Abstract
Mammalian aging is associated with decline in cognitive functions. Studies searching for a cause of cognitive aging initially focused on neuronal loss but quantitative investigations of rat, monkey, and human brain using stereology demonstrated that in normal aging, unlike in neurodegenerative disease, neurons are not lost. Instead, electron microscopic and MRI studies in normal aging monkeys revealed age-related damage to myelin sheaths, loss of axons, and reduction in white matter volume which correlates with cognitive impairments. However, little is known about the cause of myelin defects or associated axon loss. The present study investigates the effect of age on signaling pathways between oligodendroglia and neurons using a custom PCR array to assess the expression of 87 genes of interest in cortical gray matter and white matter from the inferior parietal lobe (IPL) of normal rhesus monkeys ranging in age from 4.2 to 30.4 years old. From this array data, five target genes of interest were selected for further analysis to confirm gene expression and measure protein expression. The most interesting target gene identified is brain-derived neurotrophic factor (BDNF), which was the only gene that was altered at both mRNA and protein levels. In gray matter, BDNF mRNA was decreased. While the level of the mature form of the protein was unchanged, there was a specific decrease in the precursor form of BDNF. These alterations in the BDNF in gray matter could contribute to the vulnerability and loss of the axons with age.
Electronic supplementary material
The online version of this article (10.1007/s11357-018-0006-2) contains supplementary material, which is available to authorized users.
Keywords: Aging, White matter, BDNF, Parietal cortex, Gene expression, Protein expression
Introduction
Studies of neurodegenerative diseases such as Alzheimer’s have revealed widespread loss of neurons in the cortex and other areas of the human brain (Whitehouse et al. 1982; Gómez-Isla et al. 1996). In contrast, in normal aging humans who escape frank neurodegenerative diseases, growing evidence indicates that neurons are preserved even though there is mild cognitive decline (Haug 1985; Terry et al. 1987; Freeman et al. 2008). In searching for the neurological bases of these cognitive impairments, age-related pathology in white matter has been reported in humans (Marner et al. 2003; Vernooij et al. 2008) and in the rhesus monkey model of normal aging (Peters et al. 2000; Makris et al. 2007). Studies of normal aging monkeys have been particularly illuminating as the monkey has brain structure and cognitive functions more similar to humans than other laboratory species, is long-lived with a maximal life span of over 30 years of age, lives in well-characterized and relatively uniform environmental conditions, and allows for optimal preparation of brain tissue in close proximity to behavioral assessment.
Studies of cognitively characterized rhesus monkeys have shown that age-related changes in white matter are strongly associated with age-related cognitive impairments (Sloane et al. 1999; Peters et al. 2000; Makris et al. 2007). Moreover, both electron microscopic (Bowley et al. 2010) and biochemical studies (Duce et al. 2008; Hinman et al. 2008) have shown that the white matter changes are associated with a variety of pathological changes in myelin. Some of these changes in white matter include a decrease with age of the messenger ribonucleic acid (mRNA) and protein levels of klotho, a longevity factor important for myelin homeostasis (Duce et al. 2008; Chen et al. 2013). Additionally, with age, there is an increased accumulation and fragmentation of the oligodendrocyte enzyme, 2′,3′-cyclic nucleotide phosphodiesterase (CNP) that is associated with increased ubiquitination of the protein (Hinman et al. 2008).
In addition to these biochemical changes, a variety of morphological changes are also present in the aged white matter. There is an increase of activated astrocytes and microglia in the white matter (Sloane et al. 1999; Sloane et al. 2000; Shobin et al. 2017). Axons are also lost in the white matter and can be seen in the electron microscope as degenerating axons contained within degenerating myelin sheaths in keeping with the observed decrease in density of myelinated axons (Bowley et al. 2010; Peters et al. 2010). These alterations in white matter also correlated with age-related cognitive decline.
While these data suggest that white matter is a critical nexus for age-related brain pathology, the causes underlying this white matter pathology are unknown. There are many possible mechanisms that might contribute to this pathology, but likely candidates include direct damage to myelin by age-related oxidative stress and/or inflammation, age-related decline in the capacity of oligodendroglia to maintain and repair myelin, and age-related disruptions in any of the multiple molecular signaling pathways essential for myelin homeostasis. The present study explored this latter possibility using a custom PCR gene array to examine a number of candidate gene pathways pertinent to myelin homeostasis, inflammation, and molecular signaling including neurotrophic factors, inflammatory cytokines, and neurotransmission.
Experimental methods
Subjects
A total of 27 adult rhesus monkeys (Macaca mulatta) were used in this study and are summarized in Table 1. This group consisted of 14 male and 13 female monkeys ranging in age from 4.4 to 30.4 years, effectively covering the entire adult life span (Tigges et al. 1988). Monkeys were obtained from colonies of the Yerkes National Primate Research Center (Atlanta, GA) and Labs of Virginia (Yemassee, SC). Before entering the study, they were screened for any diseases that could affect the central nervous system as well as given MRI scans to screen for occult brain damage. Once entered, the animals were housed in the Animal Science Center of Boston University Medical Campus (BUMC), which is fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care. All procedures were in accord with NIH guidelines and approved by the Institutional Animal Care and Use Committee of BUMC.
Table 1.
Subject list
| ID | Age (years) | Sex | Perfusate | Source | PCR array | qRT-PCR + WB | Behavioral testing | Dissection |
|---|---|---|---|---|---|---|---|---|
| AM081x | 4.2 | M | Krebs | YNPRC | X | No | Frozen | |
| AM147x | 4.4 | F | Krebs | YNPRC | X | X | No | Frozen |
| AM078 | 5.7 | F | Krebs | YNPRC | X | X | Yes | Frozen |
| AM146x | 6.3 | F | Krebs | YNPRC | X | X | No | Frozen |
| AM131 | 6.6 | F | Krebs | YNPRC | X | X | Yes | Frozen |
| AM14 8 h | 7.3 | M | Krebs | YNPRC | X | Yes | Frozen | |
| AM127 | 7.3 | M | Krebs | YNPRC | X | Yes | Frozen | |
| AM145x | 8.7 | M | Krebs | VA | X | No | Frozen | |
| AM092b | 8.8 | M | Krebs | YNPRC | X | Yes | Frozen | |
| AM163 | 10.1 | F | Krebs | YNPRC | X | X | Yes | Frozen |
| AM079x | 10.8 | M | Krebs | YNPRC | X | No | Frozen | |
| AM139x | 12.6 | M | Krebs | VA | X | No | Frozen | |
| AM252 | 19.9 | F | Krebs | YNPRC | X | Yes | Fresh | |
| AM160 | 20.6 | F | Krebs | YNPRC | X | X | Yes | Frozen |
| AM256 | 20.7 | F | Krebs | YNPRC | X | X | Yes | Fresh |
| AM126 | 20.9 | F | Krebs | YNPRC | X | X | Yes | Frozen |
| AM123 | 21.0 | M | Krebs | YNPRC | X | Yes | Frozen | |
| AM165 | 22.6 | M | Krebs | YNPRC | X | Yes | Frozen | |
| AM06 7 L | 24.0 | M | Krebs | YNPRC | X | Yes | Frozen | |
| AM243 | 24.4 | M | Krebs | YNPRC | X | Yes | Fresh | |
| AM164 | 24.5 | M | Krebs | YNPRC | X | Yes | Frozen | |
| AM107b | 26.0 | F | Krebs | YNPRC | X | X | Yes | Frozen |
| AM120 | 26.5 | F | Krebs | YNPRC | X | X | Yes | Frozen |
| AM098 | 28.1 | F | Krebs | YNPRC | X | X | Yes | Frozen |
| AM082 | 28.1 | F | Krebs | YNPRC | X | X | No | Frozen |
| AM04 8 L | 30.2 | M | Krebs | YNPRC | X | Yes | Frozen | |
| AM073 | 30.4 | M | Krebs | YNPRC | X | No | Frozen |
Krebs Krebs-Heinseleit buffer, YNPCR Yerkes National Primate Research Center, Atlanta, GA; VA Labs of Virginia, Yemassee, SC. Array PCR Array, qRT-PCR quantitative real-time PCR, WB western blot
Cognitive assessment
The majority of monkeys received a battery of behavioral tasks to assess cognitive performance. These tasks have been described in detail elsewhere (Herndon et al. 1997; Moore et al. 2005) but briefly included measures of learning (acquisition of the Delayed Non-Match to Sample Task—DNMS), recognition memory (delay conditions on DNMS), working memory (object and spatial conditions of the Delayed Recognition Span Task—DRST), attention, associative memory, and executive function (learning the stimulus categories and shifting set in Category Set Shifting Task—CSST). A subset of the learning and memory tasks were identified by principal components as the best predictors of cognitive decline (Herndon et al. 1997) and have been designated the Cognitive Impairment Index (CII) and used to assess associations with various neurobiological endpoints (e.g.,Peters et al. 2010).
Tissue harvest
At the conclusion of all testing, subjects were deeply anesthetized to a surgical level with sodium pentobarbital; their head fixed in a stereotactic device, and a partial craniotomy was done. The chest was then opened and they were transcardially perfused with 4 to 8 l of ice cold (4 °C) Krebs-Heinseleit buffer (6.41 mM Na2HPO4, 1.67 mM NaH2CO3, 137 mM NaCl, 2.68 mM KCl, 5.55 mM Glucose, 0.34 mM CaCl2, 2.14 mM MgCl2, pH 7.4) prepared in RNase free conditions. After all blood was cleared from the cortex, but while the cold buffer was still flowing, fresh samples of cortex and underlying white matter were removed from the dorsal half of the left inferior parietal lobule (IPL), flash frozen on dry ice, and stored at − 80 °C until processed for molecular and biochemical studies. These samples were typically frozen less than 10 min from the start of anoxia and always less than 15 min. Samples from the IPL were dissected into cortical gray matter and subcortical white matter either before freezing or while frozen as indicated in Table 1. Once the fresh samples were removed, the perfusate was switched to a mixture of 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4, 37 °C) to fix the remainder of the sampled hemisphere and the entire intact hemisphere so that they could be cryoprotected and frozen for additional studies.
