Abstract
Venous thromboembolism (VTE), the third leading cardiovascular complication, requires more understanding at molecular levels. Here, we have identified miR-145 as a key molecule for regulating thrombus formation in venous thrombosis (VT) employing network based bioinformatics approach and in vivo experiments. Levels of miR-145 showed an inverse correlation with thrombus load determined by coagulation variables. MiRNA target prediction tools and in vitro study identified tissue factor (TF) as a target gene for miR-145. The restoration of miR-145 levels in thrombotic animals via in vivo miR-145 mimic delivery resulted in decreased TF level and activity, accompanied by reduced thrombogenesis. MiR-145 levels were also reduced in VT patients and correlated with increased TF levels in patients, thereby, confirming our preclinical findings. Our study identifies a previously undescribed role of miRNA in VT by regulating TF expression. Therefore, restoration of miR-145 levels may serve as a promising therapeutic strategy for management of VT.
Keywords: Venous thrombosis, miRNA, Tissue factor, Antithrombotic therapy, Coagulation
Highlights
-
•
MiR-145 is a regulator of venous thrombus formation.
-
•
Tissue factor is a direct target of miR-145.
-
•
MiR-145 is down regulated in human venous thrombosis patients.
-
•
Restoration of miRNA-145 via in vivo miRNA mimic delivery attenuated thrombus formation.
In this study we provide an insight into the molecular mechanisms of VT regulated by miRNA. Multiple investigations conducted with animal model and VT patients showed relatively reduced miR-145 expression levels under diseased condition. MiR-145 play important role in thrombus formation by regulating TF expression which have a critical role in VT. MiR-145 overexpression in animal model of VT reduces TF levels and attenuates thrombus formation which might have important implications for miRNA based therapeutic strategies in VT. This study highlights the role of miR-145 in the etiopathology of VT and also underscores the use of miR-145 mimic as an advancement to the field.
1. Introduction
Venous thromboembolism (VTE) is a leading cardiovascular complication affecting 100–200 individuals in a population of 100,000 per year and emerging as a third largest cardiovascular disease (Malone and Agutter, 2006). A combination of genetically manipulated animal models and human epidemiological data has revealed that a variety of genetic and acquired risk factors are associated with VTE, which contribute to its multifactorial complex nature (Diaz et al., 2012, Heit et al., 2002). These complexities might be the reason for the continuation of antithrombotic drugs for decades in spite of the risk of bleeding associated with their use. Despite the extensive clinical evidence about VTE, better molecular understanding is required to explore further and identify specific therapeutic targets.
MiRNAs are non-coding RNAs modulating gene expression, either through mRNA degradation or translational repression of multitude of targets (Bartel, 2004). Each miRNA has the potential to simultaneously target multiple mRNAs, and repress genes found in similar pathways to alter biological networks (Bartel, 2009, Selbach et al., 2008). Till date, various experimental reports have highlighted the functional role of miRNAs in cardiovascular biology, physiology, and diseases (Small et al., 2010). Several miRNAs including miR-155, miR-126, miR-21 and miR-146 have been reported to regulate pathogenic signaling in the development and progression of vascular inflammation, neointimal lesion formation, atherosclerosis and coronary artery disease (Ji et al., 2007, Liu et al., 2009, Weber et al., 2010). However, to our knowledge, the precise role of miRNAs in the etiopathology of VT has not been shown yet. Keeping the essence of all these facts, the present study aimed to identify the miRNA signatures that could be of paramount importance in the understanding of the pathogenesis of thrombus formation in the venous milieu.
In the present study, using both network based bioinformatics approach and in vivo animal model system, we explored the possible role of miRNAs in VT. Importantly, our results demonstrated that the reduced levels of miR-145 were associated with enhanced thrombus formation. Furthermore, we also provided the experimental evidence for identifying tissue factor (TF), as a direct target for miR-145 with binding sites at the 3′UTR of the gene. The restoration of miR-145 levels via in vivo miRNA mimic delivery inhibited TF expression and attenuated thrombus formation in the animal model of VT, thereby indicating its antithrombotic potential. Nonetheless, our preclinical findings were also validated in human VT patients, supporting the potential translational significance of miR-145 against VT.
2. Experimental Procedures
2.1. Screening of miRNAs From Disease-miRNAs Network by Network Topology Analysis
We exploited the curated list of miRNAs with obvious importance in related vascular diseases (flow chart shown in Fig. S1A) like atherosclerosis, acute coronary syndrome, myocardial infarction as well as pathophysiological states including inflammation, endothelial cell activation, ischemia, and neoplasm. Briefly, validated miRNAs related to these diseases and pathophysiological states were obtained from miRWalk database (list shown in Supplementary excel file 1). A network of diseases and miRNAs was created. Further, the network was analyzed based on complex centrality scoring algorithms (Fig. S1B-S1D, details in Supplementary excel file 2) and a list of candidate miRNAs with higher scores was generated. The detailed method applied for network based bioinformatics is further described in Supplementary methods.
2.2. Animal Studies
All experiments were conducted in accordance with Committee for the Purpose of Control and Supervision of Experiments on Animals (IAEC-03/DIPAS/2011), Government of India. Age matched 250–300 g Sprague-Dawley rats were used. Stasis induced venous thrombosis animal model was implemented as previously described (Henke et al., 2004, Downing et al., 1998). Briefly, rats were anesthetised with ketamine (100 mg/kg), xylazine (20 mg/kg) and placed in the supine position. Following a midline laparotomy, the intestines were exteriorized and placed to the left of the animal and the inferior vena cava (IVC) was carefully separated from the surrounding tissues and then ligated just below the renal veins along with ligation of side branches. After 6 h, 12 h and 48 h of ligation, rats were euthanized, the IVC was carefully dissected. Thrombus was extracted, weight of the thrombus was measured in milligrams and length was determined in millimeters. The IVC with thrombus was fixed immediately in formalin and later stained with hematoxylin and eosin.
2.3. RNA Isolation and Quantitative RT-PCR Analysis
Total RNA was extracted from IVC tissue using TRIzol reagent (Sigma) according to the manufacturer's suggested protocol. Isolation of intimal and media RNA from IVC was modified from a previous study (Sun et al., 2012). Briefly, IVC was cut out and transferred to a dish containing ice-cold phosphate buffer saline (PBS). The tip of an insulin syringe needle was carefully inserted into one end of the IVC to facilitate a quick flush of QIAzol lysis buffer, and intima eluate was collected. The IVC leftover (media + adventitia) was washed once with PBS and snap-frozen in liquid nitrogen, for storage until total RNA extraction by TRIzol. RNA from platelets and PBMCs were isolated as previously described (Konieczna et al., 2015, Tyagi et al., 2014). The detailed method is further described in Supplementary methods. MiRNA qRT-PCR was performed with Stem-loop qRT-PCR method (Chen et al., 2005). Briefly, 500 ng of total RNA was reverse-transcribed via SuperScript-III reverse transcriptase (Invitrogen) using miRNA specific RT primers. For mRNA qRT-PCR cDNA was synthesized from 1 μg of total RNA using the Quantitect Reverse Transcription Kit (Qiagen) in accordance with the manufacturer's instructions. QRT-PCR was performed using a CFX 96 connect real-time PCR system (BioRad). Expression was normalized to the housekeeping gene. Small RNA housekeeping control was 5S ribosomal RNA for miRNA and 18S for mRNA. Primer sequences are shown in Tables S2 and S3.
