Abstract
Butterflies have evolved different colour patterns on their dorsal and ventral wing surfaces to serve different signalling functions, yet the developmental mechanisms controlling surface-specific patterning are still unknown. Here, we mutate both copies of the transcription factor apterous in Bicyclus anynana butterflies using CRISPR/Cas9 and show that apterous A, expressed dorsally, functions both as a repressor and modifier of ventral wing colour patterns, as well as a promoter of dorsal sexual ornaments in males. We propose that the surface-specific diversification of wing patterns in butterflies proceeded via the co-option of apterous A or its downstream effectors into various gene regulatory networks involved in the differentiation of discrete wing traits. Further, interactions between apterous and sex-specific factors such as doublesex may have contributed to the origin of sexually dimorphic surface-specific patterns. Finally, we discuss the evolution of eyespot number diversity in the family Nymphalidae within the context of developmental constraints due to apterous regulation.
Keywords: apterous, dorsal–ventral differentiation, butterfly wing patterns, developmental constraints, eyespot repression
1. Introduction
Butterflies are a group of organisms well known for their diverse and colourful wing patterns. Owing to the dual role these patterns play in survival and mate selection, many butterflies have evolved a signal partitioning strategy where colour patterns appearing on the hidden dorsal surfaces tend to be bright and prominent, generally functioning in sexual signalling, whereas patterns on the exposed ventral surfaces tend to be cryptically coloured and most commonly serve to ward off predators [1,2] (figure 1a). The molecular and developmental basis of individual pattern element differentiation, such as eyespots or transverse bands, has been extensively studied [3,4]. Furthermore, we have a functional understanding of the genes involved in differentiating hindwing patterns from forewing patterns [5,6]. However, the molecular mechanisms that lead to striking variations in the development of dorsal versus ventral surface-specific colour patterns remain unknown. Elucidating this process will help us understand the mechanism of diversification and specialization of wing patterns within the butterfly lineage.
Figure 1.
Dorsal-ventral surface-specific variation in butterflies. (a) Dorsal (left) and ventral (right) surfaces of Morpho menelaus and Panacea regina illustrating striking variation in colour and patterns between surfaces. (b) Dorsal (left) and ventral (right) surfaces of a male and female Bicyclus anynana. The regions boxed in red are expanded in c. (c) Magnified view of the androconial organs present only in males. Top: forewing ventral androconia with a characteristic teardrop shape surrounded by silver scales. The scales on the corresponding dorsal forewing surface are completely brown. Bottom: hindwing dorsal androconia, also surrounded by silver scales, along with two patches of hair-pencils. These traits are absent from the ventral hindwing. (Online version in colour.)
A charismatic colour pattern that is present on both dorsal and ventral wing surfaces of a family of butterflies, the nymphalids, is the eyespot. Studies on eyespot evolution through broad comparative work across 400 genera of nymphalid butterflies indicated that eyespots originated around 90 million years ago (MYA) within this family, initially restricted to the ventral hindwing surface [7,8]. The appearance of eyespots on the dorsal surfaces occurred nearly approximately 40 million years (MY) later following redeployment of eyespot gene networks from the ventral surface [8,9]. This surface-specific asymmetry in the evolution of eyespot patterns is intriguing because the molecular mechanisms leading to such evolutionary patterns are unknown and the asymmetry suggests the presence of developmental constraints that might have limited eyespot origins to ventral surfaces only.
