Abstract
Fengycin is a cyclic lipopeptide which is used as an agricultural fungicide. It is synthesized by Bacillus subtilis as an immune response against fungal infection, and functions by damaging the target’s cell membrane. Previous molecular dynamics simulations and experiments led to the hypothesis that aggregation of fengycins on the membrane surface plays a key role in cell disruption. Here, we used microsecond scale all-atom molecular dynamics simulation to understand the specificity, selectivity, and structure of fengycin oligomers. Our simulations suggest that fengycin is more likely to form stable oligomers in model fungal membranes (PC) compared to model bacterial membranes (PE:PG). Furthermore, we characterize differences in the structure and kinetics of the membrane-bound aggregates and discuss their functional implications.
Graphical Abstract
Introduction
Lipopeptides are a class of antibiotics which can effectively act against a plethora of disease-causing organisms.1 Many are expressed by bacteria and can have both antibacterial and antifungal activity. Fengycin, along with surfactins and iturins, is a member of a family of cyclic lipopeptides originally obtained from Bacillus subtilis. This bacteria grows synergistically in the roots of leguminous plants where it protects them from phytopathogens.2 Fengycin acts as a fungicide and can be used to treat various diseases in plants like clubroot disease (Plasmodiophora monoliforme), maize rot (Fusarium moniliforme), barley head blight (Fusarium graminearum), and cucurbit powdery (Podosphaera fusca).3–7 Biocontrol agents like fengycin avoid the adverse effects of chemical pesticides and are eco-friendly. Through genetic modifications, certain strains of B. subtilis are capable of over-producing lipopeptides, which led to the development of the commercially available biofungicide Serenade™(Bayer).
There are two methods by which fengycin acts against these fungi. First, it induces systemic resistance in the plants by perturbing the cell membrane of the root cells.1,8 Second, fengycin directly attacks fungi by binding to their cell membrane, causing leakage and lysis.9–12 In this work, we will focus on the latter phenomenon.
Beyond its established agricultural applications, fengycin (and related compounds) show promise as antifungal drugs. Fengycin has low hemolytic activity, and the combination of D-amino acids and cyclic structure makes it less vulnerable to degradation by peptidases compared to conventional antimicrobial peptides.3 Furthermore, membrane composition varies slowly in an evolutionary sense, slowing the advent of fungal resistance. In addition, fengycin has been found to be effective against filamentous fungi and hypothesized to be effective against localized dermatomycosis.13,14 After the approval of daptomycin, present in the drug Cubicin, increasing efforts to find lipopeptide-based drugs have increased.15 Thus, understanding how fengycin interacts with bilayers will help us understand how best to use it as a basis for future drug development.
Coarse-grained MD simulations of individual and oligomerized fengycins in various lipid bilayers suggest that they bind to all membranes with their acyl chains inserted into the hydrophobic core.16 These simulations also showed that membrane composition affected fengycin’s tendency to aggregate in the membrane; specifically, the aggregates are stable in bilayers made with phosphatidylcholine (PC) headgroups (typical of eukaryotic cells), but not in bacterium-like mixtures of phosphatidylethanolamine (PE) and phosphatidylglycerol (PG). The authors hypothesized that the aggregates, which significantly bend and distort the membrane, are the active form of the lipopeptide. These results were highly suggestive, but the limitations of a coarse-grained force field in representing detailed packing and electrostatic interactions require they be confirmed by all-atom simulations, particularly given the number of charged moieties in the system.
The goal of the present work is to clarify the composition-specific effects of fengycin on model lipid bilayers chosen to mimic eukaryotic and bacterial membranes via all-atom molecular dynamics simulations. The simulations demonstrate statistically significant differences between the lipopeptides’ behavior depending on the nature of the lipid headgroups, with resulting differences in their effects on bilayer structure.
Methods
Chemical Structure of fengycin
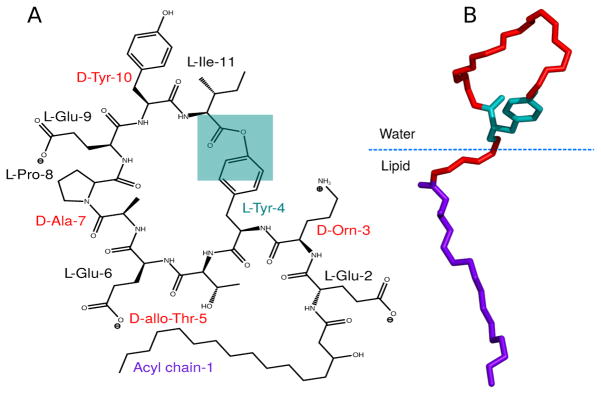
Fengycin is an amphipathic cyclic lipopeptide, with the chemical structure shown in Figure 1. It consists of an anionic cyclic decapeptide with a β-hydroxy fatty acid attached at the N-terminus.16 Fengycin naturally occurs with a number of variations in the protein sequence and acyl chain length; the specific fengycin used in the present simulations was first reported by Wu et al and characterized by Pathak et al using mass spectroscopic techniques.17,18 This particular variant has several distinctive features: (i) four D-amino acid residues (tyrosine (Tyr-10), alanine (Ala-7), non-natural ornithine (Orn-3) and threonine (Thr-5)); (ii) three negatively charged glutamate (Glu-2, Glu-6, Glu-9) residues; and one positively charged residue Orn-3, resulting in a net charge of −2; (iii) cyclic structure with the ring closure occurring between Tyr-4 and Ile-11 through an ester bond.