2.4. Analysis of Potential Targets of Differentially Expressed miRNAs
Potential targets of miRNAs were identified using online available miRNA prediction tool miRWalk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/) (Dweep et al., 2012). For accurate prediction, targets predicted by at least three tools and conserved across rat, mouse, and human were selected (Supplementary excel file 3). Selected target genes were analyzed by GeneCodis (Tabas-Madrid et al., 2012, Carmona-Saez et al., 2007, Nogales-Cadenas et al., 2009) to obtain enriched pathways with adjusted p < 0.05 as the cut-off.
2.5. In vitro Luciferase Reporter Assay
A partial TF 3′UTR sequence of 434 base pairs containing miR-145 target sites was cloned into psiCHECK-2 Vector (Promega) downstream of Renilla luciferase gene using the XhoI and NotI restriction sites. Mutagenesis of miR-145 target sites was performed by PCR splicing method, and mutated 3′UTR sequences were also cloned into psiCHECK-2 Vector. Primers used for plasmid construction (psiCHECK-2 and TF 3′UTR or mutated TF 3′UTR) are listed in table S4. HEK293 cells were seeded in 24-well plates in DMEM supplemented with 10% FBS and maintained in 5% CO2 incubator at 37 °C. The transfection was done after 24 h using Lipofectamine 3000 (Invitrogen) as per manufacturer's protocol. Cells were transfected with 100 ng of psiCHECK-2 constructs and 40 nM of Pre-miR™ miRNA precursor molecules-negative Control # 1 (non-specific/scr. mimic) and Rno-miR-145 Pre-miR™ miRNA precursor (miR-145 mimic, Ambion). Cells were harvested 24 h post transfection. Luciferase assay was performed by using Dual Luciferase Reporter Assay System (Promega) following the manufacturer's protocol. Firefly luciferase activity was normalized to Renilla luciferase activity and expressed as relative light units (RLU). The relative luciferase activity was reported as the fold change between scr. mimic and miR-145 mimic transfected cells.
2.6. In vivo miRNA Delivery
For in vivo application, miRNA mimics and inhibitors (in vivo grade, Invitrogen) were suspended in invivofectamine (Invitrogen) according to manufacturer's instructions. Invivofectamine 2.0 was used to form nanoparticles suitable for in vivo applications. mirVana miRNA mimics and inhibitors (Life Technologies) were complexed with Invivofectamine 2.0 reagent. Each animal (n = 5 per group) was administered with 200 μl mixtures containing scr. mimic/inhibitor or miR-145 mimic/inhibitor (1 nmol–10 nmol) by tail vein injection before 1 h of IVC ligation, as described in a previous study (Sun et al., 2012).
2.7. Histological Examination
After extraction IVC containing thrombus, liver, heart and kidney tissue were collected in 10% buffered formalin and embedded in paraffin. Serial cross sections (5 μm) of the IVC with thrombus were cut to analyze thrombus formation. Tissue sections were stained with hematoxylin and eosin following standard procedures. Liver, heart, and kidney tissue sections were analyzed to search for signs of any fibrosis, inflammation or toxicity. All histological images were acquired using an inverted microscope (Motic) and analyzed by Motic Images 2.0 software.
2.8. Western Blotting
Proteins from IVC tissues were extracted with RIPA lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 1% NP40, 0.25% Na-deoxycholate and 0.1% SDS) containing complete protease inhibitor cocktail. 30–50 μg of protein was loaded on SDS–polyacrylamide gel electrophoresis gel for separation, which was followed by blotting of protein on polyvinylidene fluoride membrane in semi-dry trans-blot electrophoretic transfer system (BioRad). Afterward, proteins were detected using following primary antibodies: Tissue Factor (1:1000; Abcam) and Actin (1:5000; Santacruz). HRP-conjugated secondary antibodies and chemiluminescence substrate (Sigma) was used for detection of the specific bands. Intensities of the obtained bands were quantified using ImageJ software (NIH).
2.9. Coagulation Assays and Hematology
For PT, 50 μl of plasma was incubated for 2 min at 37 °C before addition of 100 μl PT reagent (NÉOPLASTINE® CI PLUS, Stago), a recombinant TF. The clotting time, in seconds, were recorded by a semi-automated coagulation analyzer (Labitec) as per manufacturer's instruction.
For a bleeding time, animals were anesthetized, and the tail was transacted 0.5 cm from the tip using a disposable surgical blade. The tail was placed vertically in 10 ml isotonic saline at 37 °C immediately after being cut, and the bleeding time measured from the moment of transection until bleeding has stopped completely.
Blood clotting time was noted using test tube method as described previously (Lee-White method) with some modification. Briefly non-anticoagulated blood in the glass test tube was placed in a water bath at 37 °C. Glass tube was tilted every 30 s. The clotting time was measured when the blood does not flow out of the test tubes when tilted horizontally.
White blood cell (WBC), red blood cell (RBC), hemoglobin (Hb), platelet count and hematocrit (HCT) were measured with Melet Schloesing-MS4 hematology analyzer (MeletSchloesing Laboratories).
2.10. Biochemical Assays and Enzyme Immunoassay
Circulating biomarkers for liver and heart function were determined in serum samples subsequently analyzed for alanine and aspartate aminotransferases (ALT & AST) using colorimetric measurement of hydrazone formed with 2, 4 dinitrophenyl hydrazine (Reitman and Frankel, 1957), alkaline phosphatase (ALP) and lactate dehydrogenase (LDH) levels by the phenolphthalein monophosphate method (Babson and Phillips, 1965).
For TF activity assay, IVC tissue homogenate were incubated with factor VIIa, factor X, CaCl2 and spectrozyme FXa in TBS for 45 min at 37 °C. Optical density was measured at 405 nm. A standard curve was constructed from recombinant human lipidated TF. Activity was expressed in arbitrary units by reference to the standard curve.
IVC tissue homogenate supernatants were analyzed to determine the presence of rat TF antigen using sandwich enzyme immunoassay kit (Kinesis DX) according to the manufacturer's instructions. Circulating levels of rat plasma prothrombin fragment 1 + 2, TAT and D-dimer were determined by the commercially available rat prothrombin fragment 1 + 2 and D-dimer ELISA kit (Sincere Biotech).