We hypothesized that the transcription factor apterous (ap), a gene expressed on the dorsal wing surfaces of flies [10], might be implicated in differentiating dorsal from ventral wing patterns in butterflies and in constraining the evolution of novel patterns, such as eyespots, asymmetrically across these surfaces. In insects, however, this gene is often present in two copies, apA and apB, that do not necessarily share the same expression patterns, and flies are unusual for having lost one of these copies. In the beetle Tribolium castaneum, apA is expressed on the dorsal surface, whereas apB is expressed on both surfaces [11]. In the butterfly Junonia coenia, apA is expressed on the dorsal surface of larval wings [12], but the expression of apB and the role of either apA or apB in wing development and patterning is not known for this or any butterfly species. Here, we study the functions of both copies of ap during wing development in the African squinting bush brown Bicyclus anynana, which shows dorsal–ventral differences in wing patterns, including a different number of eyespots on these surfaces (figure 1b,c). To infer patterns of gene expression of both ap copies, we used in situ hybridization to localize mRNA expression during wing development. We then used targeted gene knockout using CRISPR/Cas9 to functionally verify the roles of apA and apB in surface-specific wing patterning and development.
2. Material and methods
(a). Animals
Bicyclus anynana butterflies were reared in a temperature-controlled room at 27°C with a 12 : 12 h light : dark cycle and 65% humidity. The larvae were fed on corn plants, while the adults were fed on banana.
(b). Cloning and probe synthesis
The apA sequence was obtained from [13] and the apB sequence was identified from the B. anynana genome [14]. The sequences were amplified with primers specified in electronic supplementary material, table S1, sequenced and then cloned into a PGEM-T Easy vector (Promega). Sense and antisense digoxigenin-labelled (DIG) riboprobes were synthesized in vitro using T7 and SP6 polymerases (Roche), purified by ethanol precipitation and resuspended in a 1 : 1 volume of diethyl pyrocarbonate (DEPC)-treated water : formamide.
(c). In situ hybridization
The protocol was modified slightly from [15]. Briefly, larval (last instar caterpillar) or pupal (24–28 h after pupation) wings were dissected in PBS and transferred to glass well plates containing PBST (PBS + 0.1% Tween20) at room temperature. The PBST was then immediately removed and the tissues fixed in 5% formaldehyde for 45 (larval) or 60 min (pupal) on ice, followed by five washes with cold PBST. The tissues were then incubated with 25 µg ml−1 proteinase K in cold PBST for 4 (larval) or 5 min (pupal), washed twice with 2 mg ml−1 glycine in cold PBST, followed by five washes with cold PBST. For larval wings, the peripodial membrane was then removed on ice, post-fixed for 20 min with 5% formaldehyde and washed with PBST. The wings were gradually transferred to a prehybridization buffer (5X saline sodium citrate (pH 4.5), 50% formamide, 0.1% Tween20 and 100 µg ml−1 denatured salmon sperm DNA), washed in the prehybridization buffer and incubated at 60–65°C for 1 h, followed by incubation in hybridization buffer (prehybridization buffer with 1 g l−1 glycine and 70 to 140 ng ml−1 riboprobe) for 24 h. The wings were then washed 6–10 times in prehybridization buffer at 60–65°C. They were then gradually transferred back to PBST at room temperature, washed five times in PBST and blocked overnight at 4°C (PBST + 1% BSA). The DIG-labelled probes were then detected by incubating the tissues with 1 : 3000 anti-DIG alkaline phosphatase (Roche) in block buffer for two hours, washed 10 times with block buffer, incubated in alkaline phosphatase buffer (100 mM Tris (pH 9.5), 100 mM NaCl, 5 mM MgCl2, 0.1% Tween) and finally stained with NBT/BCIP (Promega) solution at room temperature until colour developed. The reaction was stopped by washing in 2 mM EDTA in PBST and again with PBST. The samples were either mounted on slides with ImmunoHistoMount medium (Abcam) or post-fixed with 5% formaldehyde before wax embedding and sectioning (Advanced Molecular Pathology Lab, IMCB, Singapore).