RNA and protein extraction
Frozen tissue blocks from the IPL of cortical gray matter and subcortical white matter were further dissected into pieces weighing 100 mg to isolate the RNA and protein using TRIzol solution following the manufacturer’s protocol (Invitrogen). Briefly, 100 mg of tissue was added to 1 ml of TRIzol reagent and homogenized with a handheld pestle and passed through a 20-gauge needle. The RNA extract was further processed using RNeasy Mini Kit and treated with DNase I (Qiagen) to clean up the RNA and remove any DNA contamination. The integrity and the concentration of the RNA were determined by UV spectrophotometry using a NanoDrop1000 (Thermo Scientific). The protein was then isolated from the phenol-ethanol supernatant and washed with 0.3 M guanidine hydrochloride in 95% ethanol and dissolved in 1%SDS. The concentration of the protein was determined using the BCA protein assay (Pierce Biotechnology). Extracts of RNA and protein were then stored at − 80 °C until analyzed.
PCR Array
To identify potential gene targets involved in white matter deterioration and cognitive decline, samples from a subset of the total number of subjects, (seven old and five young female monkeys—identified in Table 1), were processed utilizing custom PCR gene array plates containing primers sets for 87 genes of interest, 3 housekeeping genes, and 6 internal PCR controls. The genes of interest fall into 8 main categories: (1) neurotrophins/growth factors and their receptors, (2) transcription factors, (3) GABAergic neurotransmission, (4) cytokines and receptors, (5) signal transduction, (6) oxidative stress and DNA damage, (7) cellular organization and migration, and (8) glial cell markers. The full list appears in in Table S1 located in Online Resource 1. The custom PCR arrays contained the primers for the genes of interest that were designed for us and obtained from SABiosciences (CAPX-0447; Frederick, MD) based on rhesus monkey gene sequences. Processing began using 0.5 μg total RNA in the reverse transcriptase reaction at 42 °C for 15 min to create the cDNA template. Then, the reverse transcriptase mixture containing the cDNA template was mixed with the SYBR Green detection reagents and 10 μL of the mixture was loaded into each well of the plate and the PCR was run as follows: 1 cycle of 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s and for 1 min at 60 °C using SYBR green detection on an Applied Biosystems (ABI) Prism 7900HT. The relative fold changes in gene expression were analyzed by comparing the group means of the young versus old animals using comparative CT method (2-ΔCT) and normalizing values to the average of the three housekeeping genes included in the array: beta-actin (ACTB), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and cyclophillin A (PPIA).
Quantitative real-time PCR
To verify a selected set of five genes from the PCR array results, real-time PCR was run on RNA samples from all 27 monkeys (Table 1) using commercially available TaqMan primer and probe sets designed to rhesus monkey genes using FAM-labeled MGB probes. These included the following: (1) brain-derived neurotrophic factor (BDNF) (Rh02788328_s1, Applied Biosystems, Foster City, CA); (2) meltrin-beta (ADAM19) (Rh02851168_m1, Applied Biosystems, Foster City, CA); (3) neurotrophic tyrosine kinase receptor, type 2 (NTRK2) (Rh02831791_m1, Applied Biosystems, Foster City, CA); (4) nerve growth factor receptor (NGFR) (Rh02842493_m1, Applied Biosystems, Foster City, CA); and (5) leukemia inhibitory factor-like (LOC715456; LIF) (Rh02841805_m1, Applied Biosystems, Foster City, CA). Control primers and VIC-labeled TAMARA-quenched probes to 18S rRNA (4308329, Applied Biosystems, Foster City, CA) or RPL13A (Forward: TCACGAGGTTGGCTGGAAGT, Reverse: GATCTTGGCTTTCTCCTTCCTCTT Probe: CCAGGCAGTGACAGCCACCTTGG sequences from (Noriega et al. 2010)) were used in a multiplex assay. Total RNA was tested for expression using 40 ng RNA from the gray matter, and 60 ng RNA from the subcortical white matter for BDNF, ADAM19, and NTRK2. For gene expression of NGFR, 300 ng of total white matter RNA was analyzed. Thermocycling was done on an ABI PRISM 7900 HT using the parameters of 50 °C for 30 min, 95 °C for 10 min, and 50 cycles of 95 °C for 15 s and then 60 °C for 1 min. A standard curve was calculated for each reaction with the RNA of a young subject that allowed for the determination of the quantity of the RNA expression for each gene analyzed, and the data from both the gray matter and the white matter for BDNF, ADAM19, and NTRK2 were normalized to 18S rRNA expression. The white matter data from BDNF and NGFR were normalized to RPL13A expression to minimize the difference between the threshold cycle (CT) values between the gene of interest and the normalization gene.
Antibodies
To assess protein levels, western blot analysis was done using antibodies against (1) proBDNF (Alomone Labs, ANT-006 Lot AN-05), (2) BDNF (Santa Cruz Biotechnology, sc-546), (3) tyrosine kinase receptor B (TrkB) (Millipore, AB5372), (4) p75 nerve growth factor receptor (p75 NGFR) (Novus Biologicals, M-009-100), (5) β-tubulin (Invitrogen, 32–2600), and (6) myelin oligodendrocyte glycoprotein (MOG) (Millipore, MAB5680). Secondary detection antibodies were HRP-linked goat anti-mouse and goat anti-rabbit (GE Life Sciences).
Western blot analysis
Protein samples were diluted 1:3 with a 3× sample buffer prepared fresh (240 mM Tris-HCl, pH 6.8, 6% SDS, 30% Glycerol, 16% β-mercaptoethanol) and boiled at 95 °C for 10 min before loading the protein into a 4–20% Tris-Glycine gel (Mini Protean TGX, Bio-Rad). For all blots, 20 μg of total protein was loaded on the gel. The gel was run at 200 V for 30 min in running buffer (193 mM glycine, 25 mM Tris base, 0.1% SDS). The protein was then transferred to a polyvinylidene difluoride membrane (PVDF) at 0.2 A for 2 h in transfer buffer (39 mM glycine, 48 mM Tris base, 20% methanol). After transfer, the membranes were rinsed in Tris buffered saline containing 0.1% Tween-20 (TBST) and then blocked with 5% non-fat dry milk in TBST for 1 h at room temperature (RT). Membranes were then incubated overnight at 4 °C in primary antibody (BDNF 1:200, pro-BDNF 1:500, TRKB 1:1000, NGFR 1:500). Membranes were then washed in TBST and then incubated in HRP-linked secondary antibody (1:20,000) in blocking buffer for 2 h at RT. Then, the membranes were further washed and the proteins were visualized using Amersham ECL Western Blotting Detection Reagents and Amersham Hyperfilm ECL (GE Healthcare).
The membranes were rinsed in TBST and the remaining HRP activity from the detection reagents was inactivated by rinsing the blot in 100% ethanol and then in 15% hydrogen peroxide in TBST for 15 min at 37 °C and washed again in TBST and blocked for 1 h at RT in blocking buffer. Finally, for normalization, the membranes were then probed with antibodies for β-tubulin (1:1000) for gray matter samples or MOG (0.3μg/ml) for the white matter samples and developed using the same methods described above. Quantification of the density of all bands was performed using ImageJ (NIH) and normalizing to the reference protein. Controls utilized were omission of the primary antibody as well as preabsorption of the proBDNF antibody with an 80-fold molar excess of the included antigen (Alomone Labs, Lot AG-02) to determine antibody specificity.
Statistical analysis
The PCR gene array was analyzed as the mean fold change in expression in the old (20.1–28.1 years old) versus the young (4.4–10.1 years old) animals and t tests were run for each gene on the array. For array data, the Benjamini and Hochberg false discovery rate was performed to correct for the problem of multiple comparisons. Because of the limited sample size, it eliminated significance for even the largest fold change effects. Hence, the uncorrected values are presented since we performed real-time PCR and western blots on individual genes with the largest fold changes to follow-up on the array results and eliminate the need for multiple comparison correction. The individual gene and western blot results were analyzed with Pearson correlation analysis versus age with statistical significance set at p ≤ 0.05 for two-tailed analysis using Prism (version 6; GraphPad Software Inc).
Results
Gene expression
PCR Array
The PCR array was custom designed to include genes that are related to known anatomical and functional changes that occur in the aging brain. As shown in Table 2, out of the 87 genes investigated, before correction for multiple comparisons, there were significant changes in 16 genes comparing aged and young: 9 genes in the samples of cortical gray matter and 7 genes in the subcortical white matter. These alterations in expression occurred in genes from most of the main functional categories of genes in the array, illustrating the diversity of the aging process.
Table 2.