2.11. Human Studies
Human studies were conducted in accordance with the ethical standards of Indian Council of Medical Research (01/IEC/DIPAS/12). Male patients with venous thrombosis from Army Research and Referral Hospital, Delhi, India (tertiary care hospitals), were approached for consent to participate in the study and informed consent was obtained according to the Declaration of Helsinki. Patients with pre-existing systemic diseases, malignancy, any prior surgery, or vasculitis were excluded.
2.12. Clinical Examination and Blood Sample Collection
Male patients suggestive of VT were investigated for confirmatory diagnosis. Cerebral venous sinus thrombosis (CVT) patients (n = 4) with focal neurologic deficits had headache as the presenting symptom were confirmed by magnetic resonance imaging (MRI) and magnetic resonance venogram (MRV) of the brain. Deep vein thrombosis (DVT) patients (n = 11) manifested with swelling of lower limb, confirmed by a Doppler examination. DVT associated PTE was also found in three patients. Echocardiography followed by CT pulmonary angiography was the diagnostic modalities to detect PTE. The diagnosis of portal venous thrombosis (PVT) patients (n = 2) presented with pain abdomen, hepatosplenomegaly or evidence of gastrointestinal bleed was confirmed with a Doppler examination of the portal venous system. All patients were treated with low molecular weight heparin (LMWH) and then switched over to oral anticoagulants. CVT patients were managed conservatively with cerebral decongestive measures.
The blood sampling was done by venipuncture at earliest possible time after admission to the hospitals. The hematology, biochemical and coagulation screening were done along with clinical investigations. Further, whole blood and plasma were aliquoted, immediately frozen and transported to the laboratory for molecular and genetic studies.
2.13. Laboratory Investigations
For each patient (median age, 31.5 years), the clinical profile and laboratory investigations (hemogram, PT, aPTT, Fibrinogen, D-dimer) were performed along with the complete thrombophilia screening including Protein C and S deficiency and Antithrombin-III deficiency. Protein C and Protein S activities were determined by using commercially available STACLOT kit on an automated STA compact analyzer (Diagnostica Stago). The quantification of AT III was done using STACHROM AT III kit in STA analyzer. Activated protein C resistance (APCR) was carried out using the Stago kit. PCR-RFLP method was used to perform mutational analysis for Factor V Leiden (1691G/A, rs6025), prothrombin (20210G/A, rs1799963), tissue factor plasmin inhibitor (-536C/T), fibrinogen-β (148C/T), MTHFR (677C/T) and plasminogen activator inhibitor-1 (16754G/5G) were performed by PCR-RFLP method. Total RNA from whole blood and plasma were isolated using PAXgene Blood RNA Isolation Kit (Qiagen) and TRIZOL reagent (Sigma) respectively, according to the manufacturer's instructions. miRNA expression and mRNA expression were analyzed as described earlier. Microparticle-TF activity assay was performed in human plasma using MP-TF assay kit (Aniara) according to the manufacturer's instructions.
2.14. Statistical Analysis
Data are presented as mean ± SEM. P-values were obtained using the Student t-test to compare two groups or one way ANOVA with bonferroni post hoc for multiple groups. Statistical analyses were performed with GraphPad Prism 6 software. Linear regression analysis was used to find out the correlation between parameters. For all statistical analysis, p-values are denoted as: ***p < 0.005, **p < 0.01 and *p < 0.05.
3. Results
3.1. Altered Pattern of miRNA Signatures is Associated With Progression of VT
A network-based bioinformatics approach was used to identify miRNAs that regulate multiple pathophysiological states and diseases closely related to VT (Fig. 1A and Fig. S1). The validation of miRNAs prioritized through in silico network analysis (viz. miR-1, miR-21, miR-34a, miR-125b, miR-126, miR-145, miR-146, miR-150, miR-155, and miR-210; Fig. 1B) was done using well characterized IVC ligation animal model for VT. IVC and thrombus tissue samples were harvested from animals after 24 h of IVC ligation, and expression levels for miRNAs were measured by stem-loop qRT-PCR. Compared with the sham control group, among the selected miRNAs, only miR-145 expression exhibited a significant (~ 2-fold) reduction, while other miRNAs did not show significant alterations in expression levels (Fig. 1C). Guided by these observations, our subsequent investigations were focused on miR-145. To obtain an insight about the localization of miR-145 in thrombosed IVC, analysis of miR-145 expression levels showed that miR-145 was significantly downregulated in platelets, PBMCs and the media layer of the vessel wall (VSMCs). However, the levels of miR-145 expression in intima layer of the vessel wall, which include endothelial cells, remained unaltered (Fig. S2).
Fig. 1.
Downregulation of miR-145 expression is correlated with thrombus progression in VT.
(A) Interaction network between Diseases/pathophysiological states and miRNAs involved was constructed and analyzed using Cytoscape. (B) List of top miRNAs selected from in silico analysis of miRNAs-disease network based on a scoring algorithm. (C) qRT-PCR was conducted to quantify the endogenous expression of miR-1, miR-21, miR-34a, miR-125b, miR-126, miR-145, miR-146, miR-150, miR-155, miR-210, in RNA isolated from IVC of control animals and thrombotic animals after 24 h of IVC ligation (n = 5). (D) Expression levels of Pri-miR-145 and (E) miR-145 were analyzed by qRT-PCR in control and thrombotic groups at indicated time points (6 h, 12 h, 24 h, and 48 h) post IVC ligation. Assays were performed in triplicate, and the results are presented as the fold-change in expression compared with control group. Graphs show relative expression to 5 s ribosomal RNA (n = 5). (F) Representative photographs for gross IVC with thrombus obtained from a different group of animals at indicated time point (upper panel), with their respective heat map images representing thrombus density (lower panel). (G) Thrombus weight from control and thrombotic animal groups at indicated time points showing an increase with time (n = 5). (H) Scatter plot analysis of the correlation between the miR145 expression and thrombus weight (r2 = 0.812, p < 0.0001) at 6 h, 12 h, 24 h and 48 h in the thrombotic group (n = 15). (I–K) The increase in prothrombin fragment1 + 2, D-dimer and TAT plasma levels measured by ELISA in the thrombotic group compared with control animal group (n = 5). (L) Circulatory miR-145 expression levels in plasma RNA isolated from patients (n = 20) and control subjects (n = 20). (M) ROC curve is showing an association of miR-145 with VT and the distinguished difference in patient and control. miR-1 did not show an association with VT similar to animal study. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.005. See also Figs. S1 and S2.
