylinders. Unless otherwise stated, experiments using cell lines were performed with cells in log phase of growth.
In summary, the following MDA-MB-231 cell lines were constructed:
Scrambled shRNA cell line stably transfected with scrambled shRNA as a control cell line to which other cell lines are compared.
NAT1 knockdown cell line stably transfected with NAT1 specific shRNA.
NAT1 overexpression cell line stably transfected with the NATb/NAT1*4 cassette.
The constructed MDA-MB-231 cell lines were authenticated by the ATCC Short Tandem Repeat (STR) profiling cell authentication service.
Measurement of N-acetylation in vitro
In vitro assays using the NAT1 selective substrate para-aminobenzoic acid (PABA) were conducted and the N-acetyl-PABA product was separated and quantitated as described [25]. Briefly, cells were scraped from the plate, washed in 1X phosphate-buffered saline (PBS), and lysed in 20 mM sodium phosphate pH 7.4, 1 mM ethylenediaminetetraacetic acid (EDTA), 0.2% triton X-100, 1 mM dithiothreitol (DTT), 100 μM phenylmethanesulfonyl fluoride (PMSF), 1 μg/mL aprotinin, and 2 μM pepstatin A. Cell lysate was centrifuged at 15,000 × g for 10 min and supernatant tested for NAT1 activity. Enzymatic reactions containing 50 μL suitably diluted cell lysate, PABA (300 μM) and AcCoA (1 mM) were incubated at 37°C for 10 min. Reactions were terminated by the addition of 1/10 total reaction volume of 1 M acetic acid and centrifuged at 15,000 X g for 10 min. Three independent measurements (N=3) performed in triplicate were completed for each cell line.
Characterization of recombinant human NAT1 and NAT2 inhibition by 5-HDST
Studies to characterize human NAT1 and NAT2 inhibition by 5-HDST were carried out in vitro following recombinant expression of human NAT1 (NAT1*4) and NAT2 (NAT2*4) in yeast (Schizosaccaromyces pombe) as previously described [26–28]. Suitably diluted recombinant human NAT1 was incubated with 50 μM PABA and 100 μM AcCoA in the presence of 0.005 to 7.81 μM 5-HDST for 10 min reactions. N-acetyl-PABA product was separated and quantitated as described above. Suitably diluted recombinant human NAT2 was incubated with 50 μM sulfamethazine (NAT2 selective substrate) and 100 μM AcCoA in the presence of 1.9 to 500 μM 5-HDST for 10 min reactions. N-acetyl-sulfamethazine product was separated and quantitated as previously described [26–27]. IC50 values for 5-HDST inhibition of recombinant human NAT1 and NAT2 were generated and analyzed using GraphPad Software (San Diego, CA). To further characterize the 5-HDST inhibition, suitably diluted recombinant human NAT1 was incubated with 25- to 400 μM PABA and 100 μM AcCoA in the presence of 0, 0.33, 1.0 and 3.0 μM 5-HDST for 10 min reactions. N-acetyl-PABA product was separated and quantitated as described above. Lineweaver-Burke inhibition plots were generated and analyzed using GraphPad Software.
Measurement of N-acetylation in situ
Measurement of N-acetylation in situ was determined by spiking media with a known concentration of PABA as described [29]. The cells were incubated at 37°C for 48 hours with media containing 500 μM PABA. Media were collected and 1/10 total collected media volume of 1 M acetic acid was added. Acidified media were centrifuged at 15,000 X g for 10 min. There were four independent measurements (N=4) performed in triplicate for each cell line.
Resulting supernatants from in vitro and in situ NAT1 activity assays were injected into a reverse phase C18 column (125 mm × 4 mm; 5 μM pore size). Reactants and products were eluted using a Beckman System Gold high performance liquid chromatography (HPLC) system. HPLC separation of N-acetyl-PABA was achieved using a gradient of 96:4 sodium perchlorate pH 2.5: acetonitrile at 280 nm. The data for scrambled shRNA, knockdown and overexpression cell lines are represented as mean ± SEM and analyzed statistically by a one-way ANOVA and Bonferroni post test.
Measurement of endogenous AcCoA levels
Endogenous AcCoA levels within modified MDA-MB-231 cancer cell lines (scrambled shRNA, knockdown, and overexpression) were determined by HPLC. Cell growth conditions were strictly monitored because it was observed that cellular confluence and period of growth affected the consistency of data obtained for the endogenous AcCoA concentrations. Cells were plated at 1 × 106 per 10 cm plate (3 plates per N) with fresh complete culturing media with no selection antibiotic added, and allowed to grow for 72 hours. After 72 hours, the cells were 70% confluent and collected. Media were aspirated from the plates and each plate was washed with 5 mL PBS. Then cells were dissociated from the plate with 0.5 mL trypsin [0.25% (w/v)]. Cells were suspended in 4 mL of diluted complete media (20% medium and 80% PBS) and counted in 0.5 mL aliquots. In the subsequent steps all cells and lysates were kept on ice or at 4°C. Collected cells were centrifuged, supernatant removed, and ice-cold PBS was added. Again, cells were centrifuged, supernatant removed, suspended in 1 mL of ice-cold PBS, and transferred to 1.5 mL microcentrifuge tubes. The suspended cells were collected by centrifugation and the supernatants discarded. Having removed any residual PBS, the cells were lysed in 100 μL ice-cold 5% 5-sulfosalicylic acid with a 1 mL BD Insulin Syringe with permanently attached 28 gauge needle (BD, Franklin Lakes, NJ, USA). The needle selection is important to ensure complete cell lysis without loss of volume or cellular masses within the syringe/needle. The cellular lysate was then centrifuged at 15,000 × g for 10 min. Supernatant was filtered through a syringe filter (13 mm, 0.20 μm pore size). Filtrate was collected and separated on a C18 reverse-phase HPLC column (250 mm × 4 mm; 5 μm pore size). HPLC separation and quantitation of AcCoA was achieved using a linear gradient of 100% 55 mM sodium phosphate pH 4.0: 0% methanol to 0% 55 mM sodium phosphate pH 4.0: 100% methanol over 20 min and was quantitated by absorbance at 260 nm. The data for individual determinations of scrambled shRNA, knockdown and overexpression cell lines (N= 8, 7, and 7 respectively) are represented as mean ± SEM, and were analyzed by one-way ANOVA and Bonferroni post test.