(d). Preparation of Cas9 mRNA and guide RNA
pT3TS-nCas9n was a gift from Wenbiao Chen (Addgene plasmid #46757). The plasmid was linearized with XbaI digestion and purified using a GeneJET PCR Purification Kit (Thermo Scientific). Cas9 mRNA was obtained by in vitro transcription using the mMESSAGE mMACHINE T3 kit (Ambion), tailed using the Poly(A) Tailing Kit (Ambion) and purified by lithium chloride precipitation. The guide RNA templates were prepared using a PCR-based method according to [16]. The candidate targets were manually designed by searching for a GGN18NGG sequence on the sense or antisense strand of apA and apB, preferably targeting the LIM and homeobox domains of the transcription factor (electronic supplementary material, table S1). They were blasted against the B. anynana genome on LepBase.org to check for off-target effects. The template DNA sequence was used to perform an in vitro transcription using T7 RNA polymerase (Roche) at 37°C overnight, purified by ethanol precipitation and resuspended in DEPC-treated water.
(e). Microinjections
Eggs were collected on corn leaves within one to two hours of egg laying and were arranged on thin strips of double-sided tape on a Petri dish. Cas9 mRNA and guide RNAs were mixed along with green food dye (electronic supplementary material, table S2) and injected into the eggs with a Borosil glass capillary (World Precision Instruments, 1B100F-3) using a Picospritzer II (Parker Hannifin). A piece of wet cotton was placed in the Petri dish and the eggs were allowed to develop in an incubator at 27°C and high (approx. 80%) humidity. Hatched caterpillars were placed on young corn plants using a brush. Adults that emerged were scored for their phenotypes (electronic supplementary material, table S2). A later set of injections (electronic supplementary material, table S3) was performed to test whether the three sets of guides used (together with Cas9 mRNA) impacted hatching rates relative to injections with Cas9 mRNA alone.
(f). Sequencing and genotyping mutants
Genomic DNA was extracted from leg tissues of mutant individuals using the E.Z.N.A Tissue DNA Kit (Omega Bio-tek). The region surrounding the target sequence was amplified by PCR, purified by ethanol precipitation and used to check for the presence of mutations using the T7 endonuclease I (T7EI) assay. Sequences from individuals with disruptions at the targeted regions were cloned into a PGEM-T Easy vector (Promega) and sequenced.
3. Results
(a). apA and apB are both expressed on dorsal surfaces of developing wings
We cloned both ap homologues from B. anynana and used in situ hybridization to localize apA and apB mRNA in developing larval and pupal wing discs. Both homologues of ap were localized to the dorsal surfaces of the wings (figure 2d; electronic supplementary material, figure S1b). In the last larval instar wing discs, apA was expressed uniformly on the wing surface but absent in future dorsal eyespot centres of hindwings (figure 2a) and forewings (figure 2b). In larval wing discs of the B. anynana ‘Spotty’ mutant, which develops two additional dorsal eyespots, apA was absent in the additional centres (figure 2b). Furthermore, pupal wing expression of both apA and apB was upregulated in dorsal male-specific cells that give rise to long and thin modified scales, the hair-pencils, used for dispersing pheromones during courtship (figure 2c; electronic supplementary material, figure S1c). This pattern of expression was not seen in developing female pupal wings, which lack hair-pencils (figure 2c; electronic supplementary material, figure S1c). Control sense probes for both apA and apB (electronic supplementary material, figure S1) did not show any surface-specific or hair-pencil-specific staining patterns.
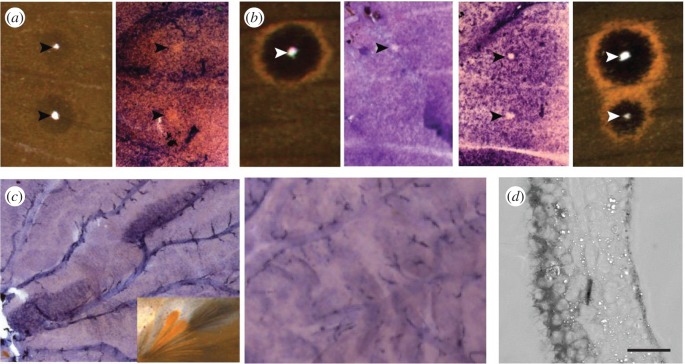
Figure 2.