To study the temporal effect of miR-145 in the progression of VT, we analyzed pri-miR-145 and miR-145 expression levels at different time points (6 h, 12 h, 24 h and 48 h post-IVC ligation). Levels of pri-miR-145 and miR-145 gradually decreased with the thrombus progression at 6 h, 12 h, and 24 h (Fig. 1D and E). An in situ inspection of gross thrombus and thrombus weight (Fig. 1F and G) showed the protracted time-dependent effect on the development of thrombi inside IVC, post ligation. Thrombus weight and miR-145 expression at stipulated time points showed significant inverse correlation (r2 = 0.8126, p < 0.0001; Fig. 1H). Moreover, prothrombin fragment 1 + 2, D-dimer and TAT levels, known biomarkers for thrombus formation were also found to be up surged till 24 h post ligation (Fig. 1I–K), suggesting that the progression of VT is related to a proportional change in miR-145 expression. Next, to extend these preclinical findings in the human disease condition, we evaluated the expression levels of miR-145 in clinically confirmed VT patients. Demographic as well as clinical characteristics of the patients are summarized in Table S1.
The expression levels of miR-145 were significantly lower in VT patients compared with control (p < 0.0001; Fig. 1L). Furthermore, the receiver operator characteristics (ROC) curve analysis of miR-145 (AUC = 0.95; p < 0.0001) was able to discriminate between patients and controls as compared to miR-1(used as control miR; AUC = 0.65; p = 0.11) depicting the association between miR-145 and disease (Fig. 1M). These data propose an association of miR-145 with thrombus formation both in an animal model and human VT patients.
3.2. Identification of Target Genes for miR-145 and Their Role in Blood Coagulation
To gain further insights into the mechanism of miR-145 mediated regulation of venous thrombogenesis, target genes were predicted for miR-145. The sequential scheme for target gene prediction and gene enrichment pathway analysis is depicted as a flowchart in Fig. 2A. This analysis showed blood coagulation as a significantly enriched pathway. Subsequently, using Cytoscape, a network was generated to visualize the target genes and enriched pathway interactions (Fig. 2B). Notably, this analysis identified several genes associated with coagulation and fibrinolytic pathway including plasminogen activator inhibitor-1 (PAI-1), TF, tissue factor pathway inhibitor (TFPI), kallikrein B plasma 1 (KLKB1), thrombomodulin (Thbd), factor-VIII (F-VIII), urokinase receptor (uPAR), and plasminogen (PLG). Expression levels of these candidate target genes were further evaluated in an animal model at different time points (6 h, 12 h, 24 h, and 48 h) post-IVC ligation. Particularly, the expression patterns for TF, PAI-1, and uPAR genes demonstrated an inverse pattern with miR-145 expression till 24 h (Fig. 2C and Fig. S3). Interestingly, increased TF expression in platelets, PBMCs, and media layer of a blood vessel (Fig. S4) showed an inverse pattern with miR-145 cellular expression pattern (Fig. S2). Nonetheless, a decrease in miR-145 expression during thrombus formation also led to a concomitant increase in TF protein expression level as reflected by Western blot analysis (Fig. 2D) and ELISA (Fig. 2E) with a subsequent rise in TF activity (Fig. 2F). To extend this miRNA – target gene association in humans, we next analyzed the TF expression in the same cohort of patient and control. Consistent with animal studies, expression of TF mRNA was significantly higher in VT patients who have shown reduced miR-145 levels (Fig. 2G and Fig. 1L). Since TF is released from circulating microparticles during thrombosis, we also tested microparticle-TF activity in plasma samples of these patients. Increased microparticle-TF activity in these samples also supported the increased TF mRNA expression data (Fig. 2H). The correlation of TF mRNA expression and microparticle-TF activity with miR-145 expression in VT patients (Fig. 2I and J) suggest a direct regulation of TF expression by miR-145. Based on these results, we hypothesized that miR-145 could play a significant role in thrombus formation, primarily by regulation of TF.
Fig. 2.
Target gene prediction of miR-145 and gene enriched pathway analysis shows enrichment of blood coagulation pathway genes.
(A) Flow diagram is depicting target gene prediction and gene enriched pathway analysis. (B) Interaction network between miR-145 target genes and target genes enriched pathway was constructed using Cytoscape. (C) TF mRNA levels were determined by qRT-PCR in RNA isolated from IVC of control and the thrombotic animal group at 6 h, 12 h, 24 h and 48 h (n = 5). (D) Representative western blot image and quantification of TF protein levels. (E) TF protein levels were determined by ELISA in tissue lysate of IVC from control and thrombotic animal group (n = 5) at 6 h, 12 h, 24 h, and 48 h. (F) TF activity in IVC of control and the thrombotic group at 6 h, 12 h, 24 h, and 48 h confirms the upregulation of TF expression (n = 5). (G) qRT-PCR analysis of target gene TF from RNA isolated from patients (n = 20) and control subjects (n = 20). Results are presented as the fold-change in expression compared with control subjects. Expression of 18 s RNA for mRNA was used as a normalization control. (H) Microparticle-TF activity in plasma from patients and control subjects (n = 20). (I) Scatter plot analysis of miR-145 expression and TF mRNA expression correlation (r2 = 0.5212, p = 0.003) in patients. (J) Scatter plot analysis of miR-145 expression and microparticle-TF activity correlation (r2 = 0.5231, p = 0.0003) in patients (n = 20). Data are shown as mean ± SEM and representative of three independent experiments. ns - Non-significant, *p < 0.05, **p < 0.01, and ***p < 0.005. See also Figs. S3 and S4.
3.3. Target Validation of miR-145 in the TF 3′-UTR
To test our hypothesis, we focused our further investigation on TF, which has a critical role in initiation and progression of thrombosis. Online available target prediction tools (RNAhybrid, PITA, and miRanda) revealed that TF has miR-145 binding sites in its 3′UTR (Fig. 3A) that is highly conserved across mammals (Fig. 3B). The “seed” sequence of miR-145 has bases that are complementary to 3′UTR of TF mRNA transcript, extending from nucleotides 450 to 456 (binding site 1) and 535 to 540 (binding site 2) (Fig. 3C). To test whether miR-145 directly binds to TF 3′UTR, we developed luciferase reporter gene constructs with either a wild-type (WT) TF 3’UTR or a miR-145 binding mutated (deleted; either with binding site 1 or binding site 1 + 2) TF 3′UTR. Co-transfection of miR-145 mimic with the WT-TF 3′UTR construct resulted in substantial inhibition of the luciferase activity, while luciferase activity of mutated TF 3′UTR constructs was not repressed (Fig. 3D and E). These results confirm a direct interaction of miR-145 with TF 3’UTR (at both binding sites -1 and 2). Altogether, these observations suggest that miR-145 plays a vital role in maintaining hemostasis by negatively regulating TF expression via direct interaction.
Fig. 3.
Tissue Factor is a direct target gene of miR-145.