Quantitation of NAT1 mRNA levels
NAT1 mRNA levels were measured as described previously [29]. Briefly, total RNA was isolated from the constructed MDA-MB-231 cell lines using the RNeasy Mini kit (Qiagen, Germantown, MD) following manufacturer’s instructions. cDNA was made using the High Capacity Reverse Transcriptase kit (Life Technologies) following manufacturer’s instructions. mRNA was quantitated via Real-time PCR using Taqman primers and probe (Life Technologies) 5′-gaattcaagccaggaagaagca-3′ 5′tccaagtccaatttgttcctagact-3′ FAM-5′-caatctgtcttctggattaa-3′-MGB and PCR conditions suggested by the manufacturer. The data for scrambled shRNA, knockdown and overexpression cell lines are represented as mean ± SEM from three separate determinations performed in triplicate (N=3), and were analyzed statistically by one-way ANOVA and Bonferroni post hoc test.
Cell morphology
Cells were plated at an initial density of 750,000 cells/10 cm-plate and allowed to grow for 96 hours at 37°C and 5% CO2. Cells were photographed with a 20× objective at 24 hour intervals on a Nikon Eclipse Ti inverted phase contrast light microscope using NIS-Elements AR Acquisition software (Nikon Instruments Inc., Melville, NY, USA).
Determination of doubling time of cell lines
Twenty-five thousand cells were plated in triplicate in 6-well plates and allowed to grow for 5 consecutive days. Cells were plated on day 1 and allowed to equilibrate for 24 hours before making the first count on day 2 followed by counts on days 3, 4, 5 and 6 using a cell counter. A cell doubling rate was calculated for each day using Equation 1 (shown below) and then rates were averaged over the 5 day time period to get a final cell doubling rate.
| Equation 1 |
In the equation, b = cell number on day n, a = cell number on day n-1, tb = hours passed since plating when cells were counted on day n, and ta = hours passed since plating when cells were counted on day n-1. This assay was performed in triplicate for each cell line (N=3). The data for the cell lines are represented as mean ± SEM from three separate determinations performed in triplicate, and were analyzed statistically by a one-way ANOVA and Bonferroni post hoc test.
Measuring the effect of a small molecule inhibitor on in situ NAT1 activity and cell growth
The ability of the small molecule inhibitor (5E)-[5-(4-hydroxy-3,5-diiodobenzylidene)-2-thioxo-1,3-thiazolidin-4-one] (5-HDST, CAS# 336171-85-8) (Ryan Scientific, Mount Pleasant, SC) to inhibit NAT1 activity was measured in situ and its effects on cell growth measured as a percentage of untreated cells. Scrambled shRNA MDA-MB-231 cells were incubated with a single dose of 5-HDST (ranging from 0–87.5 μM) for up to 6 days. Briefly, all cells were plated on day 0 and the varying concentrations of 5-HDST were added to the respective cells on day 1. Cells were plated in 12-well plates so that every successive day would have roughly the same number of cells to enable accurate measurement of PABA N-acetylation in situ and cell number. PABA (10 μM) was added to the media and incubated with the cells and 5-HDST for 24 hours prior to measurements. After 24 hours, media were removed from the cells and amounts of PABA and N-acetyl-PABA were measured by HPLC as described above. Cells were washed once with 1X PBS, scraped off the plate and counted using a cell counter. Three separate determinations performed in triplicate for PABA N-acetylation and cell number were carried out every 24 hours for 6 days (N=3). The in situ NAT1 data, represented as means ± SEM, were analyzed statistically by a one-way ANOVA and Bonferroni post hoc test on days 1 thru 6. The cell number data, represented as means ± SEM, were analyzed statistically by a two-way ANOVA and Bonferroni post- test.
Anchorage-dependent and anchorage-independent growth assays
Anchorage-dependent growth assays were performed by plating cells (300 cells/well) in triplicate in 6-well plates and allowing them to grow for 2 weeks (N=3). For small molecule NAT1 inhibitor assays, 5-HDST (ranging from 0 to 87.5 μM) was added to triplicate wells at the time of plating (N=3). Colonies were counted following staining with crystal violet. The data are represented as means ± SEM from three separate determinations performed in triplicate and were analyzed statistically by a one-way ANOVA and Bonferroni post hoc test.
The anchorage-independent growth assays were performed by plating the cells in low-melting temperature agarose. The cells were grown in normal growth media where the bottom layer was 1.5 mL (noble agar, 0.5%) and the top layer was 1.5mL (low-melting temperature agarose, 0.3%), so the total volume was 3 mL in each well of a 6 well plate. Cells (6000 cells/well) were plated in triplicate in 6-well plates and grown for 2 weeks. Colonies were counted following staining with crystal violet. For the anchorage-independent growth assay investigating effect of the small molecule NAT1 inhibitor, 5-HDST (ranging from 0 to 87.5 μM) was added at the time cells were plated. Three separate determinations performed in triplicate were completed for each assay (N=3). The data are represented as means ± SEM and were analyzed statistically by a one-way ANOVA and Bonferroni post- test.
Transwell assays to assess relative invasive ability
Transwell assays were performed by depositing 1 × 105 cells in 100 μL of DMEM containing 0.2% FBS in the top chamber of the transwell and 600 μL of complete media in the bottom chamber. 5-HDST (ranging from 0 to 100 μM) was added to media on both sides of the membrane. Cells were allowed to invade across a membrane of Matrigel (Life Technologies). After 24 hours, cells on the top side of the membrane were removed with a sterile cotton swab. Cells that invaded across the membrane were quantified using the Alamar Blue cell quantitation assay. Three separate determinations performed in triplicate were made (N=3). The data are represented as means ± SEM, and were analyzed statistically by one-way ANOVA and Bonferroni post hoc test.