apA mRNA localization in developing wing discs of Bicyclus anynana. (a) apA expression is uniform across the epidermis but absent in future dorsal eyespot centres of hindwings (n = 5). (b) apA expression is absent in the future dorsal eyespot centre of the wild-type forewing (left) (n = 3) and also in the additional eyespot centre in the B. anynana ‘Spotty’ mutant (right) (n = 7). (c) Male wings (left) (28 h after pupation) showing upregulated dorsal apA expression in the hair-pencil regions. Inset shows the hair-pencils in adult male B. anynana. Female wings (right) (25 h after pupation) show no upregulation of apA in corresponding regions of the dorsal surface. (d) Cross-sectional view of a developing wing disc showing dorsal-specific apA expression (left side of the cross section). Scale bar, 20 µm. (Online version in colour.)
(b). apA regulates dorsal surface-specific wing patterning
To functionally test the role of ap, we used the CRISPR/Cas9 system to disrupt the homeodomain and LIM domain of apA (figure 3a) and the LIM domain of apB (electronic supplementary material, figure S2a and table S2). A range of mosaic phenotypes were observed in both types of apA mutant individuals (figure 3; electronic supplementary material, figures S3 and S4). A few of these lacked wings, whose absence was visible upon pupation (electronic supplementary material, figure S3: mutant from batch#9, individual #1(M9-1)), and some adults had mosaic patches of ventral-like scales appearing on the dorsal surface (figure 3b: M9-2). In other mutants, the sex pheromone-producing organ, the androconial organ, of the ventral forewing appeared on the dorsal surface in males with its associated silver scales (figure 3b: M9-27). Males also had modified hair-pencils associated with the dorsal androconial organ of the hindwing, with loss of characteristic ultrastructure and colouration, and the absence of surrounding silver scales (figure 3b: M9-12 (bottom)). Extreme mutant individuals showed improper wing hinge formation, entire wing dorsal to ventral transformation (figure 3b: M9-3), the appearance of the ventral white band on the dorsal surface (figure 3b: M9-12 (top)), and in one case, all seven eyespots on the dorsal hindwing (figure 3b: M9-12 (bottom)), a surface that normally exhibits, on average, zero to one eyespot in males and one to two eyespots in females. apA clones also led to an enlarged outer perimeter of the gold ring in dorsal hindwing eyespots (figure 3b: M235-11). CRISPR/Cas9 disruption effects on the target sequence were verified in a few individuals, which showed the presence of deletions in the targeted regions (figure 3a).
Figure 3.
CRISPR/Cas9 mosaic wing pattern phenotypes of apA knockouts. (a) Top: regions of the apA gene in B. anynana targeted using the CRISPR/Cas9 system. Bottom: sequences of the homeodomain and LIM domain regions of mutant individuals compared with the wild-type sequence in bold. Blue is the region targeted and the PAM sequence is in red. Deletions are indicated with ‘-’. (b) A subset of the CRISPR/Cas9 apA mutant phenotypes observed in B. anynana. The left column shows the wild-type (WT) dorsal and ventral surfaces for male forewings and hindwings. M9-12 (top): the dorsal forewing of a mutant male highlighting some of the ventral-like phenotypes and defects. The boxed regions are expanded to show the appearance of ventral-like white band and silver scales. M9-3: dorsal forewing surface of a mutant female resembling the ventral surface. M9-27: mutant with the ventral teardrop shape forewing androconial organ appearing on the dorsal surface (red arrow). WT dorsal forewing androconia is shown for comparison. M9-12 (bottom): a mutant dorsal hindwing with the appearance of all seven eyespots (red arrows), normally only seen on the ventral surface. The boxed regions are expanded to show the loss of silver scales associated with the dorsal hindwing androconia and improper development of hair-pencils. WT hair-pencil is shown for comparison. M9-2: mosaic phenotype (left) on the dorsal surface with ventral-like light coloured scales. Clones are indicated with a dashed white line. Corresponding region of the other wing of the same individual (right) shows no mosaicism. M235-11: a dorsal hindwing of a mutant with the width of the gold ring resembling that of ventral eyespots. (Online version in colour.)