(A) Schematic representation of Rat TF as a target gene predicted by the miRWalk database. (B) Sequence alignment of evolutionarily conserved potential miR-145 binding sites and potential complementary residues are shown in TF 3′ UTR of different species. (C) Nucleotide locations of the two binding sites and mutated binding sites for miR-145 in Rat TF 3′ UTR are shown. (D and E) The effect of miR-145 mimic or a nonspecific control oligonucleotide (scramble) on luciferase activity in psiCHECK-2 transfected HEK293 cells expressing the wild-type or mutated 3′ UTR (binding site 1 or binding site 1 + 2) of TF. Luciferase activities were normalized to Renilla activities (n = 5. Data are shown as mean ± SEM and representative of three independent experiments. ns – Non-significant, *p < 0.05, **p < 0.01, and ***p < 0.005.
3.4. In vivo Applicability and Dose Dependent Effect of miR-145
So far, the results have suggested that loss of miR-145 is associated with VT, and the regulation of TF expression by miR-145 is compromised under the diseased state. For investigating the functional role of miR-145 in VT, we utilized gain of function approach using miR-145 mimic administration in in vivo animal model. To explore the in vivo applicability of miR-145 mimic against VT, hematological parameters, toxicity assays, histology and bleeding time were investigated after intravenous administration of miR-145. Importantly, all hematological values and bleeding time were in normal range in miR-145 mimic administered group (Fig. S5A and B). No significant difference in the levels of liver and heart function markers was found (Fig. S5C to F). Histopathological examinations revealed the normal morphological architecture of liver, heart and kidney with no indication of necrotic or other cell death events observed (Fig. S5G). Therefore, it can be concluded that administration of miR-145 mimic did not result in any detectable sub-acute toxicity and was safe for intravenous administration in animals. To determine the optimum dose of miR-145 for in vivo study, we conducted a dose-response analysis (1 nmol–10 nmol) of miR-145 mimic in a time-dependent manner. Animals were administered with either the vehicle or with scrambled negative control mimic (Scr. mimic)/miR-145 mimic, prior to IVC ligation. Thrombus weight, plasma TF activity and prothrombin time (PT) were assessed at 6 h, 12 h, and 24 h post ligation. Thrombus weight and TF activity were significantly reduced even at lower doses of 5 nmol (Fig. 4A and B), whereas PT a measurement of TF-mediated extrinsic pathway showed a statistically significant change only at higher doses (Fig. 4C). Based on this observation, the submaximal dose of 5 nmol for miR-145 mimic was selected for further in vivo studies. Interestingly, the effect of miRNA administration was found substantial from the initial time point of 6 h till 24 h at 5 nmol. It was also evident in correlation analysis between TF activity and thrombus weight at all three time points. Spearman correlation for 6 h post ligation was found significant with r = 0.96 and p = 0.0028, whereas, the respective figures for 12 h (r = 0.89; p = 0.0123) and 24 h (r = 0.89; p = 0.0125) was found comparable. These datasets are consistent with earlier findings that perturbances in miR-145 mediated TF regulation could activate coagulation cascade towards thrombogenesis. Moreover, the main target of miR-145, TF is involved in the initiation of thrombosis; therefore, all follow-up experiments were conducted at the early point of 6 h.
Fig. 4.
Optimization of miR-145 mimic dose.
For in vivo dose optimization study, animals were administered with miR-145 mimic (1 nmol–10 nmol) by tail vein injection and IVC ligation was performed. (A) Thrombus weight and (B) TF activity at 6 h, 12 h, and 24 h post ligation after miR-145 administration. (C) Increase in Prothrombin Time at 6 h, 12 h, and 24 h post ligation. Data are presented as mean ± SEM. ns – Non-significant, *p < 0.05; **p < 0.01, ***p < 0.005. See also Figs. S5 and S6.
3.5. Systemic Delivery of miR-145 Mimic Decreases Thrombus Formation by Regulation of TF Expression
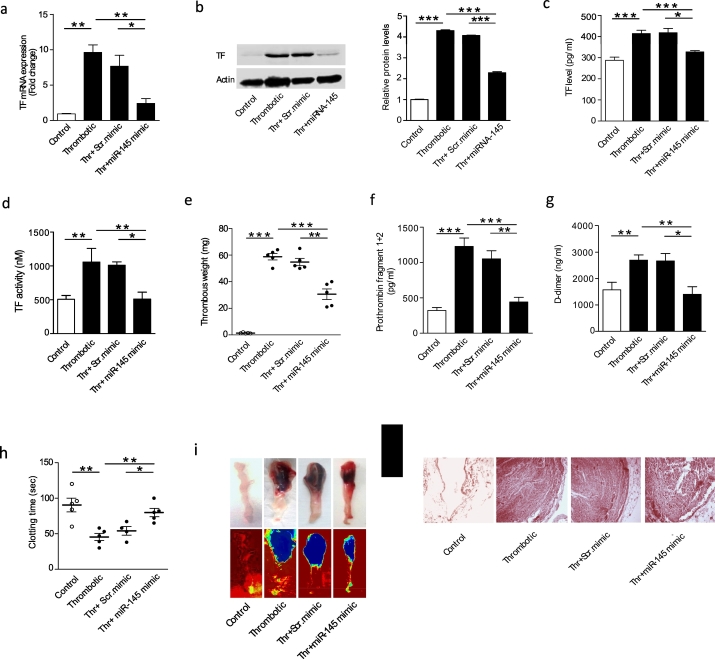
Finally, based upon our observations that the reduction of miR-145 function was linked to TF expression changes in VT patients and animal models of VT, we investigated the role of overexpression of miR-145 in vivo in an attempt to reverse these changes. Delivery of miR-145 mimic to the in vivo animal model, with tissue uptake was confirmed by qRT-PCR (Fig. S7). Pre-infusion of miR-145 mimic in thrombotic group prior to ligation led to a significant reduction in both TF mRNA levels as well as protein levels in IVC (Fig. 5A and B). TF levels in plasma were also lower in miR-145 mimic injected animals (Fig. 5C). Moreover, a reduction in TF activity after miR-145 mimic administration further confirmed the decrease in TF activity (Fig. 5D). In line with the dataset demonstrating the antithrombotic activity of miR-145, we also observed a significant reduction in thrombus weight (Fig. 5E) and length (data not shown). A delay in clotting time in the thrombotic animal group administered with miR-145 mimic was also observed (Fig. 5H) Gross thrombus examination also showed a reduction in the thrombus formation in miR-145 mimic injected animals (Fig. 5I). Further histological analysis of IVC containing thrombus confirmed that animals administered with miR-145 mimic had a less intact thrombus in their IVC when compared with either thrombotic animals or animals administered with scr. mimic (Fig. 5J). Additionally, intravenous injection of miR-145 significantly decreased the levels of prothrombin fragments 1 + 2 and D-dimers in plasma (Fig. 5F and G) suggesting a reduced thrombogenic potential among miR-145 treated animals.