Figure 1.
(A) Chemical structure of fengycin(Adapted from Horn et al).;16,17 (B) 3D orientation of one of the conformations of fengycin during our simulation. Violet sticks represent the acyl tail, red sticks show the peptide backbone and cyan sticks stand for the C-terminus and Tyr-4 ester linkage.
System Construction
Fengycin has some unusual moieties not found in the standard CHARMM36 forcefield, including the β-hydroxyl on the acyl chain and the ester bond connecting the C-terminus to Tyr-4. Parameters for these atoms were developed using the Forcefield toolkit (FFTK) plugin to VMD.19,20 A stream file (feng stream.txt) containing all new parameters is attached as supplemental information. The lipopeptide structures were built using a modeling tool, Molefacture which is found as an extension of VMD.20
We constructed a membrane-bound fengycin system by placing molecules randomly on two planes parallel to the z-axis with the acyl chains pointed towards the membrane center, producing systems with 10 lipopeptides in each leaflet. The choice to run symmetric systems is an attempt to model the long-time equilibrium behavior accessible to experiments such as the fluorescence work from Heerklotz and coworkers.10 However, this choice does mean that membrane deformations due to asymmetric binding are not accessible; some antimicrobial peptides seem to exploit that asymmetry to induce pores, with the result that at long times their activity goes away due to peptide migration to the inner leaflet.21
The Optimal Membrane Generator (OMG) package from LOOS was used to place lipids around fengycin and solvated the system.22,23 The system was electronically neutralized with excess sodium ions, with additional NaCl added to bring the free salt concentration to appromixately 100mM. We modeled water using TIP3P, as is appropriate with the CHARMM forcefield. The system was thoroughly minimized and equilibrated through a series of alternating minimizations and short dynamics runs.
Table 1 summarizes the different systems run. To model the gram-negative bacteria we used a 2:1 mixture of palmitoyl-oleoyl-phosphatidylethanolamine (PE) and palmitoyl-oleoyl-phosphatidylglycerol (PG). A pure palmitoyl-oleoyl-phosphatidylcholine (PC) bilayer was used to emulate a eukaryotic membrane. The lipopeptide-lipid systems had 10 fengycins bound to each leaflet of the lipid bilayer. As a control, we ran simulations of neat bilayers with the same compositions. For each lipopeptide system, we ran 4 independently constructed replicates, while for the pure lipid systems we ran 3 replicates of each. All the systems have 90 lipids per leaflet.
Table 1.
Summary of simulations
| System | Phospholipids | fengycins per leaflet | Length(ns) | # of replicates |
|---|---|---|---|---|
| Bacteria | PE:PG | 10 | ~5000 | 4 |
| Fungus | PC | 10 | ~4500 | 4 |
| Neat | PE:PG | 0 | ~150 | 3 |
| Neat | PC | 0 | ~150 | 3 |
Simulation protocol
All simulations were run with NAMD version 2.9.24 We used the CHARMM36 forcefield for both the peptide and lipid, including the CMAP correction for the peptide backbone; the D-amino acids were represented by defining a new atom type for the D-α carbon and transposing the matrices for the CMAP values (see the parameters for D-Orn in the supplemental information).25–28
We used Langevin dynamics for all heavy atoms with the temperature set to 310.5K, and the Langevin piston barostat, with semi-isotropic boundary conditions.29–31 We used smooth particle-mesh Ewald summation with a 96x96x96 grid to calculate long-range electrostatics.32 Van der Waals interactions were smoothly cutoff from 8 to 12 Å with the pairlist maintained at 14 Å. We used a 2 fs timestep with the bonds constrained to their equilibrium lengths using the RATTLE algorithm.33
Simulation Analysis
All analyses were done at 1 ns resolution unless otherwise specified. For equilibration purposes, we excluded the first 500 ns of the lipopeptide simulations and 100 ns of the pure membrane simulation. All analysis was performed with tools developed using LOOS, a software package for the analysis of molecular dynamics simulations, available for download from https://github.com/GrossfieldLab/loos.22,34 Unless otherwise noted, all error bars are standard errors in the mean, computed by treating each trajectory as an individual measurement.
Radial Distribution Functions
We determined the three-dimensional radial distribution functions for various pairs of atoms using the atomic rdf tool from LOOS.22,34 We also calculated the radial distribution function in the membrane plane for fengycin and the different lipid species using the xy rdf tool, also a part of LOOS; this tool operates on the molecules’ centers of mass, rather than individual atoms. The time evolution of these RDFs was also calculated by dividing the trajectory into 10 ns windows and the resulting figure can be found in the Supplemental Information.