No striking transformations of dorsal to ventral identity were observed in apB mutants (electronic supplementary material, figures S2 and S5). Some of the apB knockout phenotypes included a wing hinge defect, a missing hindwing in one case (electronic supplementary material, figure S5: B-M9-22) and disturbed margin development (electronic supplementary material, figure S2: B-M9-17), sometimes associated with wing pattern disturbances (electronic supplementary material, figure S2: B-M9-15). Sequencing showed the presence of mutations in the targeted region (electronic supplementary material, figure S2a). Injections of the three guides used (apA homeodomain, apA LIM domain, and apB LIM domain) did not significantly alter hatching rates (electronic supplementary material, table S3).
Knockdown of apA in a variety of insects from different lineages indicates that apA is necessary for wing growth and development and its function in this process seems to be highly conserved [10,11,17]. However, our experiments, in agreement with others, also indicate a varying degree of co-option of this transcription factor or one of its downstream effectors into late wing development processes such as wing patterning and exoskeletalization. In T. castaneum, RNAi knockdown of apA and apB individually shows almost no phenotypic effects, while their simultaneous knockdown leads to more dramatic phenotypes such as elytral exoskeletalization defects, depending on the developmental stage. Therefore, both apA and apB in beetles are important for early and late wing developmental processes [11]. In B. anynana, knockout of both apA and apB causes defects in early wing development, but only apA or one of its downstream targets appears to have been co-opted to control dorsal surface-specific wing patterning.
(c). apA functions both as an activator and repressor of wing traits
Interestingly, our work shows that apA has multiple different, often antagonistic functions in surface- and sex-specific development between the fore- and hindwings. For example, apA acts as a repressor of male androconial organs and silver scale development on dorsal forewings, while it promotes hair-pencil and silver scale development on the dorsal hindwings of males (figure 4a). These effects point to the likely interaction between apA and other factors such as sex-specific (doublesex) or wing-specific (Ultrabithorax) factors that together can specify sex- and wing-specific pattern development. We previously showed that Ultrabithorax (Ubx) is expressed in the hindwings but not forewings of B. anynana [20]. In addition, the presence of a gene from the sex determination pathway, doublesex (dsx), in the future androconial regions of male wings of B. anynana was also verified by in situ hybridization and semi-quantitative PCR [21]. These data support a likely combinatorial function reminiscent of the interactions between the hox gene Scr and dsx in the determination of the male-specific sex combs in the legs of D. melanogaster [22]. The presence or absence of Ubx, type of dsx splice variant, and apA may be sufficient to give each sex and wing surface a unique identity, though more work needs to be done to test this hypothesis. Given that proteins of the LIM-homeodomain subfamily, to which ap belongs, are unique in their ability to bind other proteins via their LIM domain [23], their involvement in such a large range of developmental processes, as repressors and activators, is likely.
Figure 4.