RESULTS
Effects of reduced and elevated NAT1 activity on MDA-MB-231 breast cancer cells
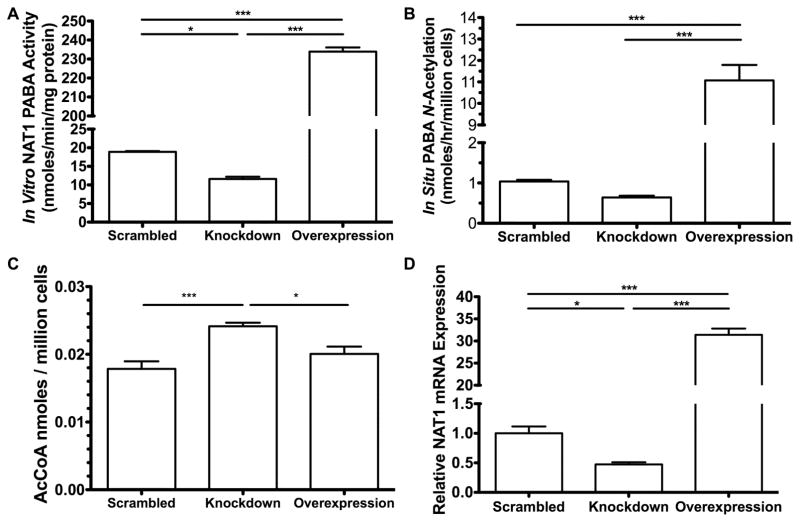
The stable partial knockdown of NAT1 reduced in vitro PABA NAT1 activity to 61% of the scrambled shRNA cell line (Figure 1A) and in situ PABA NAT1 acetylation to 62% of the scrambled shRNA cell line (Figure 1B). Stable overexpression of NAT1 increased the N-acetylation of PABA in vitro and in situ by 12-and 11-fold, respectively, compared to the scrambled shRNA cell line (Figure 1 A&B). The partial knockdown of NAT1 in NAT1 shRNA knockdown MDA-MB-231 breast cancer cells increased endogenous AcCoA levels about 35% compared to the scrambled shRNA cell line (p<0.005) (Figure 1C). Endogenous AcCoA levels in the NAT1 overexpression MDA-MB-231 cell line did not differ significantly (p>0.05) compared to the scrambled shRNA cell line but were significantly below the AcCoA levels of the partial NAT1 knockdown cell line (p<0.05) (Figure 1C). We observed that endogenous AcCoA levels were reduced in all cell lines when grown to over-confluence compared to the 70% confluent cell lines (data not shown). NAT1 mRNA expression in the stable knockdown was 47% of the scrambled shRNA cell line, and was 31-fold higher than the scrambled shRNA cell line in the stable NAT1 overexpression cell line (Figure 1D).
Figure 1. NAT1 activity and mRNA expression in stably modified MDA-MB-231 cell lines.
Each bar illustrates mean ± SEM. A. The in vitro PABA N-acetylation activity in cell line lysates (N=3). B. The in situ PABA N-acetylation in cell lines cultured in PABA spiked media (N=4). C. The endogenous AcCoA levels for scrambled shRNA (N=8), NAT1 shRNA knockdown (N=7), and NAT1 overexpression (N= 7). D. NAT1 mRNA levels in cell lines normalized to scrambled shRNA cell line (N=3). Differences between cell lines were significantly different (p* ≤ 0.05; p*** ≤ 0.001) following one-way ANOVA with a Bonferroni post hoc test.
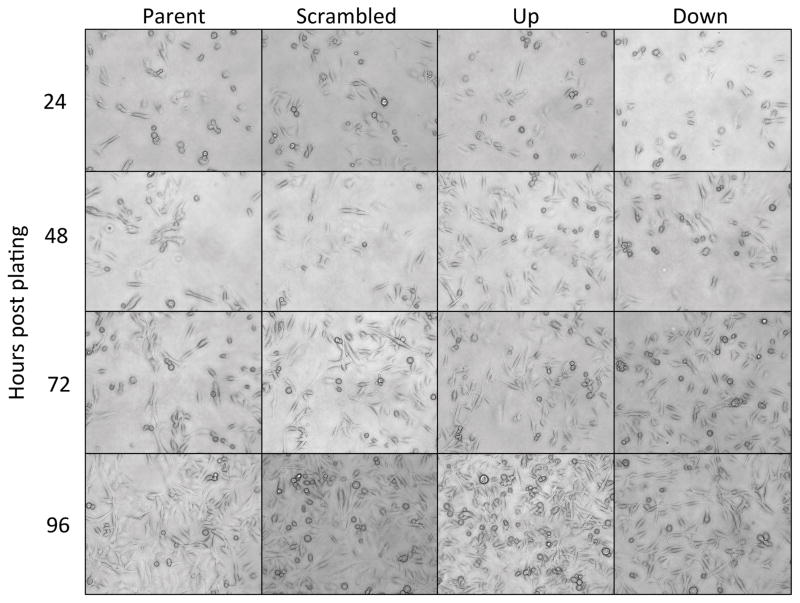
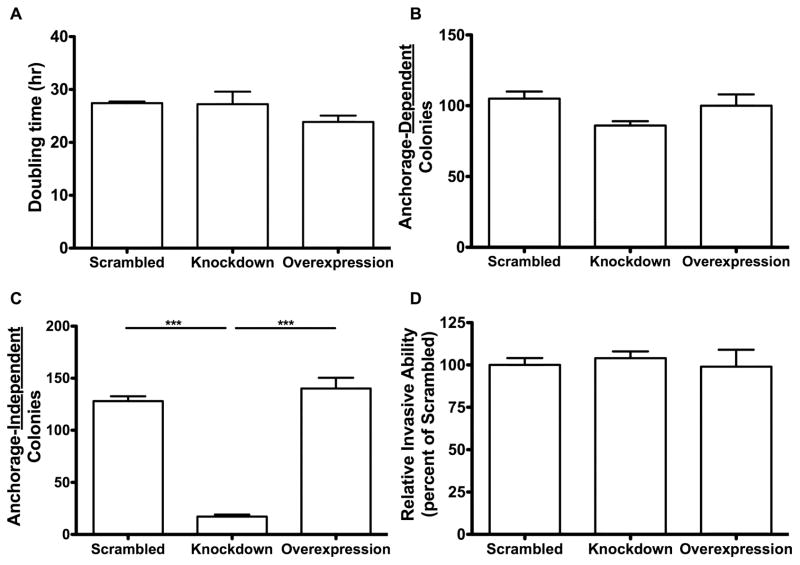
Morphological differences were not observed among the four cell lines (Parent, Scrambled, Overexpression, Knockdown) at 24, 48, 72, or 96 hours (Figure 2). Neither the knockdown nor overexpression of NAT1 significantly (p=0.26) affected the doubling time compared to the scrambled cell line. The doubling times for the scrambled shRNA cell line, NAT1 knockdown cell line and NAT1 overexpression cell line were 27.4, 27.2 and 23.4 hours respectively (Figure 3A). Neither the knockdown nor overexpression of NAT1 significantly affected anchorage-dependent growth (Figure 3B). However, the knockdown of NAT1 caused a significant ~85% decrease (p<0.0001) in anchorage-independent growth, whereas the overexpression of NAT1 had no significant effect (Figure 3C). Neither knockdown of NAT1 with shRNA nor overexpression of NAT1 had a significant effect on relative invasive ability (Figure 3D).