Quantifying Aggregation
We considered two lipopeptide molecules to be in contact if there were at least ten pairs of heavy atoms within 3 Å of each other. Using this criterion, we calculated the number of lipopeptides in each aggregate and determined the probability distribution of aggregate size. We normalized the distribution by calculating the fraction of lipopeptides that are present in different aggregate size.
Residue-Residue Contact Map
We quantify lipopeptide-lipopeptide interactions using residue contact maps. The definition of a contact is similar to that described above: two residues are in contact if they share at least ten pairs of heavy atoms within 3 Å. Fractional contacts are defined as the probability of a specific residue-residue pair being in contact given that the two molecules are in contact.
Lifetime of the aggregates
In order to characterize the kinetic stability of differently sized aggregates, we created a time series for each trajectory, assigning each frame a value of 1 when a particular aggregate was present, and 0 if it was not. We then calculated the autocorrelation function for each such time series and averaged the resulting correlation functions as a function of aggregate size.
Order parameters
To quantitate the effects of fengycin on lipid chain structure, we computed order parameters analogous to those measurable via solid-state deuterium quadrupolar splitting experiments. The order parameters are calculated using the second Legendre polynomial applied to the angle θ between a given acyl carbon-hydrogen bond and the membrane normal:
| (1) |
This calculation was performed using the order params tool from LOOS.22,34
Molecular order parameters
To characterize the length scale on which fengycin alters lipid chain structure, we calculated the molecular order parameters for lipid chains as a function of distance to the nearest fengycin, using the LOOS tool dibmops.22,34 Analogous to the deuterium order parameter shown in Eq. 1, the molecular order parameter is the second Legendre polynomial of the angle between the membrane normal and the average of the second and third principal axes of the chain.
Results and Discussion
Fengycin causes disorder in membrane
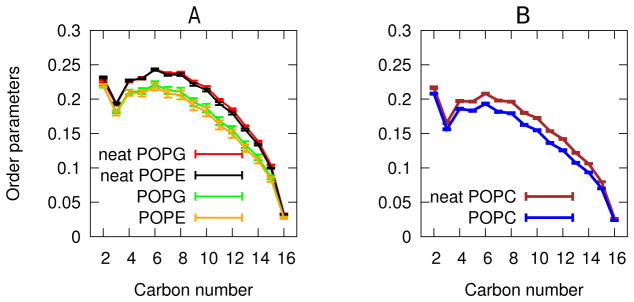
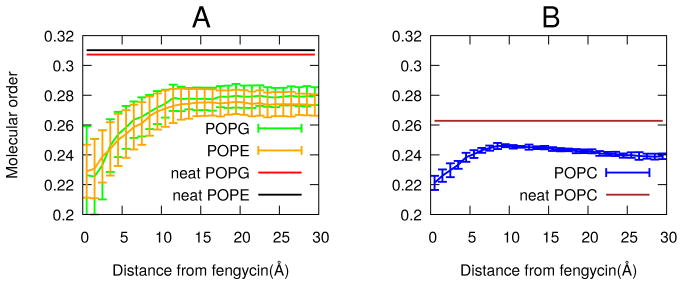
One of the most common effects of antimicrobial peptides is reducing the order of the surrounding lipids.35 To quantify this, we computed order parameter profiles for the lipid palmitoyl chains, as described in section Order parameters under Methods, with results shown in Figure 2. As expected, fengycin reduces the order parameters for all carbons between C-3 and C-15 relative to their equivalents in neat membranes. This indicates an overall disordering effect but does not indicate the length-scale on which fengycin operates. To determine this, we computed the chain order parameter as a function of distance, as discussed in section Molecular order parameters under Methods. Figure 3 also shows that the molecular order parameter for membrane bilayers containing fengycins are lower than the equivalent neat membranes, consistent with Figure 2. However, the effect is most pronounced for lipids within 10 Å of the nearest fengycin, plateauing at longer range. In addition, the order parameter remains lower than the neat membrane even at a very long range; presumably, more distant lipids are altered by their disordered neighbors and not by fengycin directly.
Figure 2.
Order parameters for the palmitoyl chain of (A)PE:PG and (B)PC. Neat indicates order parameters for membrane systems without any fengycin in them. The error bars indicate the standard error for each replicate. Carbon number for the palmitoyl chain is along the x-axis while average order parameter is along the y-axis.
Figure 3.
Molecular order parameters of palmitoyl chain as a function of distance from fengycin molecule.(A) is for PE:PG and (B) is for PC. The error bars are the standard error for each replicate. Neat membranes (no fengycins) are represented as straight lines.