Genes with altered expression in the inferior parietal lobule by qPCR array
| Gene symbol | Gene name | Accession number | Fold regulation | p value* |
|---|---|---|---|---|
| IPL cortical gray matter | ||||
| IL1β | Interleukin 1, beta | NM_001042756 | 2.45 | 0.0492 |
| GADD45G | Growth arrest and DNA-damage-inducible, gamma | NM_001193910 | 2.02 | 0.0188 |
| MYC | v-myc myelocytomatosis viral oncogene homolog | NM_001142873 | 1.82 | 0.0320 |
| STAT4 | Signal transducer and activator of transcription 4 | XM_001082561 | 1.65 | 0.0420 |
| RELN | Reelin | XM_001087819 | 1.46 | 0.0336 |
| ADAM17 | ADAM metallopeptidase domain 17 | XM_002799139 | 1.29 | 0.0073 |
| GABBR1 | Gamma-aminobutyric acid (GABA) B receptor, 1 | XM_001097474 | − 1.39 | 0.0389 |
| BDNF | Brain-derived neurotrophic factor | XM_001089568 | − 1.82 | 0.0076 |
| ADAM19 | ADAM metallopeptidase domain 19 (meltrin beta) | XM_001105246 | − 1.99 | 0.0045 |
| IPL subcortical white matter | ||||
| LOC715456 (LIF) | Leukemia inhibitory factor-like | XM_001108593 | 3.38 | 0.0147 |
| TNFα | Tumor necrosis factor | NM_001047149 | 2.35 | 0.0076 |
| NGFR | Nerve growth factor receptor | XM_001090039 | 2.14 | 0.0182 |
| GADD45G | Growth arrest and DNA-damage-inducible, gamma | NM_001193910 | 1.85 | 0.0014 |
| LOC697397 (GMFG) | Glia maturation factor gamma-like | XM_001085983 | 1.82 | 0.0026 |
| CREB1 | cAMP responsive element binding protein 1 | XM_001107192 | − 1.39 | 0.0385 |
| BDNF | Brain-derived neurotrophic factor | XM_001089568 | − 1.78 | 0.0348 |
*p value uncorrected for multiple comparisons
Genes of interest for follow-up
While alterations in many of these genes could play important roles in the changes of the aging brain, four genes were selected for further analysis with qPCR based on both the size of the fold change as well as the roles in neuronal function, axonal survival, and/or myelination as well as the availability of effective antibodies. First, BDNF was selected for follow-up because of its role as a neurotrophic factor important in the growth, survival, and differentiation of neurons (Poo 2001) and the maintenance of dendrites and axons (Gorski et al. 2003; Matsunaga et al. 2004). Interestingly, BDNF expression was reduced on the gene array in both cortical gray matter (fold change = − 1.82, p = 0.0076) and subcortical white matter (fold change = − 1.78, p = 0.0348). Second, NGFR, the low affinity receptor for BDNF and other neurotrophic factors, was selected because the gene array showed it was increased in the subcortical white matter (fold change = 2.14, p = 0.0182). Third, ADAM19 was selected since it was significantly decreased on the gene array in the gray matter (fold change = − 1.99, p = 0.0045) and approached significance in the subcortical white matter (fold change = − 1.54, p = 0.0524) and is important for the cleavage of Neuregulin-1 (Shirakabe et al. 2000), a signaling molecule for myelin sheath thickness (Michailov et al. 2004). Fourth, leukemia inhibitory factor (LIF) was also selected for further analysis since it had the highest fold change (fold change = 3.38, p = 0.0147) of any of the potential targets on the gene array and is important for oligodendrocyte function (Butzkueven et al. 2006). Finally, a fifth gene, NTRK2, was selected for PCR analysis. While it was not included on the PCR array due to design restraints, it is the gene encoding the high affinity receptor for BDNFand TrkB and could be important for understanding age-related alterations in the BDNF system.
Quantitative real-time PCR
These 5 genes (4 from the PCR array plus NTRK2) were studied in the full cohort of 27 animals shown in Table 1 with TaqMan qPCR.
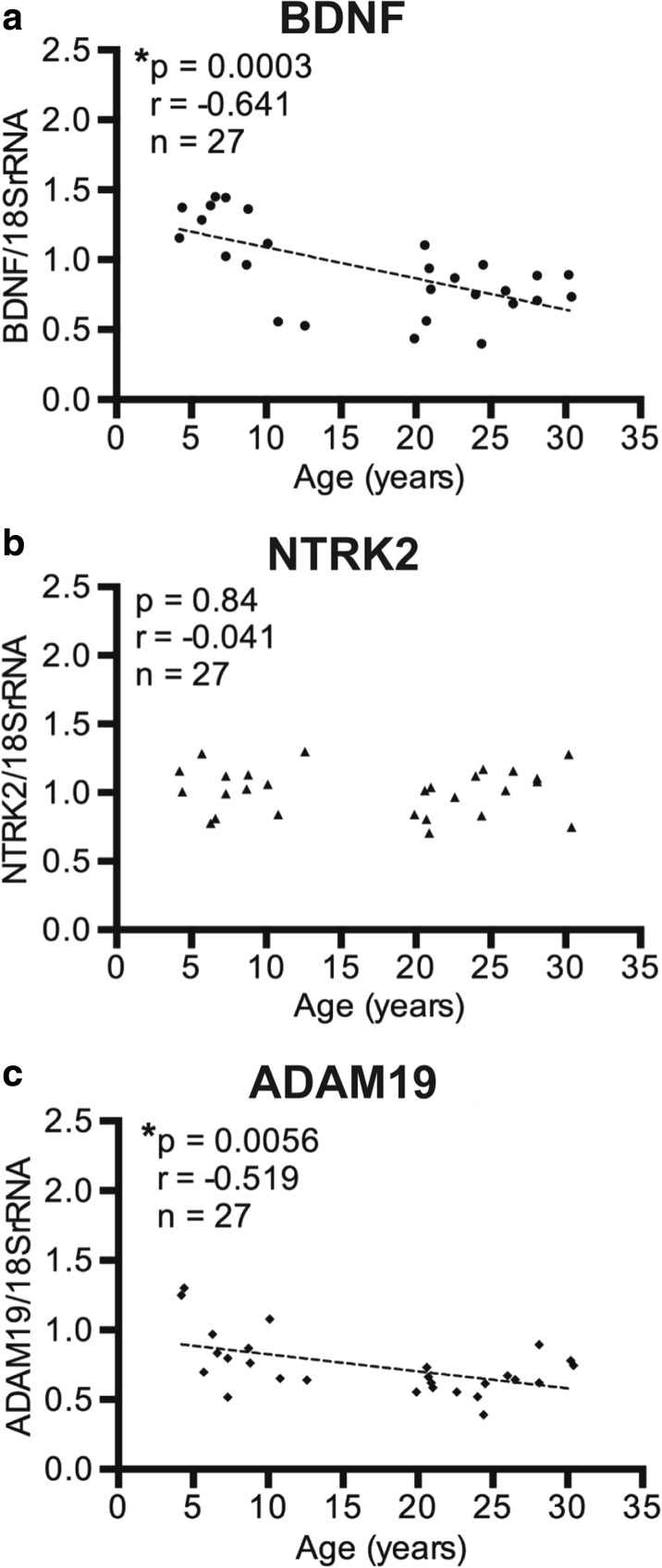
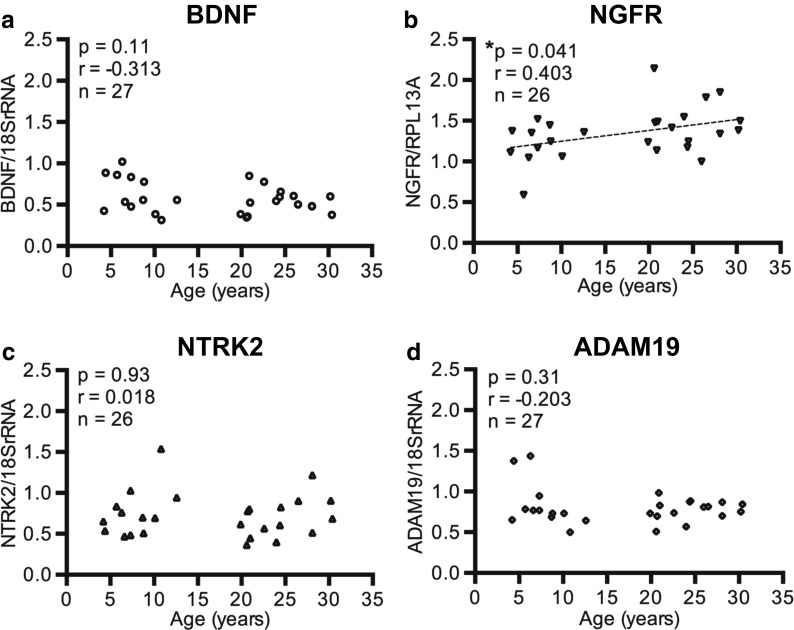
BDNF: Results demonstrated that BDNF mRNA significantly decreased with age in the cortical gray matter (r = − 0.641, p = 0.0003; Fig. 1a), confirming the array results. In contrast, in the subcortical white matter, BDNF mRNA levels only approached significance (r = − 0.313, p = 0.11; Fig. 2a).
Fig. 1.
Relative gene expression in the IPL cortical gray matter versus age. a BDNF, b NTRK2, and c ADAM19 were all standardized to the expression of 18S rRNA in 27 animals. While there was no significant change with age for NTRK2 (b), both BDNF (a), and ADAM19 (c) were significantly decreased with age in the cortical gray matter. *p < 0.05
Fig. 2.