Figure 2. Cell morphologies for MDA-MB-231 cells.
Representative photographs (20X objective) of the parent, scramble shRNA, NAT1 shRNA knockdown (down), and NAT1 overexpression (up) MDA-MB-231 cell lines were taken at 24, 48, 72, and 96 hours.
Figure 3. Cell growth properties of MDA-MB-231 cell lines stably modified with shRNA NAT1 knockdown and NAT1 overexpression.
Each bar illustrates mean ± SEM. A. Cell doubling times in the scrambled shRNA, NAT1 knockdown and NAT1 overexpression cell lines did not differ significantly following one-way ANOVA (p>0.05) (N=3). B. The anchorage-dependent growth did not differ significantly among the scrambled shRNA, NAT1 knockdown and NAT1 overexpression cell lines following one-way ANOVA (p>0.05) (N=3). C. The NAT1 knockdown cell line formed significantly less anchorage-independent colonies compared to scrambled shRNA and NAT1 overexpressing cell lines following one-way ANOVA followed by a Bonferroni post hoc test (p*** ≤ 0.001) (N=3). D. No significant differences in relative invasive ability among the scrambled shRNA, NAT1 knockdown and NAT1 overexpression cell lines were observed following one-way ANOVA (p>0.05) (N=3).
Effects of small molecule NAT1 inhibitor on MDA-MB-231 breast cancer cells
To investigate further whether the effects of NAT1 knockdown on cell growth and invasion ability were due to reduced NAT1 activity, we tested the effects of a small molecule inhibitor of NAT1.
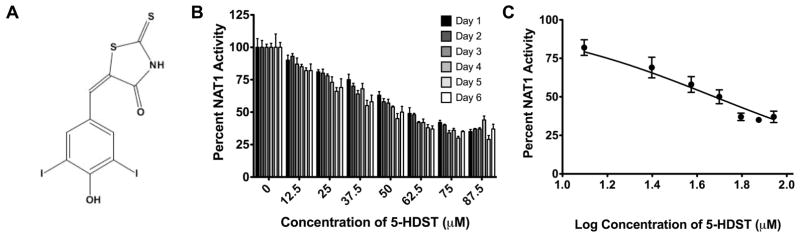
5-HDST (Figure 4A) significantly (p<0.0001) reduced PABA N-acetylation in situ in a concentration-dependent manner (Figure 4B). A single dose of 5-HDST reduced PABA N-acetylation in situ for at least 6-days (Figure 4B). The IC50 for 5-HDST inhibition of PABA N-acetylation in situ was similar on day 1 (67. 1 μM) and day 6 (47.6 ± 1.1 μM;Figure 4C).
Figure 4. Effects of a small molecular inhibitor (5-HDST) on NAT1 activity.
A. Chemical structure of 5-HDST. B. The percentage of NAT1 activity remaining after incubation with 0 to 87.5 μM 5-HDST for 1–6 days (N=3). C. Dose-response curve used to calculate the IC50 for 5-HSDT inhibition of NAT1 activity on day 6 (N=3). The IC50 is 47.6 ± 1.1 μM.
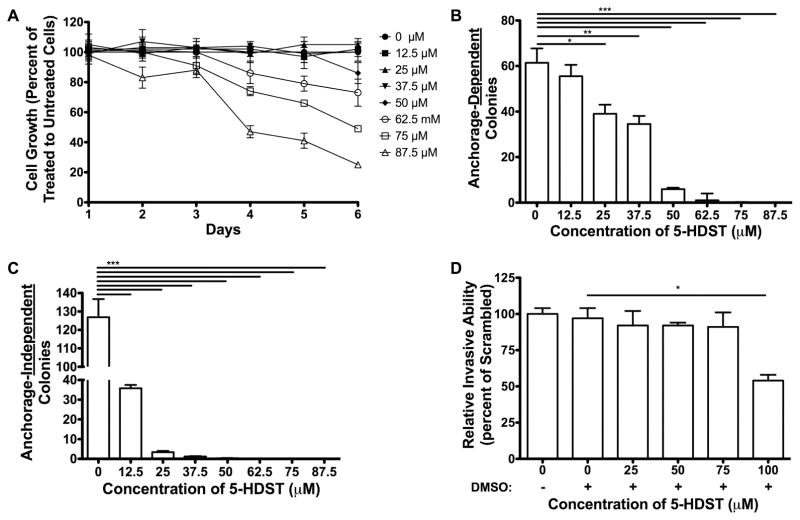
Cells treated with 5-HDST ranging from 0 to 50 μM over a period of up to 6 days produced no significant effects on cell growth (Figure 5A). Concentrations of 5-HDST greater than 50 μM caused a concentration- and time-dependent decrease in cell growth (p<0.0001) (Figure 5A).