Preferential interaction with PE
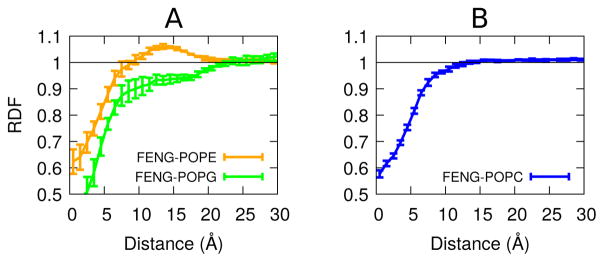
In addition to altering the lipid chain structure, antimicrobial peptides can also alter the lateral ordering of lipid headgroups.36,37 This is most easily quantified using a lateral radial distribution function, as shown in Figure 4. Using the method described in Radial Distribution Functions we calculated the radial distribution function between the center of masses of fengycin and different lipid types (PE,PG,PC). Figure 4A shows that PE is significantly enriched in the first solvation shell surrounding fengycin, while PG is equivalently depleted. By contrast, PC shows no evidence of structuring around fengycin, other than steric depletion at short range.
Figure 4.
Radial distribution function in the membrane plane between fengycin peptide ring and the lipid head group. (A) is for PE:PG and (B) is for PC. Distance between the two sets of entities is along the x-axis, while RDF is along the y-axis. The straight line at 1 represents the RDF value for a random distribution such as the ideal gas. The error bars are the standard error, treating each trajectory as a single measurement.
The apparent attraction between fengycin and PE is likely due to two factors. First, the PE amine group is a hydrogen bond donor, unlike the choline group in PC, so it can make favorable interactions with the peptide’s anionic side chains. Second, electrostatic repulsion between the peptide and PG, both of which are negatively charged, could create an apparent attraction to the zwitterionic PE headgroup.
Headgroups attract charged fengycin residues
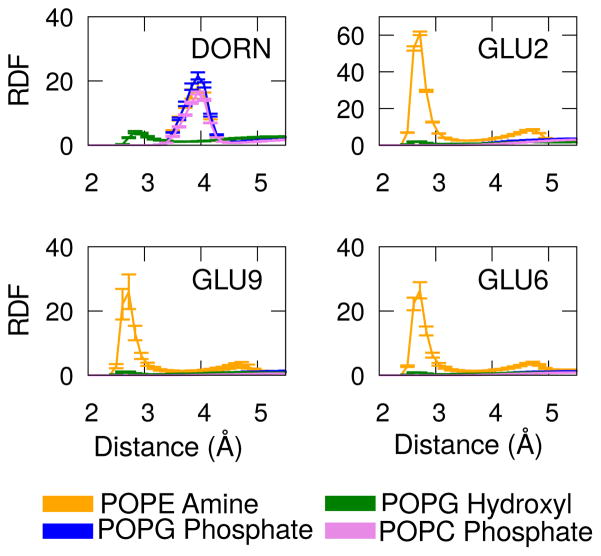
In order to test whether there are specific interactions that attract fengycin to PE head-groups, we computed a series of three-dimensional radial distribution functions as shown in Figure 5. Please note that these values are inflated by the molecules’ restriction to the membrane surface. We calculated the distribution of following specific atoms — (i)nitrogens of PE amines around oxygens in the carboxylate part of glutamates, (ii)oxygens of the hydroxyl group in PG glycerols around nitrogen of amine in Orn-3 (iii)phosphates of PG and PC around nitrogen of amines in Orn-3. These atoms were selected because they indicate most likely electrostatic attractions among themselves. Figure 5 indicates that the phosphates and hydroxyls in the lipid headgroups attract the positively charged ornithine side chains, while the three negatively charged glutamates preferentially interact with the positively charged amine of PE or with water. We do not see a net attraction between fengycin and PG because the peptide has three glutamates but only one ornithine.
Figure 5.
Three-dimensional radial distribution function between charged atoms of D-ornithine (DORN) and Glutamates (GLU) in fengycin and head groups of PE, PG and PC. RDF is along the y-axis and distance between the atoms is along the x-axis. Note the upper right panel is plotted on a different y-scale.
Qualitatively, all the glutamates show similar preference patterns, but quantitatively they are quite different. Glu-2 has a much higher contact peak with the PE amine, indicating it spends more time in contact than the other two glutamates. This difference is likely due to the covalent structure of fengycin; Glu-2 is immediately adjacent to the lipopeptide fatty acid, forcing the sidechain to remain at the membrane-water interface. By contrast, the other two glutamates are part of the ring and can either be hydrated or interact with lipid. The lateral ordering we observed for PE:PG section (Preferential interaction with PE) can be rationalized by the glutamates’ preferential packing with the PE amines.
Although PC phosphates can interact favorably with ornithine (see the upper left panel of Figure 5), overall the electrostatic forces are insufficient to create a net attraction between the peptide and PC headgroups (see Figure 4).
Fengycin aggregation depends on membrane composition
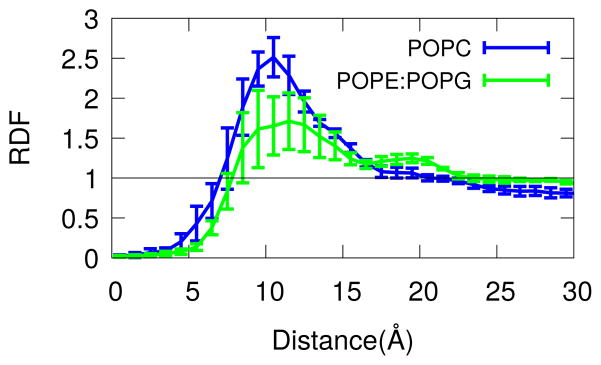
Our working hypothesis in performing these simulations was that the ability of fengycin to damage target membranes was related to its aggregation.16 This in turn suggests that aggregation should vary with the bilayer’s lipid composition, so that fungal (eurkaryotic) membranes are damaged while the native bacterial membranes are not. To test this hypothesis, we first computed the fengycin-fengycin lateral radial distribution function, shown in Figure 6.