Relative gene expression in the IPL subcortical white matter versus age. a BDNF standardized to 18S rRNA in 27 animals, b NGFR standardized to RPL13A in 26 animals (AM079x did not amplify), c NTRK2 standardized to 18S rRNA in 26 animals (AM107b did not amplify), and d ADAM19 standardized to 18S rRNA in 27 animals. Only the expression of NGFR was significantly changed, showing an increase with age. *p < 0.05
BDNF receptors NGFR and NTRK2: Two receptors for BDNF were then examined with qPCR to determine whether their expression was altered as well. Due to the low expression of NGFR mRNA and the need to increase the starting amount of mRNA, we used an alternate housekeeping gene, RPL13A, to decrease the difference between the threshold cycle (CT) values for NGFR and the housekeeping gene. NGFR was significantly increased in the subcortical white matter (r = 0.403, p = 0.041; n = 26 Fig. 2b), confirming the result from the PCR array. Note that one subject was omitted as the RNA for the housekeeping gene was amplified but NGFR RNA failed to reach the level of detection. In the gray matter, NGFR was not detectable in any of the 27 subjects and was not pursued further. In contrast to NGFR, the high affinity BDNF receptor, NTRK2, was found to be unaltered in both the cortical gray matter (r = − 0.041, p = 0.84; Fig. 1b) and in the subcortical white matter (r = − 0.018, p = 0.93; Fig. 2c).
ADAM19: The mRNA levels of ADAM19 were decreased with age in the cortical gray matter (r = − 0.519, p = 0.0056; Fig. 1c) but were not altered in the subcortical white matter (r = − 0.203, p = 0.31; Fig. 2d), confirming the array results.
LIF: The level of expression of LIF mRNA using qPCR was below the level of detection in the current experiment. Hence, like NGFR in the gray matter, the data are not presented and LIF was not examined further.
Sex Differences: Differences between the expression in males and females were examined in two ways. First, the effect of sex on qPCR expression of each gene in the total population of males was compared to expression in the total population of females. Second, a similar comparison of males versus females was done after stratifying the subjects into young (≤ 12.6 years old) and aged (≥ 19.9 years old) groups and looking at the difference of males and females in each age group. When looking at the total population, there was no significant differences between males and females in gene expression levels of BDNF, NTRK2, or ADAM19 in the gray matter (Online Resource 2, Fig. S1a). When stratified by age, there were also no differences between males and females in the gene expression levels of BDNF, NTRK2, or ADAM19 in the gray matter of the young animals (Online Resource 2, Fig. S1b), or the aged animals (Online Resource 2, Fig. S1c). In the white matter, there were no differences between males and females in BDNF, NGFR, NTRK2, and ADAM19 in the total population (Online Resource 2, Fig. S2a), within the young group (Online Resource 2, Fig. S2b), or the aged group (Online Resource 2, Fig. S2c).
Behavior: In the 6 young and 13 aged animals that had data from behavioral testing, there was no correlation between the gene expression levels and the overall cognitive impairment index (CII). When examining the individual tasks (e.g., DNMS Acquisition, perseverative errors on the set shifting task (CSST)), there were scattered correlations significant at p ≤ 0.05. However, there was no pattern to these significant findings and they did not survive correction for multiple comparisons.
Protein expression
To determine if age-related differences in the mRNA levels assayed with qPCR translate into changes at the protein level, the genes where the mRNA expression was confirmed (BDNF, NTRK2, and ADAM19 in the gray matter, and BDNF, NTRK2, ADAM19, and NGFR in the white matter) were investigated by western blot. The protein levels were standardized to the expression of β-tubulin in the gray matter and to MOG in the subcortical white matter.
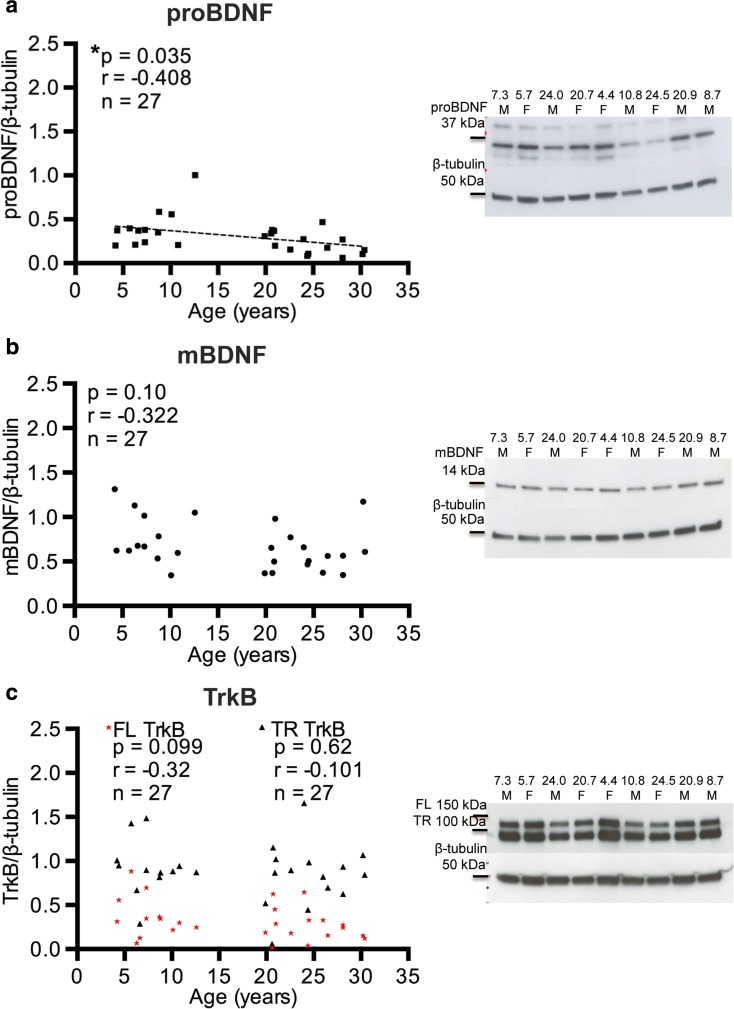
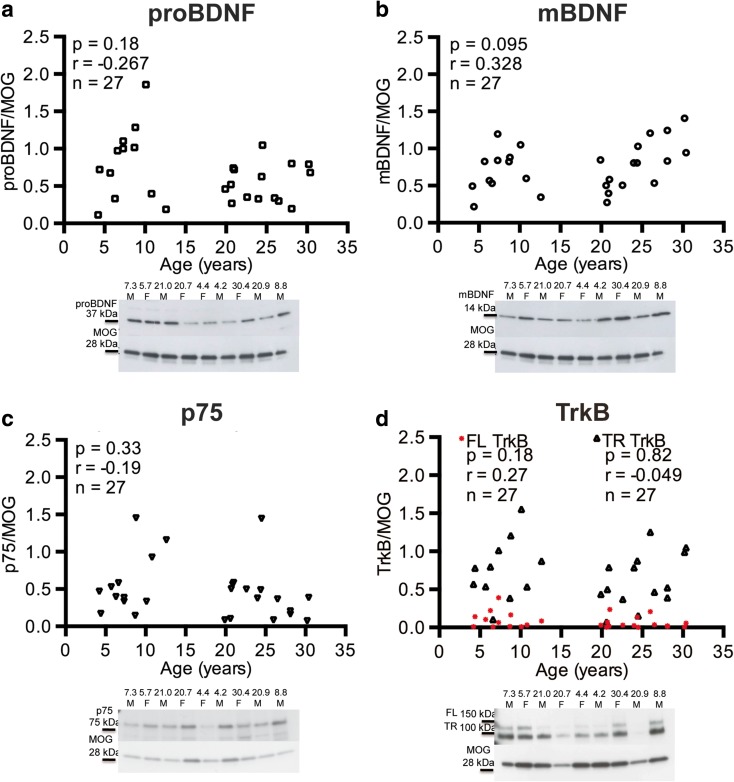
BDNF: BDNF exists in two forms at the protein level, the precursor protein proBDNF and the cleaved product, mature BDNF (mBDNF). ProBDNF was detected as a 34 kDa band by western blot, while mBDNF was detected as a 14 kDa band. ProBDNF was significantly decreased in the cortical gray matter (r = − 0.408, p = 0.035; Fig. 3a), but not in the subcortical white matter (r = − 0.267, p = 0.18; Fig. 4a). Interestingly, there was no change with age in protein levels of mBDNF in the cortical gray matter (r = − 0.019, p = 0.97; Fig. 3b) or in the subcortical white matter (r = − 0.183, p = 0.39; Fig. 4b).
Fig. 3.
Protein expression in the IPL gray matter versus age. The levels of a proBDNF, b mature BDNF (mBDNF), and c TrkB full length (FL TrkB) and truncated (TR TrkB) forms are shown standardized to β-tubulin. Representative blots are shown for each antibody. Only proBDNF showed a significant change with age in protein expression. *p < 0.05
Fig. 4.
Protein expression in the IPL white matter versus age. The levels of a mature BDNF, b proBDNF, c NGFR, and d TrkB full length (FL TrkB) and truncated (TR TrkB) forms standardized to myelin oligodendrocyte glycoprotein (MOG) are shown. Representative blots for each antibody are below each graph. There are no significant changes in any of the proteins with age in the subcortical white matter
BDNF receptors p75 NGFR and TrkB: For the BDNF receptors, protein expression of the 75 kDa band representing the low affinity p75 NGFR receptor was not significantly altered with age in the subcortical white matter (r = 0.19, p = 0.33; Fig. 4c). The high affinity BDNF receptor, TrkB, arises from the NTRK2 gene and exists as both the full length receptor (FL TrkB) and a truncated receptor lacking the kinase domain (TR TrkB). There was also no change in the protein expression with age of the either the FL TrkB receptor identified as a 148-kDa band in the gray matter (r = − 0.32, p = 0.099; Fig. 3c) or subcortical white matter (r = − 0.27, p = 0.18; Fig. 4d) or the TR TrkB represented as a 95-kDa band in the gray matter (r = − 0.101, p = 0.62; Fig. 3c) or the subcortical white matter (r = − 0.049, p = 0.82; Fig. 4d).