Figure 5. Cell growth properties of MDA-MB-231 cells incubated with 5-HDST.
Bars/symbols represent means ± SEM from three separate determinations performed in triplicate (N=3). A. Cell number (percent of vehicle) measured after incubation (1–6 days) with 5-HDST. B. Anchorage-dependent colony formation following incubation with 5-HDST for 14 days. C. Anchorage-independent colony formation following incubation with 5-HDST for 14 days. D. Transwell assays to measure migratory ability of scrambled shRNA cell line incubated with 5-HDST. One-way ANOVA was conducted to compare relative invasive ability between groups (p<0.05). The scrambled shRNA cell line incubated with 100 μM 5-HDST had significantly (p<0.05) less invasive ability compared to scrambled shRNA cell line incubated with vehicle following one-way ANOVA and Bonferroni post hoc test (p<0.05).
Incubation with 5-HDST reduced anchorage-dependent cell growth, but the effect was marginal below 50 μM (Figure 5B). Incubation with 5-HDST markedly reduced anchorage-independent cell growth at 12.5 μM (Figure 5C). Incubation with 5-HDST had no effect on relative invasion ability until concentrations of 5-HDST exceeded 75 μM (p<0.05)(Figure 5D).
Characterization of recombinant human NAT1 and NAT2 inhibition by 5-HDST
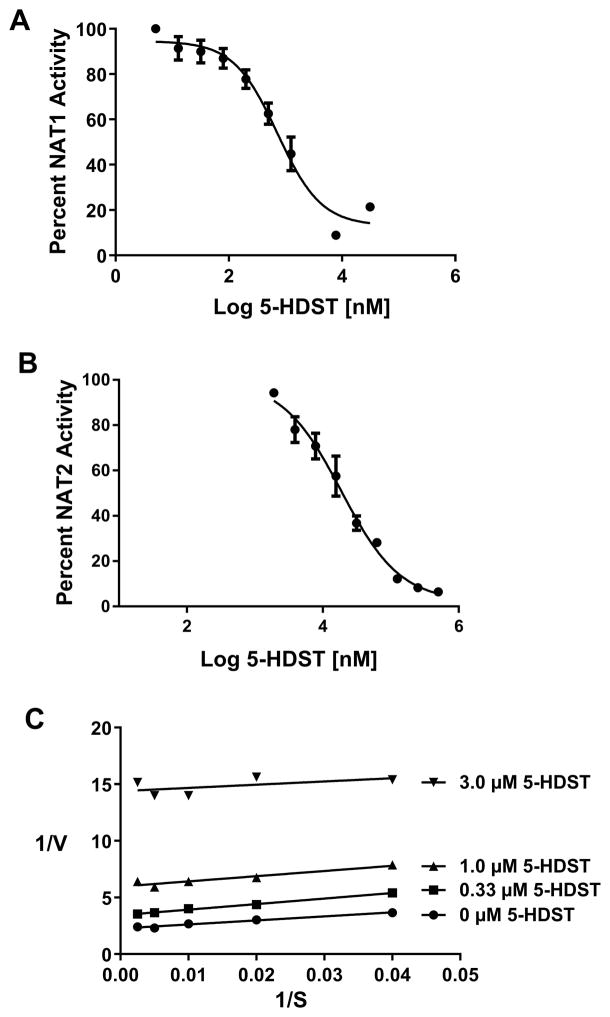
5-HDST exhibited 25-fold higher selectivity towards inhibition of recombinant human NAT1 (IC50 =0.732 ± 0.012 μM; Figure 6A) than human NAT2 (IC50 =18.7 ± 0. 2 μM; Figure 6B). 5-HDST inhibition of recombinant human NAT1 was uncompetitive (Figure 6C).
Figure 6. Characterization of recombinant human NAT1 and NAT2 inhibition by 5-HDST.
A. Dose-response curve used to calculate the IC50 for 5-HSDT inhibition of recombinant human NAT1. Circles illustrate Mean ± SEM (N=3). The IC50 is 0.732 ± 0.012 μM. B. Dose-response curve used to calculate the IC50 for 5-HSDT inhibition of recombinant human NAT2. Circles illustrate Mean ± SEM (N=3). The IC50 is 18.7 ± 0. 2 μM. C. Lineweaver-Burke plot for 5-HDST (various concentrations indicated on the figure) inhibition of recombinant human NAT1. Values on the abscissa (1/S) are μM PABA concentration. Analysis of the plot is consistent with uncompetitive inhibition of recombinant human NAT1 by 5-HDST.
DISCUSSION
We investigated whether differences in human NAT1 expression affect breast cancer cell growth and metastatic properties. The novelty of our approach included NAT1 knockdown and overexpression with a FRT system, which facilitates investigations of decreased or increased NAT1 expression independent of off-target effects because all the expression cassettes were inserted into the same genomic location.
NAT1 knockdown using shRNA resulted in approximately 39% reduction in NAT1 in vitro activity, a comparable 35% increase in cellular AcCoA levels, and a ~85% decrease in anchorage-independent growth compared to the scrambled shRNA cell line. However, the NAT1 shRNA knockdown did not yield significant changes (p>0.05) in cell doubling time, anchorage-dependent growth or relative invasion ability. Previous studies, using the same NAT1 shRNA knockdown plasmid in a colon adenocarcinoma cell line achieved an 80–90% knockdown of NAT1 [10] with significant changes in cell growth and invasion but only a 15% reduction in anchorage-independent cell growth. The differences observed between the present study and the previous NAT1 knockdown study [10] could be due to the different cell line used, our use of the FRT system to minimize the possibilities of off-target effects, the media and culture conditions utilized, or the level of NAT1 knockdown. It is possible there is a threshold knockdown of NAT1 required to observe effects on cell growth and cell invasion that is tissue or cell-type specific.