Figure 6.
Lateral radial distribution function between fengycins. Lipopeptide separation is the x-axis, while the y-axis is probability density. The straight line at 1 represents the RDF value for a random distribution.
The fengycin-fengycin RDF in PC shows a single large peak at roughly 10 Å, indicating significant peptide-peptide attraction and the presence of some aggregates. Visual inspection of the trajectories confirms that aggregates begin to appear after the first 500 ns to 1 μs. By contrast, the first peak is significantly smaller in PE:PG membrane, but there is a small secondary peak around 20 Å. This indicates that while there is less aggregation overall in the “bacterial membrane”, there is a small tendency to form more elongated structures.
Characterizing aggregate structure
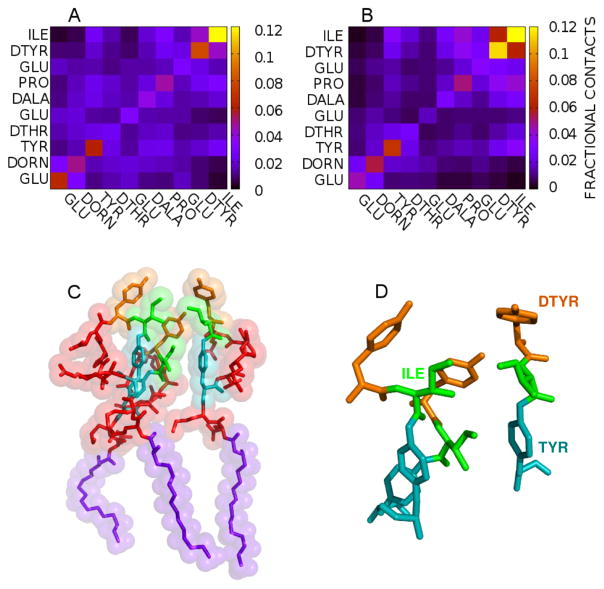
Experimental work has shown that small variations in the sequence of fengycin-like molecules can have significant effects on their activity that cannot be easily explained by changes in membrane affinity.38 However, if aggregation occurred via specific structures, the mutations could plausibly disrupt the packing and reduce its favorability. Accordingly, we examined the fengycin aggregates and their tendency to form specific residue-residue contacts. Figures 7A and 7B show the residue-residue contact probabilities in PC and PE:PG bilayers, respectively. Each box in the heat map represents the likelihood of specific sidechain-side chain contacts within lipopeptide oligomers.
Figure 7.
Fractional contacts between fengycin residues which are in contact at (A) PE:PG and (B)PC membranes. (C) shows three fengycins which are in contact while (D) shows the residues in fengycins that have higher fractional contact value in both (A) and (B). Yellow, Green and Cyan sticks represents the connectivity in heavy atoms of D-Tyr, Ile and Ring Tyr respectively. Violet sticks show the acyl tail in fengycin and red represent the peptide bonds.
The results indicate there is significant diversity in the ensemble of oligomeric structures; the most likely pairing (between Ile residues on each peptide) is only present in roughly 10% of the dimers. That said, there are some interesting features. The ring-closing residues (Ile and Tyr) form the strongest contacts in both membrane environments. These hydrophobic residues are far from the fatty acid moiety, and thus are less likely to be buried in the membrane; the exposed hydrophobic surface is a natural conduit for peptide-peptide interactions. This would also explain the prevalence of DTyr-DTyr contacts and Ile-DTyr contacts as well. Tyr-Tyr contacts were also found to be higher in our all-atom simulations and in the previous coarse-grained simulation results.25 Figure 7C shows an example of three fengycins in close proximity and Figure 7D zooms in to focus on the three hydrophobic residues (Tyr-4, Tyr-10 and Ile-11) which are most likely to be in contact. This indicates that tyrosines and isoleucines from adjacent fengycins can clump together. And this is what is observed as high contacts along the diagonal in Figure 7A–B. The other likely pairings include Glu-2 and Orn. Here the phenomenon is largely the reverse — these charged moieties are very hydrophilic, but are constrained to remain at the membrane-water interface because they are adjacent to the fatty acid. Forming a charge pair could stabilize the partial dehydration required by their proximity to the membrane.