ADAM19: Due to the lack of a specific antibody that would work with samples from the rhesus monkey, the protein levels of ADAM19 could not be determined.
LIF: The protein level of LIF in the IPL could not be determined due to the lack of an antibody that worked in the rhesus monkey samples.
Sex differences: In the total population in the gray matter, there was significantly less mBDNF expression in females than males (p = 0.014) but there were no differences between males and females in proBDNF, TrkB TR, or TrkB FL (Online Resource 2, Fig. S3a). In the young animals in the gray matter, there were no sex differences for any of the proteins examined (Online Resource 2, Fig. S3b). In the gray matter of aged group, there was significantly more proBDNF in females and significantly less mBDNF in females than males with no change in TrkB TR or TrkB FL (Online Resource 2, Fig. S3c). In the white matter, there was no difference between males and females for any of the proteins examined in the total population (Online Resource 2, Fig. S4a), young population (Online Resource 2, Fig. S4b), or aged population (Online Resource 2, Fig. S4c).
Behavior: There was no relationship between protein expression and our overall cognitive impairment index (CII).
Discussion
Overview and summary
The object of this study was to explore changes in the expression of genes related to the maintenance of axons and myelin in the normal aging monkey brain in order to identify factors that might contribute to age-related white matter pathology. Previous microarray studies in normal aging rhesus monkey model utilized human whole genome arrays (Duce et al. 2008). In contrast, the current approach utilized an array based on the now available rhesus monkey genome and targeted a limited number of primers directed towards genes likely to be involved in white matter dysregulation. Additionally, this study separately analyzed the cortical gray matter and the subcortical white matter even though the pathology of interest is located in the white matter. This is important as the status of neurons in the gray matter is critical to the maintenance of axons that interact with the glial cells that myelinate them in the white matter.
A custom PCR-based gene array was used to screen potential target genes in the pathways of neuronal and glial survival that are altered in normal aging. The PCR array identified nine candidate genes in the gray matter and seven in the white matter of the IPL that had large fold changes with age. Of these, five were selected for analysis with quantitative PCR and age-related changes were confirmed for three of these genes as significant decreases in the expression of BDNF and ADAM19 mRNA in the gray matter and a significant increase in NGFR in the white matter. To determine if the gene expression changes resulted in a change in the expression at the protein level, western blot analysis was conducted for proBDNF, mBDNF, TrkB, and p75 NGFR. This analysis confirmed a significant decrease in the precursor of BDNF (proBDNF) in the gray matter while mBDNF and the two receptors were stable.
Gene expression
The PCR array identified several mRNAs that were altered with age while others were stable, confirming the heterogeneity of the aging process as reported in previous microarray studies (Duce et al. 2008). With the small sample size used (seven aged, five young), power was not sufficient for the observed difference to meet the Benjamini and Hochberg false discovery rate correction for the 90 genes examined with custom array, so to confirm expression changes, genes with the largest fold change were further investigated in follow-up studies using qPCR in a larger sample of subjects.
Quantitative real-time PCR
Not all of the genes that were identified as significantly altered with age by the array were confirmed by qPCR. In the cortical gray matter, BDNF and ADAM19 both decreased but did not exhibit the highest fold changes in the array and yet in the expanded sample (15 aged, 12 young) changed most significantly by qPCR. In contrast, while BDNF in subcortical white matter was decreased with age in the array, follow-up qPCR showed no significant effect of age on BDNF expression in the subcortical white matter. The age-related increase in NGFR in the subcortical white matter shown by the array was also confirmed by qPCR while ADAM19 was not significantly altered in the subcortical white matter on the array and this too was confirmed by qPCR. Finally, the gene with the highest fold change on the array, LIF, was not confirmed by qPCR because the expression level in the young animals in the expanded subject list fell below the level of detection. So, while the primer and probe set did work to amplify the gene, the sensitivity of the reaction at the level of starting RNA was not sufficient to allow for the successful analysis of the experimental comparison. Of the genes that were able to be successfully analyzed by qPCR, all of the results of the PCR array were confirmed except for BDNF expression in the subcortical white matter.
Technical considerations
The failure to confirm changes in BDNF in the subcortical white matter could be due to several technical differences between the two experiments. First, the array results could simply be a false positive as the sample size was too small to adjust for multiple comparisons. Secondly, the two experiments utilized different normalization factors that could alter the relationship with age of the gene of interest. The PCR array was normalized to the average expression of three separate housekeeping genes (ACTB, GAPDH, PPIA) as separate reactions in the calculation of the fold change of the aged versus the young animals. In contrast, the qPCR experiments were run as a multiplex reaction normalized to a single housekeeping gene, either 18S rRNA or RPL13A. Since this multiplex reaction requires a stable housekeeping gene,18S rRNA was selected for most analyses. However, since 18S rRNA is very abundant, the less abundant RPL13A, which has been shown to be stable in the brain of rhesus monkey (Noriega et al. 2010), was used for the least abundant transcripts, NGFR, and LIF.
Finally, another factor that may affect the ability to confirm the changes in the expression of the genes is the presence of multiple isoforms of the gene from differential splicing and alternate transcription start sites. All of the genes that were successfully analyzed by qPCR have differentially spliced isoforms of mRNA, and the qPCR experiments utilized primers to amplify all isoforms of the gene. In contrast, the primers utilized in the PCR array were selected by SABiosciences and the location of these primers in the gene sequence is unknown. Hence, it is possible that the two experiments looked at different isoforms of the genes and this contributed to differing results. However, since the results from the PCR array were confirmed for most genes including BDNF in the gray matter, it is not likely that this was a major confound for most results.
Protein expression
While the level of gene expression is important, it is essential to determine if the level of gene expression is representative of the functional level of the protein level. Western blot analysis demonstrated that the decrease in BDNF mRNA expression in the gray matter of the IPL is accompanied by a decrease in the level of proBDNF. In contrast, there was no decrease with age in the mature form (mBDNF) which is cleaved from proBDNF. This finding is similar to that reported in rat brain where aging had a specific effect on the level of proBDNF without affecting mBDNF levels (Perovic et al. 2013). In the subcortical white matter, there were no significant changes in either proBDNF or mBDNF which is consistent with the lack of significant alteration at the mRNA level.
The other significant gene expression change was an increase in the nerve growth factor receptor (NGFR) with age in the subcortical white matter. However, the expression of the p75 NGFR receptor protein was not altered with age. In addition, protein levels of the other BDNF receptor (not included in the array), NTRK2 (TrkB), did not change significantly with age in either the gray matter or the subcortical white matter for either the full-length or the truncated receptor lacking the kinase domain. This too is in parallel with the mRNA results.
Limitations
One limitation of this study is that no subjects in the middle age range from 13 to 19 years of age were available. Hence, we were unable to determine if the trends and changes in mRNA and protein expression are truly linear across the entire life span or if or where there may be a middle age break point. This is important because in the rat, studies have found a transient decrease in the p75 NGFR receptor protein at 12 and 18 months of age with recovery of expression at 24 months to levels similar to 6 months of age (Perovic et al. 2013). Hence, we cannot determine if regulation of the p75 NGFR protein in the rhesus monkey is similar to that in the rat. In addition, alterations in NTRK2 expression in the aging cortex have been reported by in situ hybridization in human cerebral cortex, but it was a layer specific with significant decreases in full-length NTRK2 mRNA limited to layers 2 and 3 in the prefrontal cortex (Romanczyk et al. 2002). If this specific change occurs in the aging monkey, it could be missed because the extracts from the gray matter used in this study included all six layers of cortex.
BDNF and normal aging
The most interesting and important result from this study is the specific decrease of proBDNF with age. It has been reported that in both mild cognitive impairment (MCI) and Alzheimer’s disease (AD), levels of both proBDNF and mature BDNF are decreased in the parietal cortex (Michalski and Fahnestock 2003; Peng et al. 2005), suggesting a relationship with cognitive impairment. In contrast to these human studies where comparisons were with age-matched controls, the present study compared young and old across the full adult age range from 4 to 30 years old and found a decrease in proBDNF. This age range equates to humans from about 12 to 90 years of age (a 1:3 age ratio with humans (Tigges et al. 1988). Since BDNF is normally expressed in both neurons and glial cells, especially astrocytes and microglia (Elkabes et al. 1996; Wu et al. 2004), the exact source of the decreased expression is unknown. However, as the rhesus monkey does not develop Alzheimer’s disease (Sloane et al. 1997) and does not lose forebrain neurons (Roberts et al. 2012; Giannaris and Rosene 2012), this study shows that there is a decrease in the level of BDNF mRNA and proBDNF protein in the absence of the neuron loss that occurs in AD and other neurodegenerative diseases.
ProBDNF activates the low affinity receptor, p75 NGFR, which in turn can activate apoptosis pathways (Teng et al. 2005). ProBDNF can also inhibit neurite outgrowth and reduce spine density (Koshimizu et al. 2009). In contrast, mBDNF activates the high affinity receptor, TrkB, which promotes neuronal survival and synaptic plasticity (Poo 2001). Since there is preservation of neuron number with age, sublethal changes in signaling and neuron function have been suggested to contribute to the cognitive impairments seen with normal aging (Morrison and Hof 1997). Reduction in levels of proBDNF without a change in mBDNF could reflect increased cleavage of proBDNF as a compensatory response to preserve levels of mBDNF that promote neuronal survival and maintenance of the dendritic tree and axon (Gorski et al. 2003; Matsunaga et al. 2004). In contrast, proBDNF signals through the p75 NGFR and leads to neurite retraction and loss of spines (Koshimizu et al. 2009), so reduced levels of proBDNF would reduce this deleterious effect. These opposite actions of the mature and pro forms of BDNF make it essential to identify the post-translational status of each form in the aging brain as total BDNF expression cannot differentiate between the state of the protein and the differences between the two signaling pathways. Hence, the specific reduction seen in proBDNF in the IPL gray matter may reflect a compensatory response that maintains levels of mBDNF despite decreased mRNA expression, helping to preserve spines and axons.