Fig. 5.
miR-145 mimic administration reduces thrombus formation by regulating TF expression in an animal model of VT.
Animals were administered with scr. mimic or miR-145 mimic by intravenous injection and divided into four groups- Control, thrombotic, thrombotic + scr. mimic and thrombotic + miR-145 mimic then sacrificed after 6 h post IVC ligation. (A–D) qRT-PCR for TF mRNA expression, representative western blot with quantification of TF protein expression, TF ELISA and TF activity in IVC tissue showing restoration of miR-145 significantly reduces the TF levels (n = 5). (E) Thrombus weight decreases in the thrombotic group administered with miR-145 mimic compared to scr. mimic/thrombotic group (n = 5). (F and G) The decrease in prothrombin F1 + 2, D-dimer plasma levels in plasma samples of thrombotic animal group administered with miR-145 mimic compared with scr. mimic/thrombotic group (n = 5). (H) Increase in clotting time in the thrombotic animal group administered with miR-145 mimic compared with scr. mimic/thrombotic group (n = 5). (I) Representative photographs of gross IVC with thrombus (upper panel) and representative heat map images of IVC with thrombus (lower panel) presenting thrombus density. (J) Representative photomicrographs of Hematoxylin-eosin-stained sections of thrombus with the vessel wall (200 ×). Data are presented as mean ± SEM. *p < 0.05; **p < 0.01, ***p < 0.005. See also Fig. S7.
3.6. Systemic Delivery of miR-145 Inhibitor has Procoagulant Effect
Next to further confirm the effect of miR-145 in VT; animals were administered with miR-145 inhibitor or negative control inhibitor prior to IVC ligation. Administration of miR-145 inhibitor in thrombotic animals resulted in decreased miR-145 expression (Fig. S7). Furthermore, increased TF expression, TF activity in miR-145 inhibitor administered group also confirmed the regulation of TF expression by miR-145 (Fig. 6A and B). Decrease in thrombus formation, prothrombin time (PT) and clotting time further reinforced the pro-coagulant effect of miR-145 inhibitor in thrombotic animals (Fig. 6C–F). A slight decrease in bleeding time was found in miR-145 inhibitor administered group though change was not significant (Fig. S8). Altogether, these observations confirmed that miR-145 poses an antithrombotic potential by regulating TF expression.
Fig. 6.
miR-145 inhibitor administration increases thrombogenesis.
Animals were administered with miR-145 inhibitor by intravenous injection prior to IVC ligation then euthanized after 24 h. (A) qRT-PCR for TF mRNA expression. (B) TF activity showing inhibition of miR-145 significantly increases the TF levels (n = 5). (C) Thrombus weight increases in the thrombotic group administered with miR-145 inhibitor compared to thrombotic group (n = 5). (D) Representative photographs of gross IVC with thrombus (Upper panel) and representative heat map images of IVC with thrombus (lower panel) showing thrombus density. Reduced (E) clotting time and (F) prothrombin time (PT) in the thrombotic animal group administered with miR-145 inhibitor compared with a thrombotic group (n = 5). Data are presented as mean ± SEM. *p < 0.05; **p < 0.01, ***p < 0.005. See also Figs. S7 and S8.
4. Discussion
In this study, we blended the network-based bioinformatics approach together with in vivo animal model as well as human studies and identified miR-145 as a critical regulatory factor for the development of VT. More importantly, this regulation was found to be mediated by regulating the expression of TF, an important trigger for initiation of the extrinsic pathway of the coagulation cascade. The present study explicitly demonstrated intravenous administration of miR-145 mimic suppresses thrombosis.
We used in silico network theory to decipher the additional system-level regulatory mechanisms of miRNAs in the complex disease-miRNAs network. This approach provides support for the application of network theory to understand the integrative origins of disease and could be extended to other cardiovascular diseases. Our use of network theory and experimental validation in an animal model as well as in human predicted and confirmed miRNA-based disease network in VT. Moreover, we also established the association between the miR-145 and thrombosis both in in vivo animal model and human diseased state (Fig. 1). We used stasis animal model of VT that provides a total stasis environment and results in the most severe vein wall reaction to thrombosis as compared to the other models. IVC ligation model is a combination of stasis-induced vein wall injury and enhanced TF expression in endothelial cells and leukocytes induce thrombosis (Zhou et al., 2009). Moreover, this model resembles more with clinical VT conditions as compared to other animal models e.g. flow models or partial stasis model known as IVC stenosis model and ferric chloride injury model. A significant disadvantage of these models is the large variation in the size of the thrombus with even absence of thrombi in few animals.
Although, dating back to the previous reports which showed that miR-145 modulates the balance of VSMCs proliferation and contraction/differentiation in vascular diseases (Rangrez et al., 2011), in our study, the assumed role of miR-145 was further investigated by prediction of target genes. The predicted target genes, when subjected to gene enrichment pathway analysis, identified genes with potential regulatory roles in coagulation. Identification of TF, a key upstream regulator of coagulation as a target of miR-145 could be an important step towards elucidation of etiopathology of VT. Since a significant negative correlation was observed between TF expression and miR-145 that generated additional support to our findings and prompted further investigations into the regulatory role of miR-145 in TF expression. Literature evidences are available where different miRNAs have been shown to regulate TF expression. For instance, the down-regulation of miR-19b and miR-20a have been observed in patients with systemic lupus erythematosus and antiphospholipid syndrome, where increased TF expression results in a hypercoagulable state (Teruel et al., 2011). Similarly, miR-223 also suppresses TF expression in thrombogenesis during atherosclerotic plaque rupture (Li et al., 2014). However, these miRNAs (miR-19b, miR-20a, and miR-223) were eliminated during the in silico disease-network analysis for any association with VT. We did not emphasize to analyze expression of these miRNAs in our study. However, their possible involvement in other vascular disorders cannot be overlooked and could be exploited in future investigations. Nevertheless a recent study showed that circulating miR-126 exhibits antithrombotic properties via regulating post-transcriptional TF expression, thereby impacting the hemostatic balance of the vasculature in diabetes mellitus (Witkowski et al., 2016), however, in our study, we could not find any significant change in miR-126 expression in thrombosis. Different disease conditions as well as inter species variation might be the explanation for this variance. Since inflammation is a key pathological feature of thrombosis therefore, it is worthwhile noting that enrichment analysis using predicted target genes of miR-145 resulted in enrichment of “inflammation mediated by chemokine and cytokine signalling pathway”. This is in agreement with the previous study identifying miR-145 to negatively regulate pro-inflammatory cytokine release from airway smooth muscle cells in chronic obstructive pulmonary disease by targeting SMAD3 (O'Leary et al., 2016).