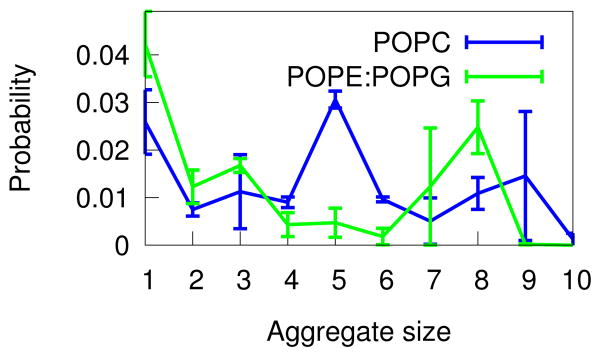
Aggregate size is lipid-dependent
If the tendency to aggregate is to explain selective targeting of specific membranes, we should see a difference in the distribution of aggregate sizes between ”bacteria-like” PE:PG membranes and ”eukaryotic” PC membranes. Using the Quantifying Aggregation method described in Methods, we calculated the probability distribution based on aggregate size using fengycins as the selection shown in Figure 8. Surprisingly, it looks like there is a stronger tendency to form larger aggregates (specifically, 7–8mers) in PE:PG membranes than in PC, in apparent contradiction with Figure 6 which overall indicates a higher probability of fengycin-fengycin pairs in PC.
Figure 8.
Probability of a specific size fengycin aggregate existing in either of the two membrane systems.
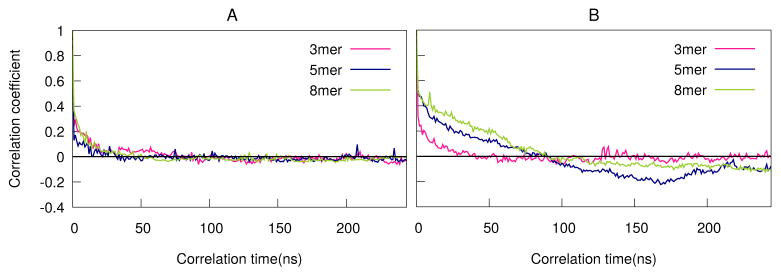
This contradiction is resolved by looking at the lifetimes for different aggregate sizes, plotted in Figure 9 (see section Lifetime of the aggregates in Methods for discussion of how the lifetimes were computed). Figure 9 shows the autocorrelation curve for fengycin trimers(3-mers), pentamers(5-mers), and octamers(8-mers) in two different membrane systems, PE:PG and PC. In PC, trimers have the shortest lifetimes, while pentamers have longer lifetimes than octamers. By contrast, all three aggregates sizes have comparable (and very short) lifetimes in PE:PG. This indicates that small aggregates like trimers are only transient in both membranes, typically falling apart within 15 ns or so. In PE:PG, the lifetimes have gotten even shorter, with all three aggregates surviving perhaps 10 ns or so. By contrast, pentamers are longer-lived in PC membranes, at roughly 100 ns. Figure 9A shows octamers are transient structures in PE:PG membranes, but are metastable in PC as indicated by Figure 9B. Visual inspection of trajectories clarifies the difference: when larger aggregates are present in PE:PG, they are nearly always a result of “collisions” between smaller aggregates without much in the way of stabilizing interactions; as a result, they diffuse apart promptly. By contrast, when larger aggregates form in PC, they are typically stabilized by a combination of polar interactions near the membrane and hydrophobic ones far from it, as discussed above. The relative lack of significantly favorable sidechain interactions with PC headgroups (compared to PE:PG) also likely contributes to the increased kinetic stability.
Figure 9.
Lifetime of 3-mer,5-mer and 8-mer in the two membrane systems (A)PE:PG and (B)PC. Time delay is along the x-axis while auto-correlation coefficient is along the y-axis.
Going forward, we see several important questions to answer. First, the present simulations place fengycins on both membrane leaflets, essentially representing equilibrium conditions accessible in biophysical experiments. However, biologically the fengycins would “attack” from outside the cell, initially binding exclusively to one leaflet. It is possible that the aggregation differences would be made more significant by this assymmetry, as suggested by our previous coarse-grained work.16
Second, it is extremely difficult to quantify the thermodynamics of aggregate formation on the timescales accessible to all-atom membrane simulations, because equilibrium simulations would have to run long enough for aggregates to form and break up multiple times. Quantitatively understanding this thermodynamics — necessary to test the hypothesis that aggregation controls function — will instead require some form of enhanced sampling in order to obtain quantitative accuracy.
Finally, we know that fengycins damage fungal membranes, but do far less damage to mammalian membranes, even though both are eukaryotic. Given the interest in using fengycin-like molecules as potential antifungal medicines it would be valuable to understand why they target one organism but not the other.39 One likely hypothesis would be to attribute the differences to the sterol present in the fungal vs. mammalian membranes (ergosterol and cholesterol respectively). Some experiments indicate that cholesterol’s tendency to increase membrane order may protect mammalian membranes, but the mechanisms are unclear and could likely be revealed by future simulations.38 Similarly, the presence of different anionic lipids in fungal membranes (e.g. phosphatidylinositol) may also play a role, particularly if their behavior is different from anionic lipids found in bacteria such as PG.