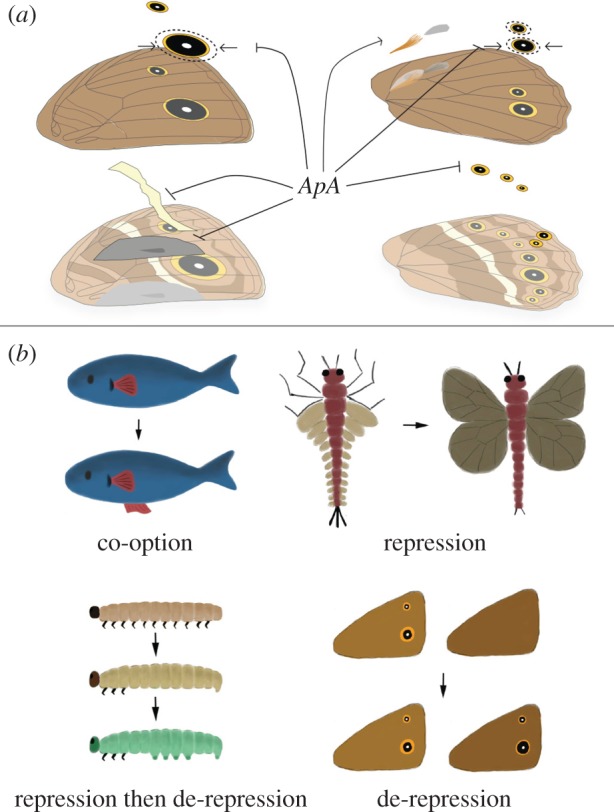
The role of apterous in surface-specific wing patterning in B. anynana and evolution of serial homologues in butterflies. (a) A schematic of the different functions of apA on the dorsal surface of B. anynana. apA acts as a repressor of ventral traits such as the white transversal band, forewing androconia, hindwing eyespots and the outer perimeter of the gold ring, and acts as an activator of hindwing hair-pencils and silver scales. (b) Different modes of serial homologue evolution involving the co-option of a (fin) gene network to a novel body location [18], repression of the ancestrally repeated (wing) network in a subset of body segments (adapted from [19]), repression followed by de-repression of the (limb) network in certain body segments [5] and de-repression of a never-expressed (eyespot) network at a novel body location. (Online version in colour.)
4. Discussion and conclusion
Mutations in apA point to this gene functioning as a dorsal surface selector in B. anynana butterflies. Selector genes comprise a small set of developmental genes that are critical for specifying cell, tissue, segment or organ identities in organisms [24]. The wing selector hox gene Ubx allows hindwings to have a different identity from forewings. For example, the restricted expression of Ubx in hindwings of most insects examined so far is required for membranous wing formation in beetles and bugs [25], haltere formation in flies [26] and hindwing-specific colour patterns in butterflies [6]. When Ubx is mutated, in all the examples described above, hindwings acquire the identity of forewings, and when Ubx is overexpressed in forewings, these acquire a more hindwing-like identity [20]. In B. anynana, apA functions in a similar manner along the dorsal–ventral axis of each wing—mutations in this gene make dorsal wing surfaces acquire a ventral identity. This type of homeotic mutation was also observed in a limited way, in bristles along the margin of the wings of D. melanogaster, where ap mutant clones developed bristles with a ventral identity [27]. Bicyclus anynana, however, appears to have made inordinate use of apA for surface-specific colour patterning and sexual trait development across the entire wing, which is a novel described role for this gene across insects.
Further, this work highlights the possible role of apA in restricting the origin and early evolution of serial homologues such as eyespots in nymphalid butterflies to the ventral surface of the wings only. The appearance of additional eyespots on the dorsal surface of hindwings in apA mutants, and the absence of apA mRNA at the precise position where a few dorsal eyespots develop in both fore- and hindwings at the stage of eyespot centre differentiation, implicates apA as a repressor of eyespot development. The additional gaps in apA expression observed in Spotty mutants further suggests that genetic mechanisms of eyespot number evolution on the dorsal surface proceeded via local repression of apA. We propose, thus, that the original ventral restriction of eyespots was due to the ancestral presence of apA on dorsal wing surfaces, and that eyespots' later appearance on these surfaces was due to local apA repression.