In neurons, both proBDNF and mBDNF are trafficked to the dendrites and axon terminals and the mBDNF is released at synapses in an activity-dependent manner (Yang et al. 2009; Matsuda et al. 2009). The released mBDNF is able to act on both the pre-synaptic and post-synaptic cells to support the axon and neuron (Sobreviela et al. 1996; Gärtner et al. 2006). Impaired action potential conduction due to myelin damage would likely reduce this activity-dependent release. If transport of the protein to or from the synapse in the axons and dendrites with age was altered, signaling resulting from the activity-dependent regulated release of the protein would be decreased even more. Such a decrease in signaling could then underlie the observed reductions in spines and atrophy of dendrites (for review see (Dickstein et al. 2013)) as well as the loss of long axons (Bowley et al. 2010; Peters et al. 2010) observed in the normal aging monkey brain. There are conflicting reports as to whether proBDNF is also released from the neuron in an activity dependent manner to exert a biological effect through the binding to the p75 NGFR receptor or if it is simply an intermediary protein in the formation of mBDNF with little significance for signaling (Matsumoto et al. 2008; Yang et al. 2009). Regardless of whether the proBDNF is released or is an intermediate protein in the formation of mBDNF, the decrease in proBDNF with age would shift the balance of the signal to mBDNF. On one hand, if proBDNF is released, it would bind to p75 NGFR causing spine loss, while mBDNF would bind to TrkB promoting spine stability. On the other hand, if proBDNF is the intermediate protein, the reduced proBDNF would be a result of the neuron compensating to preserve mBDNF levels and signaling to preserve the cell and synapse. However, the decreased action potential conduction due to myelin damage would decrease the ability of the mBDNF to act on the synapse leading to the loss of the spines and axon (Kohama et al. 2012).
In addition to trophic support of synapses, mBDNF promotes myelination by oligodendrocytes (Du et al. 2006). However, myelination is inhibited through the interaction of myelin components such as Nogo-66, myelin-associated glycoprotein (MAG), and oligodendrocyte myelin glycoprotein (OMgp) with the receptor complex of p75 NGFR and the Nogo receptor (NgR1) independent of neurotrophin binding to p75 NGFR (Wang et al. 2002). With the increase in NGFR mRNA in the subcortical white matter found in this study and the loss of proBDNF in the gray matter, this balance between the inhibition of myelination through the p75 NGFR receptor and the promotion of myelination by mBDNF may be important for abnormal remyelination and axonal loss seen in normal aging (Peters et al. 2000). While the specific contribution to the age-related myelin pathology of the levels of proBDNF and mBDNF reported in this study is not clear, these results point to the importance of considering the entire BDNF pathway in future studies of myelin pathology.
Sex differences in BDNF expression
The only differences between males and females were a decrease in mBDNF protein expression in females in the total population and a decrease of mBDNF and increase in proBDNF in females in only the aged population in the gray matter. Sex differences in BDNF have been previously reported in humans with an increase in total BDNF expression in both the CSF and plasma of females (Lommatzsch et al. 2005; Komulainen et al. 2008; Li et al. 2009; Driscoll et al. 2012). These studies did not differentiate between the two species of BDNF. In the aged population, the contrasting finding of decreased mBDNF and increased proBDNF in the females compared to the males point to changes in the processing of proBDNF into mBDNF. The conversion of proBDNF to mBDNF is regulated intracellularly by the proprotein convertase furin (Seidah et al. 1996; Chao 2003) and extracellularly by plasmin (Pang et al. 2004; Gray and Ellis 2008) and matrix metalloproteinase 9 (MMP-9) (Mizoguchi et al. 2011; Yoshida et al. 2012; Yamamori et al. 2013). The expression of furin has been shown to be negatively regulated by the by estrogen binding to the estrogen receptor beta (Stygar et al. 2007). Since we were unable to assess activity of these enzymes, we cannot exclude that the changes seen in the expression of mBDNF and proBDNF in the IPL gray matter of females could be due to changes in the expression or activity of the conversion enzymes between males and females.
Other candidate genes
Two genes that were identified as candidates in the array and which could be important for myelin homeostasis could not be fully analyzed and confirmed. ADAM19 was examined at the mRNA level confirming the result from the PCR array that there was an age-related decrease in the gray matter and no change in the subcortical white matter. However, due to a lack of a specific antibody that would work with the rhesus monkey samples, the protein levels of ADAM19 could not be determined. The other gene, LIF, which was increased in white matter could not be verified by qPCR due to technical problems with the primer sets not amplifying the mRNA in the young animals in the larger cohort. The amount of RNA used in the multiplex reactions was less than in the PCR array potentially explaining why LIF was not able to be confirmed. Follow-up on protein levels was also prevented due to a lack of a specific antibody for LIF. However, these results do not rule out that either of these genes could contribute to the age-related white matter pathology and both deserve further study.
Finally, several of the genes identified as potential targets by the PCR array were not selected for follow-up due to the availability of the samples and reagents but could also contribute to brain aging. One was ADAM 17 which encodes for the tumor necrosis factor alpha (TNFα)-convertase enzyme which releases the proinflammatory cytokine TNFα from the cell membrane (Black et al. 1997). The other was growth arrest and DNA-damage-inducible, gamma (GADD45G), a marker of the activation of the tumor suppressor protein p53 leading to the repair of DNA damage (Ying et al. 2005).
Conclusions
This study is the first to broadly examine mRNA expression and protein levels of genes important to myelin homeostasis across the life span of the normal aging rhesus monkey. While the results confirm that aging is a multivariate process, one strong result was that the neurotrophic factor BDNF and its receptor show significant age-related changes that could contribute to age-related vulnerability of white matter. The findings of this study are consistent with studies in aging rats and humans that show that neurotrophic factor signaling is altered in the aging brain. While white matter changes are the main neuropathology present in the aging monkey brain, this study provides evidence of heterogeneous changes in genes in both the gray and white matter with age where subtle changes in BDNF signaling in cortical gray matter may contribute to white matter dysfunction and axon loss (for review see (Peters and Kemper 2012)) that reflect a cascade of events contributing to the pathology.
One well-documented contributor to this cascade is oxidative stress. This increases with age and contributes to activation of astrocytes and microglia and may exacerbate changes in myelin (Sloane et al. 1999; Sloane et al. 2000; Shobin et al. 2017). For example, as myelin damage accumulates, oligodendrocytes try to remyelinate axons, but the resulting sheaths can be either thinner than in young adults or show abnormal redundant loops of myelin around the axons (Peters et al. 2000; Peters and Sethares 2003). It also appears that even well formed sheaths become shorter with age as assessed by increases in number of paranodes (Peters and Sethares 2003). Such changes in the myelin sheath could alter axonal conduction leading to action potential failure. This in turn could decrease trophic factor feedback. Since BDNF supports the axon (for review see (Poo 2001)), decreased trophic feedback from the postsynaptic cell to the axon could cause the retraction of the axon and the axonal loss visible in the white matter as discussed by Kohama et al. 2012. The decreased trophic feedback could also be reflected in reported decreases in the number of dendritic spines and synapses on the post-synaptic cells (Dickstein et al. 2013). The results of this study identify age-related decreases in BDNF mRNA and the proBDNF protein as potential contributors to the age-related development of myelin plasticity and associated cognitive aging.
Electronic supplementary material
(PDF 124 kb)
(PDF 4346 kb)
Acknowledgements
We thank Dr. Shelley Russek for allowing the use of the RT-PCR machine in her laboratory for this study as well as helpful discussions about the pro-BDNF antibody.
Author contributions
AAR and DLR designed the study and drafted the manuscript. AAR, CRA, and DLR determined genes included on the PCR array. AAR completed all experiments and analyses.
Funding
This work was supported by National Institute of Health Grants: P01-AG000001, P51-RR00165, R01-AG043640, R01-AG042512, T32-GM008541.
Compliance with ethical standards
All procedures were in accord with NIH guidelines and approved by the Institutional Animal Care and Use Committee of BUMC.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s11357-018-0006-2) contains supplementary material, which is available to authorized users.