Conclusions
Fengycins operate as biological fungicides primarily by targeting and damaging the fungus’ outer membranes, while leaving their plant hosts and the bacteria that produce them unharmed. We used all-atom molecular dynamics simulations of fengycin molecules bound to two membrane compositions (PE:PG and PC) chosen as simple mimics of bacterial and eukaryotic membranes in order to explain this functional selectivity. In particular, we showed that while fengycin binding disorders the lipid hydrophobic region independent of headgroup type, its aggregation propensities varies with lipid composition. Although aggregates form in both membrane types, larger aggregates are only stable in the eukaryotic-like membranes representative of their target fungi.
In addition, fengycin perturbs the membrane irrespective of the lipid composition, but the extent of membrane leakage is traced back to formation of aggregates. Hence the aggregation process itself depends on the tug of war between two favoring interactions. One is the hydrophobic interactions between Tyr-10, Tyr-4 and Ile-11 of adjacent fengycins which lead to stable aggregates and the other is the favorable electrostatic interactions between fengycin’s charged residues (glutamates and ornithine) and the lipid head groups. These two attractive and repulsive forces determine whether fengycin will form aggregates on membrane surfaces or not and thus regulate the membrane selectivity of fengycin.
Supplementary Material
References
- 1.Ongena M, Jacques P. Bacillus lipopeptides: versatile weapons for plant diseases biocontrol. Trends Microbiol. 2007;16:115–124. doi: 10.1016/j.tim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 2.Ongena M, Jacques P, Touré Y, Destain J, Jabrane A, Thonart P. Involvement of fengycin-type lipopeptides in the multifaceted biocontrol potential of Bacillus subtilis. Appl Microbiol Biotechnol. 2005;69:29–38. doi: 10.1007/s00253-005-1940-3. [DOI] [PubMed] [Google Scholar]
- 3.Vanittanakom N, Loeffler W, Koch U, Jung G. Fengycin–a novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F-29-3. J Antibiot (Tokyo) 1986;39:888–901. doi: 10.7164/antibiotics.39.888. [DOI] [PubMed] [Google Scholar]
- 4.Li XY, Mao ZC, Wang YH, Wu YX, He YQ, Long CL. Diversity and active mechanism of fengycin-type cyclopeptides from Bacillus subtilis XF-1 against Plasmodiophora brassicae. J Microbiol Biotechnol. 2013;23:313–321. doi: 10.4014/jmb.1208.08065. [DOI] [PubMed] [Google Scholar]
- 5.Hu LB, Shi ZQ, Zhang T, Yang ZM. Fengycin antibiotics isolated from B-FS01 culture inhibit the growth of Fusarium moniliforme Sheldon ATCC 38932. FEMS Microbiol Lett. 2007;272:91–98. doi: 10.1111/j.1574-6968.2007.00743.x. [DOI] [PubMed] [Google Scholar]
- 6.Chan YK, Savard ME, Reid LM, Cyr T, McCormick WA, Seguin C. Identification of lipopeptide antibiotics of a Bacillus subtilis isolate and their control of Fusarium graminearum diseases in maize and wheat. BioControl. 2009;54:567–574. [Google Scholar]
- 7.Romero D, de Vicente A, Olmos JL, Dávila JC, Pérez-García A. Effect of lipopeptides of antagonistic strains of Bacillus subtilis on the morphology and ultra-structure of the cucurbit fungal pathogen Podosphaera fusca. J Appl Microbiol. 2007;103:969–976. doi: 10.1111/j.1365-2672.2007.03323.x. [DOI] [PubMed] [Google Scholar]
- 8.Ongena M, Jourdan E, Adam A, Paquot M, Brans A, Joris B, Arpigny JL, Thonart P. Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ Microbiol. 2007;9:1084–1090. doi: 10.1111/j.1462-2920.2006.01202.x. [DOI] [PubMed] [Google Scholar]
- 9.Heerklotz H, Seelig J. Leakage and lysis of lipid membranes induced by the lipopeptide surfactin. Eur Biophys J. 2007;36:305–314. doi: 10.1007/s00249-006-0091-5. [DOI] [PubMed] [Google Scholar]
- 10.Patel H, Tscheka C, Edwards K, Karlsson G, Heerklotz H. All-or-none membrane permeabilization by fengycin-type lipopeptides from Bacillus subtilis {QST713} Biochim Biophys Acta (BBA), Biomembr. 2011;1808:2000–2008. doi: 10.1016/j.bbamem.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Deleu M, Paquot M, Nylander T. Effect of fengycin, a lipopeptide produced by Bacillus subtilis, on model biomembranes. Biophys J. 2008;94:2667–2679. doi: 10.1529/biophysj.107.114090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deleu M, Paquot M, Nylander T. Fengycin interaction with lipid monolayers at the air-aqueous interface-implications for the effect of fengycin on biological membranes. J Colloid Interface Sci. 2005;283:358–365. doi: 10.1016/j.jcis.2004.09.036. [DOI] [PubMed] [Google Scholar]
- 13.Eeman M, Deleu M, Paquot M, Thonart P, Dufrêne Y. Nanoscale properties of mixed fengycin/ceramide monolayers explored using atomic force microscopy. Langmuir. 2005;21:2505–2511. doi: 10.1021/la0475775. [DOI] [PubMed] [Google Scholar]