The ancestral presence of a repressor (apA) of a gene regulatory network in a specific body location, followed by repression of the repressor, seems to represent a novel mode of serial homologue diversification (figure 4b). This mode of serial homologue diversification is similar but also distinct from the mechanism previously proposed to lead to the reappearance of abdominal appendages in lepidopteran larvae—via local repression of the limb repressor hox protein, Abdominal-A (Abd-A) [5,28]. In contrast to eyespots, when arthropod appendages first originated, they were probably present in every segment of the body [29]. Limbs were later repressed in abdominal segments, and finally they were de-repressed in some of these segments in some insect lineages [5]. So, while the last steps of abdominal appendage and eyespot number diversification are similar (de-repression of a repressed limb/eyespot network), the early stages are different.
Comparative work across nymphalid butterflies also showed that the origin of dorsal eyespots was dependent on the presence of corresponding ventral eyespots in ancestral lineages [9]. This implies that the extant diversity of eyespot patterns is biased/limited due to developmental constraints, probably imposed by apA. Interestingly, while approximately 99% of the species in our database display such constraints, i.e., dorsal eyespots always having ventral counterparts, a few butterflies (such as Argyrophenga antipodium or Cassionympha cassius) display dorsal eyespots that lack ventral counterparts. The molecular basis for these rare patterns remains to be explored.
In summary, we uncovered a key transcription factor, apA, that due to its restricted expression on dorsal wing surfaces allowed B. anynana butterflies to develop and evolve their strikingly different dorsal and ventral wing patterns under natural and sexual selection. The interaction of apA with other sex- and wing-specific factors may explain the surface-specific pattern diversity we see across this as well as other butterfly species, but future comparative work is needed to further test these hypotheses. Additionally, our work has identified a new system to examine how developmental constraints, via apA repression of eyespot development, have shaped eyespot number biodiversity.
Supplementary Material
Acknowledgements
We thank Arjen van't Hof and Luqman Aslam for their help in retrieving sequence information from the B. anynana genome, Mainak Das Gupta for his help in the in situ hybridization protocols and Monteiro lab members for their support.
Ethics
No animal experimentation permits were required for the experiments conducted here. Bicyclus anynana butterflies have been reared in the laboratory since 1988 and were imported under an AVA permit to the laboratory in Singapore.
Data accessibility
All data supporting this article are provided in the main text or as part of the electronic supplementary material.
Authors' contributions
A.P. and A.M. designed the study. A.P. performed the research and analysed the data. A.P. and A.M wrote the manuscript. All the authors gave their final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
This work was funded by Ministry of Education, Singapore grant R-154-000-602-112, the National University of Singapore grant R-154-000-587-133 and by the Department of Biological Sciences, NUS.
References
- 1.Kemp DJ. 2007. Female butterflies prefer males bearing bright iridescent ornamentation. Proc. R. Soc. B 274, 1043–1047. ( 10.1098/rspb.2006.0043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prudic KL, Stoehr AM, Wasik BR, Monteiro A. 2015. Eyespots deflect predator attack increasing fitness and promoting the evolution of phenotypic plasticity. Proc. R. Soc. B 282, 20141531 ( 10.1098/rspb.2014.1531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monteiro A. 2015. Origin, development, and evolution of butterfly eyespots. Annu. Rev. Entomol. 60, 253–271. ( 10.1146/annurev-ento-010814-020942) [DOI] [PubMed] [Google Scholar]