Contributor Information
Amy A. Robinson, Phone: 352-284-3642, Email: amyarobinson@gmail.com
Carmela R. Abraham, Email: cabraham@bu.edu
Douglas L. Rosene, Email: drosene@bu.edu
References
- Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, Nelson N, Boiani N, Schooley KA, Gerhart M, Davis R, Fitzner JN, Johnson RS, Paxton RJ, March CJ, Cerretti DP. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385(6618):729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- Bowley MP, Cabral H, Rosene DL, Peters A. Age changes in myelinated nerve fibers of the cingulate bundle and corpus callosum in the rhesus monkey. J Comp Neurol. 2010;518(15):3046–3064. doi: 10.1002/cne.22379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butzkueven H, Emery B, Cipriani T, Marriott MP, Kilpatrick TJ. Endogenous leukemia inhibitory factor production limits autoimmune demyelination and oligodendrocyte loss. Glia. 2006;53(7):696–703. doi: 10.1002/glia.20321. [DOI] [PubMed] [Google Scholar]
- Chao MV. Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat Rev Neurosci. 2003;4(4):299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- Chen C-D, Sloane JA, Li H, Aytan N, Giannaris EL, Zeldich E, Hinman JD, Dedeoglu A, Rosene DL, Bansal R, Luebke JI, Kuro-o M, Abraham CR. The antiaging protein Klotho enhances oligodendrocyte maturation and myelination of the CNS. J Neurosci. 2013;33(5):1927–1939. doi: 10.1523/JNEUROSCI.2080-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickstein DL, Weaver CM, Luebke JI, Hof PR. Dendritic spine changes associated with normal aging. Neuroscience. 2013;251:21–32. doi: 10.1016/j.neuroscience.2012.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll I, Martin B, An Y, Maudsley S, Ferrucci L, Mattson MP, Resnick SM. Plasma BDNF is associated with age-related white matter atrophy but not with cognitive function in older, non-demented adults. PLoS One. 2012;7(4):e35217. doi: 10.1371/journal.pone.0035217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Fischer TZ, Clinton-Luke P, Lercher LD, Dreyfus CF. Distinct effects of p75 in mediating actions of neurotrophins on basal forebrain oligodendrocytes. Mol Cell Neurosci. 2006;31(2):366–375. doi: 10.1016/j.mcn.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Duce JA, Podvin S, Hollander W, Kipling D, Rosene DL, Abraham CR. Gene profile analysis implicates Klotho as an important contributor to aging changes in brain white matter of the rhesus monkey. Glia. 2008;56(1):106–117. doi: 10.1002/glia.20593. [DOI] [PubMed] [Google Scholar]
- Elkabes S, DiCicco-Bloom EM, Black IB. Brain microglia/macrophages express neurotrophins that selectively regulate microglial proliferation and function. J Neurosci. 1996;16(8):2508–2521. doi: 10.1523/JNEUROSCI.16-08-02508.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman SH, Kandel R, Cruz L, Rozkalne A, Newell K, Frosch MP, Hedley-Whyte ET, Locascio JJ, Lipsitz LA, Hyman BT. Preservation of neuronal number despite age-related cortical brain atrophy in elderly subjects without Alzheimer disease. J Neuropathol Exp Neurol. 2008;67(12):1205–1212. doi: 10.1097/NEN.0b013e31818fc72f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gärtner A, Polnau DG, Staiger V, Sciarretta C, Minichiello L, Thoenen H, Bonhoeffer T, Korte M. Hippocampal long-term potentiation is supported by presynaptic and postsynaptic tyrosine receptor kinase B-mediated phospholipase Cgamma signaling. J Neurosci. 2006;26(13):3496–3504. doi: 10.1523/JNEUROSCI.3792-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannaris EL, Rosene DL. A stereological study of the numbers of neurons and glia in the primary visual cortex across the lifespan of male and female rhesus monkeys. J Comp Neurol. 2012;520(15):3492–3508. doi: 10.1002/cne.23101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Isla T, Price JL, McKeel DW, et al. Profound loss of layer II entorhinal cortex neurons occurs in very mild Alzheimer’s disease. J Neurosci. 1996;16(14):4491–4500. doi: 10.1523/JNEUROSCI.16-14-04491.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JA, Zeiler SR, Tamowski S, Jones KR. Brain-derived neurotrophic factor is required for the maintenance of cortical dendrites. J Neurosci. 2003;23(17):6856–6865. doi: 10.1523/JNEUROSCI.23-17-06856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray K, Ellis V. Activation of pro-BDNF by the pericellular serine protease plasmin. FEBS Lett. 2008;582(6):907–910. doi: 10.1016/j.febslet.2008.02.026. [DOI] [PubMed] [Google Scholar]
- Haug H. Are neurons of the human cerebral cortex really lost during aging? A morphometric examination. In: Traber J, Gispen WH, editors. Senile dementia of the azheimer type. Berlin: Springer Verlag; 1985. pp. 150–163. [Google Scholar]
- Herndon JG, Moss MB, Rosene DL, Killiany RJ. Patterns of cognitive decline in aged rhesus monkeys. Behav Brain Res. 1997;87(1):25–34. doi: 10.1016/S0166-4328(96)02256-5. [DOI] [PubMed] [Google Scholar]
- Hinman JD, Chen C-D, Oh S-Y, Hollander W, Abraham CR. Age-dependent accumulation of ubiquitinated 2′,3′-cyclic nucleotide 3′-phosphodiesterase in myelin lipid rafts. Glia. 2008;56(1):118–133. doi: 10.1002/glia.20595. [DOI] [PubMed] [Google Scholar]
- Kohama SG, Rosene DL, Sherman LS. Age-related changes in human and non-human primate white matter: from myelination disturbances to cognitive decline. Age (Dordr) 2012;34(5):1093–1110. doi: 10.1007/s11357-011-9357-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komulainen P, Pedersen M, Hänninen T, Bruunsgaard H, Lakka TA, Kivipelto M, Hassinen M, Rauramaa TH, Pedersen BK, Rauramaa R. BDNF is a novel marker of cognitive function in ageing women: the DR’s EXTRA study. Neurobiol Learn Mem. 2008;90(4):606–613. doi: 10.1016/j.nlm.2008.07.014. [DOI] [PubMed] [Google Scholar]
- Koshimizu H, Kiyosue K, Hara T, Hazama S, Suzuki S, Uegaki K, Nagappan G, Zaitsev E, Hirokawa T, Tatsu Y, Ogura A, Lu B, Kojima M. Multiple functions of precursor BDNF to CNS neurons: negative regulation of neurite growth, spine formation and cell survival. Mol Brain. 2009;2(1):27. doi: 10.1186/1756-6606-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Peskind ER, Millard SP, Chi P, Sokal I, Yu CE, Bekris LM, Raskind MA, Galasko DR, Montine TJ. Cerebrospinal fluid concentration of brain-derived neurotrophic factor and cognitive function in non-demented subjects. PLoS One. 2009;4(5):e5424. doi: 10.1371/journal.pone.0005424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommatzsch M, Zingler D, Schuhbaeck K, Schloetcke K, Zingler C, Schuff-Werner P, Virchow JC. The impact of age, weight and gender on BDNF levels in human platelets and plasma. Neurobiol Aging. 2005;26(1):115–123. doi: 10.1016/j.neurobiolaging.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Makris N, Papadimitriou GM, van der Kouwe A, Kennedy DN, Hodge SM, Dale AM, Benner T, Wald LL, Wu O, Tuch DS, Caviness VS, Moore TL, Killiany RJ, Moss MB, Rosene DL. Frontal connections and cognitive changes in normal aging rhesus monkeys: a DTI study. Neurobiol Aging. 2007;28(10):1556–1567. doi: 10.1016/j.neurobiolaging.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Marner L, Nyengaard JR, Tang Y, Pakkenberg B. Marked loss of myelinated nerve fibers in the human brain with age. J Comp Neurol. 2003;462(2):144–152. doi: 10.1002/cne.10714. [DOI] [PubMed] [Google Scholar]
- Matsuda N, Lu H, Fukata Y, Noritake J, Gao H, Mukherjee S, Nemoto T, Fukata M, Poo M. Differential activity-dependent secretion of brain-derived neurotrophic factor from axon and dendrite. J Neurosci. 2009;29(45):14185–14198. doi: 10.1523/JNEUROSCI.1863-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Rauskolb S, Polack M, Klose J, Kolbeck R, Korte M, Barde YA. Biosynthesis and processing of endogenous BDNF: CNS neurons store and secrete BDNF, not pro-BDNF. Nat Neurosci. 2008;11(2):131–133. doi: 10.1038/nn2038. [DOI] [PubMed] [Google Scholar]
- Matsunaga W, Shirokawa T, Isobe K. BDNF is necessary for maintenance of noradrenergic innervations in the aged rat brain. Neurobiol Aging. 2004;25(3):341–348. doi: 10.1016/S0197-4580(03)00093-9. [DOI] [PubMed] [Google Scholar]
- Michailov GV, Sereda MW, Brinkmann BG, Fischer TM, Haug B, Birchmeier C, Role L, Lai C, Schwab MH, Nave KA. Axonal neuregulin-1 regulates myelin sheath thickness. Science. 2004;304(5671):700–689. doi: 10.1126/science.1095862. [DOI] [PubMed] [Google Scholar]
- Michalski B, Fahnestock M. Pro-brain-derived neurotrophic factor is decreased in parietal cortex in Alzheimer’s disease. Brain Res Mol Brain Res. 2003;111(1-2):148–154. doi: 10.1016/S0169-328X(03)00003-2. [DOI] [PubMed] [Google Scholar]