Despite a 12-fold overexpression of NAT1 in vitro activity, significant (p>0.05) changes in endogenous AcCoA levels, cell doubling time, anchorage-dependent growth, anchorage-independent cell growth, or relative invasion ability were not observed. In contrast, a previous study using HB4a cells reported that NAT1 overexpression resulted in enhanced growth properties [3]. The MDA-MB-231 cells used in the present study are a claudin-low, triple-negative breast cancer cell line whereas the HB4a cell line is derived from normal luminal mammary epithelial cells. Thus, the lack of differences between scrambled shRNA and NAT1 overexpression MDA-MB-231 cell lines may be a result of the cell line utilized.
Recent reports that NAT1 catalyzes the hydrolysis of AcCoA in the presence of folate suggest that NAT1 may affect endogenous levels of AcCoA [1]. Consistent with this concept, we found that partial NAT1 knockdown by 39% elevated endogenous AcCoA levels by 35% (Figure 1C). Although this finding is consistent with AcCoA hydrolysis catalyzed by NAT1, it does not provide evidence this is the primary or sole mechanism for the elevation of AcCoA. Rat embryonic fibroblasts from rapid acetylator congenic rats (high levels of rat NAT2 which is orthologous to human NAT1) have lower levels of AcCoA than rat embryonic fibroblasts derived from slow acetylator congenic rats, which have low levels of rat NAT2 [30]. Human NAT1 knockout MDA-MB-231 and MCF-7 breast cancer cell lines exhibit elevated cellular AcCoA levels compared to the parental cell lines [31]. Thus while a direct connection for physiological differences in AcCoA levels cannot be drawn to NAT1 AcCoA hydrolysis, accumulating data suggest that NAT1 may influence endogenous AcCoA levels. Emerging evidence reveals that cells monitor AcCoA levels as a key indicator of metabolic state, including growth versus survival, fed versus fasted, and normal versus tumorigenic metabolic states [32]. Differences in AcCoA levels could provide an explanation for differences observed in metabolites of fatty acid synthesis and amino acids [33]. The effects of NAT1 knockdown on anchorage-independent cell growth and endogenous AcCoA levels may provide insight into why congenic rapid NAT2 (ortholog to human NAT1) rats develop more mammary tumors than congenic slow NAT2 rats following administration of mammary carcinogens that are not metabolized by rat NAT2 [30].
A second objective of this study was to investigate inhibition of NAT1 via a small molecule inhibitor to NAT1 knockdown using shRNA. We selected a novel small molecule inhibitor (5-HDST) similar to one identified and characterized previously [22]. The scrambled shRNA cell line was utilized as the control in both the small molecular inhibitor and shRNA knockdown experiments to facilitate this comparison.
5-HDST exhibited 25-fold higher selectivity in vitro towards the inhibition of recombinant human NAT1 (IC50 0.732 ± 0.012 μM) than human NAT2 (IC50 18.7 ± 0. 2 μM). Further characterization of the 5-HDST inhibition of recombinant human NAT1 was consistent with uncompetitive inhibition. 5-HDST also decreased the ability of the breast cancer cell line to N-acetylate PABA in situ with an IC50 value of 47.6 ± 1.1 μM. A previous study determined an IC50 value of 101 ± 11 μM for (Z)-5-(2-hydroxybenzylidene)-2- thioxothiazolidin-4-one (Rhod-o-hp), which is structurally similar to 5-HDST, also determined by in situ PABA N-acetylation inhibition assay [22].
Cell growth was reduced in a concentration dependent manner above 50 μM (Figure 5A). The concentration-dependent effects on NAT1 activity and cell growth were similar in trend on day six. In the scrambled shRNA cell line incubated with 5-HDST, we observed a decrease in both anchorage-dependent and anchorage-independent cell growth. It is possible that the reduction in anchorage-dependent growth reflects cellular toxicity of the small molecule inhibitor. The reduction in anchorage-independent growth reflects a combination of cellular toxicity and NAT1 inhibition in the breast cancer cell line MDA-MB-231. We assessed relative invasion ability using a transwell assay and observed no change in the scrambled shRNA cell line treated with the small molecule inhibitor 5-HDST until the concentration exceeded 75 μM. Concentrations of 5-HDST as high as 87.5 μM showed no effects on cell viability up to 24 hours.
The results of this study complement but do not fully replicate previous reports regarding the effect of NAT1 knockdown on cancer cell properties [8,10,34]. In the present studies, approximately 40% decrease of NAT1-catalyzed N-acetylation in the MDA-MB-231 breast cancer cell line by either shRNA knockdown or 5-HDST inhibition resulted in a significant decrease in anchorage-independent growth without affecting overall cell growth and relative invasion ability. NAT1 shRNA partial knockdown increased endogenous AcCoA levels 35% compared to the scrambled shRNA and NAT1 overexpression. Based on results of this study, our laboratory and others are utilizing CRISPR/Cas9 to make permanent deletions at the NAT1 gene open reading frame causing 100% knockout of NAT1. In preliminary studies, our laboratory has reported that NAT1 knockout in MDA-MB-231 breast cancer cell lines with two different guide RNAs replicates the results reported here with shRNA partial NAT1 knockdown [31,35]. In summary, inhibition of NAT1 by either partial knockdown (shRNA) or small molecule inhibition reduced anchorage-independent growth in the MDA-MB-231 human breast cancer cell line. These findings suggest that human NAT1 could serve as a target for the prevention and/or treatment of breast cancer.
Acknowledgments
Portions of this work constituted partial fulfillment of a PhD in Pharmacology and Toxicology at the University of Louisville for Marcus W. Stepp [35].
Funding
This work was supported in part by the National Institute of Environmental Sciences [Grant USPHS T32-ES011564].