In our study, downregulation of miR-145 in VSMCs, PBMCs and platelets and their association with TF expression were found in thrombotic animals that dissected the cellular origin of miR-145. Though, the previous report, related to atherosclerosis, demonstrated that secretion of miR-145 by endothelial cells influences the function of VSMCs and reduces the progression of atherosclerosis (Hergenreider et al., 2012). It was also suggested that the transfer of miR-145 between vascular cells within the atherosclerotic plaques might influence gene expression in the recipient cells. Complying with such reports, and looking at miR-145 and TF expression in our model system, the potential sources of both miR-145 and TF included the VSMCs in vessel wall, along with circulating cells like platelets, and leukocytes as shown by a previous study (Zhou et al., 2009), However, additional studies might provide evidence into the transfer and secretion of miR-145 during thrombus formation.
Interestingly, we also observed decreased expression levels of miR-145 in VT patients, with an inverse trend between miR-145 and TF expression levels (Fig. 2). The circulating or blood-borne TF in humans has been studied in the recent past, and hematopoietic cell-derived microparticles with TF on their outer surface have been known for their role in thrombosis (Zwicker et al., 2011). As microparticles are also the major carrier of TF in circulation (Diehl et al., 2012) we measured the microparticle-associated TF activity in patients that exhibited a negative correlation with miR-145 expression whereas levels of TFPI remained similar (data not shown). These data strongly implicate the selective regulation of TF expression by miR-145 could be an important regulatory mechanism for maintaining hemostasis both in humans as well as animals.
The administration of miRNA mimics to normal tissue is unlikely to induce adverse events as no sign of inflammation (data not shown) or any alterations in toxicity markers (Fig. S5) were observed. Nonetheless, the reduced thrombus size (Fig. 5) in animals administered with miR-145 mimic, further supported the notion that restoration of miR-145 levels might be used as an effective strategy to manage the burden of VT and post-thrombotic syndrome. Our findings are also consistent with previous studies, suggesting TF as a potential therapeutic target for thrombosis, where anti-TF antibodies and small molecule inhibitors of the TF: FVIIa complex have shown to reduce thrombus size in arterial and venous models of thrombosis using rabbits and nonhuman primates (Pawashe et al., 1994, Szalony et al., 2003). In a previous study, a monoclonal antibody directed against TF, known as ALT836, has developed which binds to TF at its FX binding site. In patients with stable coronary artery disease enrolled in the PROXIMATE-TIMI 27 trial, this antibody had an interesting anticoagulant effect without any significant side effect such as bleeding (Morrow et al., 2005). Likewise, in our study; the antithrombotic effect was shown by inhibiting TF expression through miRNA mimic administration. The current antithrombotic strategies which involve targeting the coagulation factor-Xa and thrombin, although showed a remarkable success to manage VT, yet it is highly desirable to find a molecule lying upstream of the hemostatic pathway for the designing of an efficient and safer anticoagulant. Our dataset representing the role of miR-145 as an upstream regulator of TF is a definite foot forward in this regard.
In summary, we demonstrate a regulatory mechanism consisting of TF and miR-145 expression. Functional studies indicated that the modulation of miR-145 reduced thrombus formation via downregulation of TF. This study also provides an insight into the molecular mechanism of VT. Thus, our study highlights the role of miR-145 in the etiopathology of VT and also underscores the use of miR-145 mimic as a promising advancement to the field.
Source of Funding
This study was funded by Defence Research and Development Organization, project SL-10/DIP-255. A.S is a Senior Research Fellow of the University Grant Commission.
Conflict of Interest
There are no conflicts of interest to this study.
Author Contribution
A.S. performed the experiments, analyzed the data, and wrote the manuscript; P.K.J. performed bioinformatics analysis, D.H. participated in miRNA expression study, N.G. performed animal experiments; T.T helped in analyzing the data and edited the manuscript; V·N and T.C. participated in the human study; A.P. conducted the ELISA experiments and helped in statistical analysis; S·S performed coagulation assays; B.K. carried out SNP analysis; S·S participated in toxicity assays; S.G. participated in miRNA studies and edited the manuscript, M.Z.A. designed the study, interpreted the data, and drafted the manuscript.
Acknowledgments
The authors are extremely thankful to all the volunteers who participated in the study. We are grateful to Dr. Shashi Bala Singh (Director, DIPAS), Directorate General Armed Forces Medical Sciences. Dr. MA Qadar Pasha and Dr. Perwez Alam, CSIR-Institute of Genomics and Integrative Biology, Delhi, India for their help in cloning. Dr. Iti Garg, Dr. R. J. Tirpude, Ms. Aatira Vijay and Mr. Bhagwat Singh for their support during the study.
The following are the supplementary data related to this article.
Supplementary material 1
Supplementary material 2
Supplementary material 3
Supplementary material 4
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2017.11.022.
References
- Babson A.L., Phillips G.E. A rapid colorimetric assay for serum lactic dehydrogenase. Clin. Chim. Acta. 1965;12:210–215. doi: 10.1016/0009-8981(65)90032-x. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Saez P., Chagoyen M., Tirado F., Carazo J.M., Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8:R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Ridzon D.A., Broomer A.J. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz J.A., Obi A., Myers D.D., Wrobleski S.K., Henke P.K., Mackman N., Wakefield T.W. Critical review of mouse models of venous thrombosis. Arterioscler. Thromb. Vasc. Biol. 2012;32:556–562. doi: 10.1161/ATVBAHA.111.244608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl P., Fricke A., Sander L., Stamm J., Bassler N., Htun N., Ziemann M., Helbing T., El-Osta A., Jowett J.B., Peter K. Microparticles: major transport vehicles for distinct microRNAs in circulation. Cardiovasc. Res. 2012;93:633–644. doi: 10.1093/cvr/cvs007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing L.J., Strieter R.M., Kadell A.M., Wilke C.A., Austin J.C., Hare B.D., Burdick M.D., Greenfield L.J., Wakefield T.W. IL-10 regulates thrombus-induced vein wall inflammation and thrombosis. J. Immunol. 1998 Aug 1;161(3):1471–1476. [PubMed] [Google Scholar]