- 14.Eeman M, Olofsson G, Sparr E, Nasir MN, Nylander T, Deleu M. Interaction of fengycin with stratum corneum mimicking model membranes: a calorimetry study. Colloids Surf B. 2014;121:27–35. doi: 10.1016/j.colsurfb.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 15.Raja A, LaBonte J, Lebbos J, Kirkpatrick P. Daptomycin. Nat Rev Drug Discovery. 2003;2:943–944. doi: 10.1038/nrd1258. [DOI] [PubMed] [Google Scholar]
- 16.Horn JN, Romo TD, Grossfield A. Simulating the mechanism of antimicrobial lipopeptides with all-atom molecular dynamics. Biochemistry. 2013;52:5604–5610. doi: 10.1021/bi400773q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu CY, Chen CL, Lee YH, Cheng YC, Wu YC, Shu HY, Götz F, Liu ST. Nonribosomal synthesis of fengycin on an enzyme complex formed by fengycin synthetases. J Biol Chem. 2007;282:5608–5616. doi: 10.1074/jbc.M609726200. [DOI] [PubMed] [Google Scholar]
- 18.Pathak KV, Keharia H, Gupta K, Thakur SS, Balaram P. Lipopeptides from the banyan endophyte, Bacillus subtilis K1: mass spectrometric characterization of a library of fengycins. J Am Soc Mass Spectrom. 2012;23:1716–1728. doi: 10.1007/s13361-012-0437-4. [DOI] [PubMed] [Google Scholar]
- 19.Mayne CG, Saam J, Schulten K, Tajkhorshid E, Gumbart JC. Rapid parameterization of small molecules using the Force Field Toolkit. J Comput Chem. 2013;34:2757–2770. doi: 10.1002/jcc.23422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–8. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 21.Krauson AJ, He J, Wimley WC. Determining the mechanism of membrane permeabilizing peptides: Identification of potent, equilibrium pore-formers. Biochim Biophys Acta, Biomembr. 2012;1818:1625–1632. doi: 10.1016/j.bbamem.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romo TD, Grossfield A. LOOS: an extensible platform for the structural analysis of simulations. Conf Proc IEEE Eng Med Biol Soc. 2009;2009:2332–2335. doi: 10.1109/IEMBS.2009.5335065. [DOI] [PubMed] [Google Scholar]
- 23.Lin D, Grossfield A. Thermodynamics of micelle formation and micelle-membrane fusion modulate the activity of antimicrobial lipopeptides. Biophys J. 2015;109:750–759. doi: 10.1016/j.bpj.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horn JN, Cravens A, Grossfield A. Interactions between fengycin and model bilayers quantified by coarse-grained molecular dynamics. Biophys J. 2013;105:1612–1623. doi: 10.1016/j.bpj.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang J, MacKerell AD., Jr CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J Comput Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Best RB, Zhu X, Shim J, Lopes PE, Mittal J, Feig M, MacKerell AD., Jr Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. J Chem Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klauda JB, Venable RM, Freites JA, OConnor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Jr, Pastor RW. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider T, Stoll E. Molecular-dynamics study of a three-dimensional one-component model for distortive phase transitions. Phys Rev B. 1978;17:1302. [Google Scholar]
- 30.Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: the Langevin piston method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 31.Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- 32.Ewald PP. The calculation of optical and electrostatic grid potential. Ann Phys. 1921;64:253–287. [Google Scholar]
- 33.Andersen HC. RATTLE: A velocity version of the SHAKE algorithm for molecular dynamics calculations. J Comput Phys. 1983;52:24–34. [Google Scholar]
- 34.Romo TD, Leioatts N, Grossfield A. Lightweight object oriented structure analysis: tools for building tools to analyze molecular dynamics simulations. J Comput Chem. 2014;35:2305–2318. doi: 10.1002/jcc.23753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heerklotz H, Wieprecht T, Seelig J. Membrane perturbation by the lipopeptide surfactin and detergents as studied by deuterium NMR. J Phys Chem B. 2004;108:4909–4915. [Google Scholar]
- 36.Wadhwani P, Epand R, Heidenreich N, Bürck J, Ulrich A, Epand R. Membrane-active peptides and the clustering of anionic lipids. Biophys J. 2012;103:265–274. doi: 10.1016/j.bpj.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Epand RM, Epand RF. Lipid domains in bacterial membranes and the action of antimicrobial agents. Biochim Biophys Acta, Biomembr. 2009;1788:289–294. doi: 10.1016/j.bbamem.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 38.Fiedler S, Heerklotz H. Vesicle leakage reflects the target selectivity of antimicrobial lipopeptides from Bacillus subtilis. Biophys J. 2015;109:2079–2089. doi: 10.1016/j.bpj.2015.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pirri G, Giuliani A, Nicoletto SF, Pizzuto L, Rinaldi AC. Lipopeptides as anti-infectives: a practical perspective. Cent Eur J Biol. 2009;4:258–273. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.