- Mizoguchi H, Nakade J, Tachibana M, Ibi D, Someya E, Koike H, Kamei H, Nabeshima T, Itohara S, Takuma K, Sawada M, Sato J, Yamada K. Matrix metalloproteinase-9 contributes to kindled seizure development in pentylenetetrazole-treated mice by converting pro-BDNF to mature BDNF in the hippocampus. J Neurosci. 2011;31(36):12963–12971. doi: 10.1523/JNEUROSCI.3118-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore TL, Killiany RJ, Herndon JG, Rosene DL. A non-human primate test of abstraction and set shifting: an automated adaptation of the Wisconsin card sorting test. J Neurosci Methods. 2005;146(2):165–173. doi: 10.1016/j.jneumeth.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Morrison JH, Hof PR. Life and death of neurons in the aging brain. Science. 1997;278(5337):412–419. doi: 10.1126/science.278.5337.412. [DOI] [PubMed] [Google Scholar]
- Noriega NC, Kohama SG, Urbanski HF. κMicroarray analysis of relative gene expression stability for selection of internal reference genes in the rhesus macaque brain. BMC Mol Biol. 2010;11(1):47. doi: 10.1186/1471-2199-11-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang PT, Teng HK, Zaitsev E, Woo NT, Sakata K, Zhen S, Teng KK, Yung WH, Hempstead BL, Lu B. Cleavage of proBDNF by tPA/plasmin is essential for long-term hippocampal plasticity. Science. 2004;306(5695):487–491. doi: 10.1126/science.1100135. [DOI] [PubMed] [Google Scholar]
- Peng S, Wuu J, Mufson EJ, Fahnestock M. Precursor form of brain-derived neurotrophic factor and mature brain-derived neurotrophic factor are decreased in the pre-clinical stages of Alzheimer’s disease. J Neurochem. 2005;93(6):1412–1421. doi: 10.1111/j.1471-4159.2005.03135.x. [DOI] [PubMed] [Google Scholar]
- Perovic M, Tesic V, Mladenovic Djordjevic A, Smiljanic K, Loncarevic-Vasiljkovic N, Ruzdijic S, Kanazir S. BDNF transcripts, proBDNF and proNGF, in the cortex and hippocampus throughout the life span of the rat. Age (Dordr) 2013;35(6):2057–2070. doi: 10.1007/s11357-012-9495-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Kemper T. A review of the structural alterations in the cerebral hemispheres of the aging rhesus monkey. Neurobiol Aging. 2012;33(10):2357–2372. doi: 10.1016/j.neurobiolaging.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Sethares C. Is there remyelination during aging of the primate central nervous system? J Comp Neurol. 2003;460(2):238–254. doi: 10.1002/cne.10639. [DOI] [PubMed] [Google Scholar]
- Peters A, Moss MB, Sethares C. Effects of aging on myelinated nerve fibers in monkey primary visual cortex. J Comp Neurol. 2000;419(3):364–376. doi: 10.1002/(SICI)1096-9861(20000410)419:3<364::AID-CNE8>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Peters A, Sethares C, Moss MB. How the primate fornix is affected by age. J Comp Neurol. 2010;518(19):3962–3980. doi: 10.1002/cne.22434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poo MM. Neurotrophins as synaptic modulators. Nat Rev Neurosci. 2001;2(1):24–32. doi: 10.1038/35049004. [DOI] [PubMed] [Google Scholar]
- Roberts DE, Killiany RJ, Rosene DL. Neuron numbers in the hypothalamus of the normal aging rhesus monkey: stability across the adult lifespan and between the sexes. J Comp Neurol. 2012;520(6):1181–1197. doi: 10.1002/cne.22761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanczyk TB, Weickert CS, Webster MJ, Herman MM, Akil M, Kleinman JE. Alterations in trkB mRNA in the human prefrontal cortex throughout the lifespan. Eur J Neurosci. 2002;15(2):269–280. doi: 10.1046/j.0953-816x.2001.01858.x. [DOI] [PubMed] [Google Scholar]
- Seidah NG, Benjannet S, Pareek S, Chrétien M, Murphy RA. Cellular processing of the neurotrophin precursors of NT3 and BDNF by the mammalian proprotein convertases. FEBS Lett. 1996;379(3):247–250. doi: 10.1016/0014-5793(95)01520-5. [DOI] [PubMed] [Google Scholar]
- Shirakabe K, Wakatsuki S, Kurisaki T, Fujisawa-Sehara A. Roles of meltrin beta /ADAM19 in the processing of neuregulin. J Biol Chem. 2000;276(12):9352–9358. doi: 10.1074/jbc.M007913200. [DOI] [PubMed] [Google Scholar]
- Shobin E, Bowley MP, Estrada LI, Heyworth NC, Orczykowski ME, Eldridge SA, Calderazzo SM, Mortazavi F, Moore TL, Rosene DL. Microglia activation and phagocytosis: relationship with aging and cognitive impairment in the rhesus monkey. GeroScience. 2017;39(2):199–220. doi: 10.1007/s11357-017-9965-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloane JA, Pietropaolo MF, Rosene DL, Moss MB, Peters A, Kemper T, Abraham CR. Lack of correlation between plaque burden and cognition in the aged monkey. Acta Neuropathol. 1997;94(5):471–478. doi: 10.1007/s004010050735. [DOI] [PubMed] [Google Scholar]
- Sloane JA, Hollander W, Moss MB, Rosene DL, Abraham CR. Increased microglial activation and protein nitration in white matter of the aging monkey. Neurobiol Aging. 1999;20(4):395–405. doi: 10.1016/S0197-4580(99)00066-4. [DOI] [PubMed] [Google Scholar]
- Sloane JA, Hollander W, Rosene DL, Moss MB, Kemper T, Abraham CR. Astrocytic hypertrophy and altered GFAP degradation with age in subcortical white matter of the rhesus monkey. Brain Res. 2000;862(1-2):1–10. doi: 10.1016/S0006-8993(00)02059-X. [DOI] [PubMed] [Google Scholar]
- Sobreviela T, Pagcatipunan M, Kroin JS, Mufson EJ. Retrograde transport of brain-derived neurotrophic factor (BDNF) following infusion in neo- and limbic cortex in rat: relationship to BDNF mRNA expressing neurons. J Comp Neurol. 1996;375(3):417–444. doi: 10.1002/(SICI)1096-9861(19961118)375:3<417::AID-CNE6>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Stygar D, Masironi B, Eriksson H, Sahlin L. Studies on estrogen receptor (ER) and responses on gene regulation in peripheral blood leukocytes in vivo using selective ER agonists. J Endocrinol. 2007;194(1):101–119. doi: 10.1677/JOE-06-0060. [DOI] [PubMed] [Google Scholar]
- Teng HK, Teng KK, Lee R, Wright S, Tevar S, Almeida RD, Kermani P, Torkin R, Chen ZY, Lee FS, Kraemer RT, Nykjaer A, Hempstead BL. ProBDNF induces neuronal apoptosis via activation of a receptor complex of p75NTR and sortilin. J Neurosci. 2005;25(22):5455–5463. doi: 10.1523/JNEUROSCI.5123-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry RD, DeTeresa R, Hansen LA. Neocortical cell counts in normal human adult aging. Ann Neurol. 1987;21(6):530–539. doi: 10.1002/ana.410210603. [DOI] [PubMed] [Google Scholar]
- Tigges J, Gordon TP, McClure HM, Hall EC, Peters A. Survival rate and life span of rhesus monkeys at the Yerkes Regional Primate Research Center. Am J Primatol. 1988;15(3):263–273. doi: 10.1002/ajp.1350150308. [DOI] [PubMed] [Google Scholar]
- Vernooij MW, de Groot M, van der Lugt A, Ikram MA, Krestin GP, Hofman A, Niessen WJ, Breteler MMB. White matter atrophy and lesion formation explain the loss of structural integrity of white matter in aging. NeuroImage. 2008;43(3):470–477. doi: 10.1016/j.neuroimage.2008.07.052. [DOI] [PubMed] [Google Scholar]
- Wang KC, Kim JA, Sivasankaran R, Segal R, He Z. P75 interacts with the Nogo receptor as a co-receptor for Nogo, MAG and OMgp. Nature. 2002;420(6911):74–78. doi: 10.1038/nature01176. [DOI] [PubMed] [Google Scholar]
- Whitehouse P, Price D, Struble R, Clark A, Coyle J, Delon M. Alzheimer’s disease and senile dementia: loss of neurons in the basal forebrain. Science. 1982;215(4537):1237–1239. doi: 10.1126/science.7058341. [DOI] [PubMed] [Google Scholar]
- Wu H, Friedman WJ, Dreyfus CF. Differential regulation of neurotrophin expression in basal forebrain astrocytes by neuronal signals. J Neurosci Res. 2004;76(1):76–85. doi: 10.1002/jnr.20060. [DOI] [PubMed] [Google Scholar]
- Yamamori H, Hashimoto R, Ishima T, Kishi F, Yasuda Y, Ohi K, Fujimoto M, Umeda-Yano S, Ito A, Hashimoto K, Takeda M. Plasma levels of mature brain-derived neurotrophic factor (BDNF) and matrix metalloproteinase-9 (MMP-9) in treatment-resistant schizophrenia treated with clozapine. Neurosci Lett. 2013;556:37–41. doi: 10.1016/j.neulet.2013.09.059. [DOI] [PubMed] [Google Scholar]
- Yang J, Siao C-J, Nagappan G, Marinic T, Jing D, McGrath K, Chen ZY, Mark W, Tessarollo L, Lee FS, Lu B, Hempstead BL. Neuronal release of proBDNF. Nat Neurosci. 2009;12(2):113–115. doi: 10.1038/nn.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying J, Srivastava G, Hsieh WS, Gao Z, Murray P, Liao SK, Ambinder R, Tao Q (2005) The stress-responsive gene GADD45G is a functional tumor suppressor, with its response to environmental stresses frequently disrupted epigenetically in multiple tumors. Clin Cancer Res 11(18):6442–6449. 10.1158/1078-0432.CCR-05-0267 [DOI] [PubMed]
- Yoshida T, Ishikawa M, Niitsu T, Nakazato M, Watanabe H, Shiraishi T, Shiina A, Hashimoto T, Kanahara N, Hasegawa T, Enohara M, Kimura A, Iyo M, Hashimoto K. Decreased serum levels of mature brain-derived neurotrophic factor (BDNF), but not its precursor proBDNF, in patients with major depressive disorder. PLoS One. 2012;7(8):e42676. doi: 10.1371/journal.pone.0042676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 124 kb)
(PDF 4346 kb)