- Dweep H., Sticht C., Pandey P., Gretz N. miRWalk–database: prediction of possible miRNA binding sites by "walking" the genes of three genomes. J. Biomed. Inform. 2012;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Heit J.A., O'Fallon W.M., Petterson T.M., Lohse C.M., Silverstein M.D., Mohr D.N., Melton L.J. Relative impact of risk factors for deep vein thrombosis and pulmonary embolism: a population-based study. Arch. Intern. Med. 2002;162:1245–1248. doi: 10.1001/archinte.162.11.1245. [DOI] [PubMed] [Google Scholar]
- Henke P.K., Varga A., De S., Deatrick C.B., Eliason J., Arenberg D.A., Sukheepod P., Thanaporn P., Kunkel S.L., Upchurch G.R.Jr., Wakefield T.W. Deep vein thrombosis resolution is modulated by monocyte CXCR2-mediated activity in a mouse model. Arterioscler. Thromb. Vasc. Biol. 2004;24:1130–1137. doi: 10.1161/01.ATV.0000129537.72553.73. [DOI] [PubMed] [Google Scholar]
- Hergenreider E., Heydt S., Treguer K., Boettger T., Horrevoets A.J., Zeiher A.M., Scheffer M.P., Frangakis A.S., Yin X., Mayr M., Braun T., Urbich C., Boon R.A., Dimmeler S. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell Biol. 2012;14:249–256. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- Ji R., Cheng Y., Yue J., Yang J., Liu X., Chen H., Dean D.B., Zhang C. Microrna expression signature and antisense-mediated depletion reveal an essential role of microrna in vascular neointimal lesion formation. Circ. Res. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- Konieczna J., Sánchez J., Palou M., Picó C., Palou A. Blood cell transcriptomic-based early biomarkers of adverse programming effects of gestational calorie restriction and their reversibility by leptin supplementation. Sci. Rep. 2015;5:9088. doi: 10.1038/srep09088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Chen H., Ren J. MicroRNA-223 inhibits tissue factor expression in vascular endothelial cells. Atherosclerosis. 2014;237:514–520. doi: 10.1016/j.atherosclerosis.2014.09.033. [DOI] [PubMed] [Google Scholar]
- Liu X., Cheng Y., Zhang S., Lin Y., Yang J., Zhang C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ. Res. 2009;104:476–487. doi: 10.1161/CIRCRESAHA.108.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone P.C., Agutter P.S. The etiology of deep venous thrombosis. QJM. 2006;99:581–593. doi: 10.1093/qjmed/hcl070. [DOI] [PubMed] [Google Scholar]
- Morrow D.A., Murphy S.A., McCabe C.H., Mackman N., Wong H.C., Antman E.M. Potent inhibition of thrombin with a monoclonal antibody against tissue factor: results of the PROXIMATE-TIMI 27 trial. Eur. Heart J. 2005;26:682–688. doi: 10.1093/eurheartj/ehi094. [DOI] [PubMed] [Google Scholar]
- Nogales-Cadenas R., Carmona-Saez P., Vazquez M., Vicente C., Yang X., Tirado F., Carazo J.M., Pascual Montano A. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37:W317–W322. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary L., Sevinç K., Papazoglou I.M., Tildy B., Detillieux K., Halayko A.J., Chung K.F., Perry M.M. Airway smooth muscle inflammation is regulated by microRNA-145 in COPD. FEBS Lett. 2016;590:1324–1334. doi: 10.1002/1873-3468.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawashe A.B., Golino P., Ambrosio G., Migliaccio F., Ragni M., Pascucci I., Chiariello M., Bach R., Garen A., Konigsberg W.K. A monoclonal antibody against rabbit tissue factor inhibits thrombus formation in stenotic injured rabbit carotid arteries. Circ. Res. 1994;74:56–63. doi: 10.1161/01.res.74.1.56. [DOI] [PubMed] [Google Scholar]
- Rangrez A.Y., Massy Z.A., Metzinger-Le Meuth V., Metzinger L. miR-143 and miR-145: molecular keys to switch the phenotype of vascular smooth muscle cells. Circ. Cardiovasc. Genet. 2011;4:197–205. doi: 10.1161/CIRCGENETICS.110.958702. [DOI] [PubMed] [Google Scholar]
- Reitman S., Frankel S. A colorimetric method for the determination of serum glutamic oxalacetic and glutamic pyruvic transaminases. Am. J. Clin. Pathol. 1957;28:56–63. doi: 10.1093/ajcp/28.1.56. [DOI] [PubMed] [Google Scholar]
- Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Small E.M., Frost R.J., Olson E.N. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–1032. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Icli B., Wara A.K., Belkin N., He S., Kobzik L., Hunninghake G.M., Vera M.P., MICU Registry, Blackwell T.S., Baron R.M., Feinberg M.W. MicroRNA-181b regulates NF-κBmediated vascular inflammation. J. Clin. Invest. 2012;122:1973–1990. doi: 10.1172/JCI61495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalony J.A., Suleymanov O.D., Salyers A.K. Administration of a small molecule tissue factor/factor VIIa inhibitor in a non-human primate thrombosis model of venous thrombosis: effects on thrombus formation and bleeding time. Thromb. Res. 2003;112:167–174. doi: 10.1016/j.thromres.2003.10.017. [DOI] [PubMed] [Google Scholar]
- Tabas-Madrid D., Nogales-Cadenas R., Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012;40:W478–W483. doi: 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teruel R., Pérez-Sánchez C., Corral J., Herranz M.T., Pérez-Andreu V., Saiz E., García-Barberá N., Martínez-Martínez I., Roldán V., Vicente V., López-Pedrera C., Martínez C. Identification of miRNAs as potential modulators of tissue factor expression in patients with systemic lupus erythematosus and antiphospholipid syndrome. J. Thromb. Haemost. 2011;9:1985–1992. doi: 10.1111/j.1538-7836.2011.04451.x. [DOI] [PubMed] [Google Scholar]
- Tyagi T., Ahmad S., Gupta N., Sahu A., Ahmad Y., Nair V., Chatterjee T., Bajaj N., Sengupta S., Ganju L., Singh S.B., Ashraf M.Z. Altered expression of platelet proteins and calpain activity mediate hypoxia-induced prothrombotic phenotype. Blood. 2014;123:1250–1260. doi: 10.1182/blood-2013-05-501924. [DOI] [PubMed] [Google Scholar]
- Weber C., Schober A., Zernecke A. Micrornas in arterial remodelling, inflammation and atherosclerosis. Curr. Drug Targets. 2010;11:950–956. doi: 10.2174/138945010791591377. [DOI] [PubMed] [Google Scholar]
- Witkowski M., Weithauser A., Tabaraie T., Steffens D., Kränkel N., Witkowski M., Stratmann B., Tschoepe D., Landmesser U., Rauch-Kroehnert U. Micro-RNA-126 reduces the blood thrombogenicity in diabetes mellitus via targeting of tissue factor. Arterioscler. Thromb. Vasc. Biol. 2016;36:1263–1271. doi: 10.1161/ATVBAHA.115.306094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., May L., Liao P., Gross P.L., Weitz J.I. Inferior vena cava ligation rapidly induces tissue factor expression and venous thrombosis in rats. Arterioscler. Thromb. Vasc. Biol. 2009;29:863–869. doi: 10.1161/ATVBAHA.109.185678. [DOI] [PubMed] [Google Scholar]
- Zwicker J.I., Trenor C.C., Furie B.C., Furie B. Tissue factor-bearing microparticles and thrombus formation. Arterioscler. Thromb. Vasc. Biol. 2011;31:728–733. doi: 10.1161/ATVBAHA.109.200964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2
Supplementary material 3
Supplementary material 4