List of Abbreviations
- 5-HDST
(5E)-[5-(4-hydroxy-3,5-diiodobenzylidene)-2-thioxo-1,3-thiazolidin-4-one]
- AcCoA
acetyl-Coenzyme A
- NAT1
N-acetyltransferase 1
- PABA
para-aminobenzoic acid
- PBS
phosphate-buffered saline
- EDTA
ethylenediaminetetraacetic acid
- DMEM
Dulbecco’s Modified Eagles Medium
- HPLC
high performance liquid chromatography
- DTT
dithiothreitol
- PMSF
phenylmethanesulfonyl fluoride
- STR
Short Tandem Repeat
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Participated in research design: Doll, States, and Hein
Conducted experiments: Stepp and Doll.
Contributed new reagents or analytic tools: Stepp and Doll.
Performed data analysis: Stepp and Doll.
Wrote or contributed to the writing of the manuscript: Stepp, Doll, Carlisle, States, and Hein.
All authors read and approved the final manuscript
References
- 1.Laurieri N, Dairou J, Egleton JE, et al. From Arylamine N-Acetyltransferase to Folate-Dependent Acetyl CoA Hydrolase: Impact of Folic Acid on the Activity of (HUMAN)NAT1 and Its Homologue (MOUSE)NAT2. PLoS One. 2014;9(5):e96370. doi: 10.1371/journal.pone.0096370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stepp MW, Mamaliga G, Doll MA, States JC, Hein DW. Folate-dependent hydrolysis of acetyl-coenzyme A by recombinant human and rodent arylamine N-acetyltransferases. Biochemistry and biophysics reports. 2015;3:45–50. doi: 10.1016/j.bbrep.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adam PJ, Berry J, Loader JA, et al. Arylamine N-Acetyltransferase-1 Is Highly Expressed in Breast Cancers and Conveys Enhanced Growth and Resistance to Etoposide in Vitro. Mol Cancer Res. 2003;1(11):826–835. [PubMed] [Google Scholar]
- 4.Abba MC, Hu Y, Sun H, et al. Gene expression signature of estrogen receptor alpha status in breast cancer. BMC Genomics. 2005;6:37. doi: 10.1186/1471-2164-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endo Y, Yamashita H, Takahashi S, et al. Immunohistochemical determination of the miR-1290 target arylamine N-acetyltransferase 1 (NAT1) as a prognostic biomarker in breast cancer. BMC Cancer. 2014;14:990. doi: 10.1186/1471-2407-14-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savci-Heijink CD, Halfwerk H, Koster J, van de Vijver MJ. A novel gene expression signature for bone metastasis in breast carcinomas. Breast cancer research and treatment. 2016;156(2):249–259. doi: 10.1007/s10549-016-3741-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smid M, Wang YX, Klijn JGM, et al. Genes associated with breast cancer metastatic to bone. J Clin Oncol. 2006;24(15):2261–2267. doi: 10.1200/JCO.2005.03.8802. [DOI] [PubMed] [Google Scholar]
- 8.Tiang JM, Butcher NJ, Minchin RF. Small molecule inhibition of arylamine N-acetyltransferase Type I inhibits proliferation and invasiveness of MDA-MB-231 breast cancer cells. Biochem Biophys Res Commun. 2010;393(1):95–100. doi: 10.1016/j.bbrc.2010.01.087. [DOI] [PubMed] [Google Scholar]
- 9.Bonifas J, Scheitza S, Clemens J, Biomeke B. Characterization of N-Acetyltransferase 1 Activity in Human Keratinocytes and Modulation by para-Phenylenediamine. J Pharmacol Exp Ther. 2010;334(1):318–326. doi: 10.1124/jpet.110.167874. [DOI] [PubMed] [Google Scholar]
- 10.Tiang JM, Butcher NJ, Cullinane C, Humbert PO, Minchin RF. RNAi-Mediated Knock-Down of Arylamine N-acetyltransferase-1 Expression Induces E-cadherin Up-Regulation and Cell-Cell Contact Growth Inhibition. PLoS One. 2011;6:e17031. doi: 10.1371/journal.pone.0017031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim SJ, Kang HS, Chang HL, et al. Promoter hypomethylation of the N-acetyltransferase 1 gene in breast cancer. Oncol Rep. 2008;19(3):663–668. [PubMed] [Google Scholar]
- 12.Butcher NJ, Arulpragasam A, Minchin RF. Proteasomal degradation of N-acetyltransferase 1 is prevented by acetylation of the active site cysteine - A mechanism for the slow acetylator phenotype and substrate-dependent down-regulation. J Biol Chem. 2004;279(21):22131–22137. doi: 10.1074/jbc.M312858200. [DOI] [PubMed] [Google Scholar]
- 13.Butcher NJ, Ilett KF, Minchin RF. Substrate-dependent regulation of human arylamine N-acetyltransferase-1 in cultured cells. Mol Pharmacol. 2000;57(3):468–473. doi: 10.1124/mol.57.3.468. [DOI] [PubMed] [Google Scholar]
- 14.Butcher NJ, Minchin RF. Arylamine N-acetyltransferase 1 gene regulation by androgens requires a conserved heat shock element for heat shock factor-1. Carcinogenesis. 2010;31(5):820–826. doi: 10.1093/carcin/bgq042. [DOI] [PubMed] [Google Scholar]
- 15.Bonamassa B, Ma YJ, Liu DX. Glucocorticoid Receptor-Mediated Transcriptional Regulation of N-acetyltransferase 1 Gene Through Distal Promoter. AAPS J. 2012;14(3):581–590. doi: 10.1208/s12248-012-9370-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dairou J, Atmane N, Rodrigues-Lima F, Dupret JM. Peroxynitrite irreversibly inactivates the human xenobiotic-metabolizing enzyme arylamine N-acetyltransferase 1 (NAT1) in human breast cancer cells - A cellular and mechanistic study. J Biol Chem. 2004;279(9):7708–7714. doi: 10.1074/jbc.M311469200. [DOI] [PubMed] [Google Scholar]
- 17.Atmane N, Dairou J, Paul A, Dupret JM, Rodrigues-Lima F. Redox regulation of the human xenobiotic metabolizing enzyme arylamine N-acetyltransferase 1 (NAT1) - Reversible inactivation by hydrogen peroxide. J Biol Chem. 2003;278(37):35086–35092. doi: 10.1074/jbc.M303813200. [DOI] [PubMed] [Google Scholar]
- 18.Ragunathan N, Dairou J, Pluvinage B, et al. Identification of the xenobiotic-metabolizing enzyme arylamine N-acetyltransferase 1 as a new target of cisplatin in breast cancer cells: Molecular and cellular mechanisms of inhibition. Mol Pharmacol. 2008;73(6):1761–1768. doi: 10.1124/mol.108.045328. [DOI] [PubMed] [Google Scholar]
- 19.Kukongviriyapan V, Phromsopha N, Tassaneeyakul W, et al. Inhibitory effects of polyphenolic compounds on human arylamine N-acetyltransferase 1 and 2. Xenobiotica. 2006;36(1):15–28. doi: 10.1080/00498250500489901. [DOI] [PubMed] [Google Scholar]
- 20.Chen YS, Ho CC, Cheng KC, et al. Curcumin inhibited the arylamines N-acetyltransferase activity, gene expression and DNA adduct formation in human lung cancer cells (A549) Toxicol In Vitro. 2003;17(3):323–333. doi: 10.1016/s0887-2333(03)00020-1. [DOI] [PubMed] [Google Scholar]
- 21.Malka F, Dairou J, Ragunathan N, Dupret JM, Rodrigues-Lima F. Mechanisms and kinetics of human arylamine N-acetyltransferase 1 inhibition by disulfiram. FEBS J. 2009;276(17):4900–4908. doi: 10.1111/j.1742-4658.2009.07189.x. [DOI] [PubMed] [Google Scholar]
- 22.Russell AJ, Westwood IM, Crawford MHJ, et al. Selective small molecule inhibitors of the potential breast cancer marker, human arylamine N-acetyltransferase 1, and its murine homologue, mouse arylamine N-acetyltransferase 2. Biorg Med Chem. 2009;17(2):905–918. doi: 10.1016/j.bmc.2008.11.032. [DOI] [PubMed] [Google Scholar]
- 23.Millner LM, Doll MA, Cai J, States JC, Hein DW. NATb/NAT1*4 promotes greater arylamine N-acetyltransferase 1 mediated DNA adducts and mutations than NATa/NAT1*4 following exposure to 4-aminobiphenyl. Mol Carcinog. 2012;51(8):636–646. doi: 10.1002/mc.20836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metry KJ, Zhao S, Neale JR, et al. 2-amino-1-methyl-6-phenylimidazo [4,5-b] pyridine-induced DNA adducts and genotoxicity in Chinese hamster ovary (CHO) cells expressing human CYP1A2 and rapid or slow acetylator N-acetyltransferase 2. Mol Carcinog. 2007;46(7):553–563. doi: 10.1002/mc.20302. [DOI] [PubMed] [Google Scholar]
- 25.Hein DW, Doll MA, Nerland DE, Fretland AJ. Tissue distribution of N-acetyltransferase 1 and 2 catalyzing the N-acetylation of 4-aminobiphenyl and O-acetylation of N-hydroxy-4-aminobiphenyl in the congenic rapid and slow acetylator Syrian hamster. Mol Carcinog. 2006;45(4):230–238. doi: 10.1002/mc.20164. [DOI] [PubMed] [Google Scholar]
- 26.Leff MA, Fretland AJ, Doll MA, Hein DW. Novel human N-acetyltransferase 2 alleles that differ in mechanism for slow acetylator phenotype. J Biol Chem. 1999;274(49):34519–34522. doi: 10.1074/jbc.274.49.34519. [DOI] [PubMed] [Google Scholar]
- 27.Fretland AJ, Leff MA, Doll MA, Hein DW. Functional characterization of human N-acetyltransferase 2 (NAT2) single nucleotide polymorphisms. Pharmacogenetics. 2001;11(3):207–215. doi: 10.1097/00008571-200104000-00004. [DOI] [PubMed] [Google Scholar]
- 28.Fretland AJ, Doll MA, Leff MA, Hein DW. Functional characterization of nucleotide polymorphisms in the coding region of human N-acetyltransferase 1. Pharmacogenetics. 2001;11(6):511–520. doi: 10.1097/00008571-200108000-00006. [DOI] [PubMed] [Google Scholar]
- 29.Millner LM, Doll MA, Stepp MW, States JC, Hein DW. Functional analysis of arylamine N-acetyltransferase 1 (NAT1) NAT1*10 haplotypes in a complete NATb mRNA construct. Carcinogenesis. 2012;33(2):348–355. doi: 10.1093/carcin/bgr273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stepp MW, Doll MA, Samuelson DJ, Sanders MA, States JC, Hein DW. Congenic rats with higher arylamine N-acetyltransferase 2 activity exhibit greater carcinogen-induced mammary tumor susceptibility independent of carcinogen metabolism. BMC Cancer. 2017;17(1):233. doi: 10.1186/s12885-017-3221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stepp MW, Doll MA, Hein DW. The Toxicologist: Supplement to Toxicological Sciences. 1. Vol. 150. Society of Toxicology; Baltimore, MD: Mar 13, 2017. Decreased Human Arylamine N-Acetyltransferase 1 (NAT1) Elevates Cellular Acetyl-Coenzyme A Levels [abstract number 1488] [Google Scholar]
- 32.Shi L, Tu BP. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Curr Opin Cell Biol. 2015;33:125–131. doi: 10.1016/j.ceb.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlisle SM, Trainor PJ, Yin XM, et al. Untargeted polar metabolomics of transformed MDA-MB-231 breast cancer cells expressing varying levels of human arylamine N-acetyltransferase 1. Metabolomics. 2016;7:111. doi: 10.1007/s11306-016-1056-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiang JM, Butcher NJ, Minchin RF. Effects of human arylamine N-acetyltransferase I knockdown in triple-negative breast cancer cell lines. Cancer medicine. 2015;4(4):565–574. doi: 10.1002/cam4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stepp MW. Doctoral dissertation. University of Louisville; 2017. Role of Human Arylamine N-acetyltransferase 1 in Tumorigenesis and Cancer Biology. [Google Scholar]