- 4.Martin A, Reed RD. 2010. Wingless and aristaless2 define a developmental ground plan for moth and butterfly wing pattern evolution. Mol. Biol. Evol. 27, 2864–2878. ( 10.1093/molbev/msq173) [DOI] [PubMed] [Google Scholar]
- 5.Warren R, Nagy L, Selegue J, Gates J, Carroll S. 1994. Evolution of homeotic gene regulation and function in flies and butterflies. Nature 372, 458–461. ( 10.1038/372458a0) [DOI] [PubMed] [Google Scholar]
- 6.Weatherbee SD, Nijhout HF, Grunert LW, Halder G, Galant R, Selegue J, Carroll S. 1999. Ultrabithorax function in butterfly wings and the evolution of insect wing patterns. Curr. Biol. 9, 109–115. ( 10.1016/S0960-9822(99)80064-5) [DOI] [PubMed] [Google Scholar]
- 7.Oliver JC, Tong X, Gall LF, Piel WH. 2012. A single origin for Nymphalid butterfly eyespots followed by widespread loss of associated gene expression. PLoS Genet. 8, e1002893 ( 10.1371/journal.pgen.1002893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oliver JC, Beaulieu JM, Gall LF, Piel WH, Monteiro A. 2014. Nymphalid eyespot serial homologues originate as a few individualized modules. Proc. R. Soc. B 281, 20133262 ( 10.1098/rspb.2013.3262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schachat SR, Oliver JC, Monteiro A. 2015. Nymphalid eyespots are co-opted to novel wing locations following a similar pattern in independent lineages. BMC Evol. Biol. 15, 20 ( 10.1186/s12862-015-0300-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen B, McGuffin ME, Pfeifle C, Segal D, Cohen SM. 1992. Apterous, a gene required for imaginal disc development in Drosophila encodes a member of the LIM family of developmental regulatory proteins. Genes Dev. 6, 715–729. ( 10.1101/gad.6.5.715) [DOI] [PubMed] [Google Scholar]
- 11.Tomoyasu Y, Arakane Y, Kramer KJ, Denell RE. 2009. Repeated co-options of exoskeleton formation during wing-to-elytron evolution in beetles. Curr. Biol. 19, 2057–2065. ( 10.1016/j.cub.2009.11.014) [DOI] [PubMed] [Google Scholar]
- 12.Carroll SB, Gates J, Keys DN, Paddock SW, Panganiban GE, Selegue JE, Williams JA. 1994. Pattern formation and eyespot determination in butterfly wings. Science 265, 109–114. ( 10.1126/science.7912449) [DOI] [PubMed] [Google Scholar]
- 13.Conceicao IC, Long AD, Gruber JD, Beldade P. 2011. Genomic sequence around butterfly wing development genes: annotation and comparative analysis. PLoS ONE 6, e23778 ( 10.1371/journal.pone.0023778) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nowell RW, et al. 2017. A high-coverage draft genome of the mycalesine butterfly Bicyclus anynana. Gigascience 6, 1–7. ( 10.1093/gigascience/gix035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramos D, Monteiro A. 2007. In situ protocol for butterfly pupal wings using riboprobes. J. Vis. Exp. 4, 208 ( 10.3791/208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bassett AR, Tibbit C, Ponting CP, Liu JL. 2013. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 4, 220–228. ( 10.1016/j.celrep.2013.06.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu FZ, Li KY, Li J, Hu DB, Zhao J, He YP, Zou YL, Feng YN, Hua HX. 2015. Apterous a modulates wing size, bristle formation and patterning in Nilaparvata lugens. Sci. Rep. 5, 1–12. ( 10.1038/srep10526) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruvinsky I, Gibson-Brown JJ. 2000. Genetic and developmental bases of serial homology in vertebrate limb evolution. Development 127, 5233–5244. (doi:11076746) [DOI] [PubMed] [Google Scholar]
- 19.Carroll SB, Weatherbee SD, Langeland JA. 1995. Homeotic genes and the regulation and evolution of insect wing number. Nature 375, 58–61. ( 10.1038/375058a0) [DOI] [PubMed] [Google Scholar]
- 20.Tong X, Hrycaj S, Podlaha O, Popadic A, Monteiro A. 2014. Over-expression of Ultrabithorax alters embryonic body plan and wing patterns in the butter fly Bicyclus anynana. Dev. Biol. 394, 357–366. ( 10.1016/j.ydbio.2014.08.020) [DOI] [PubMed] [Google Scholar]
- 21.Bhardwaj S, Prudic KL, Bear A, Das Gupta M, Wasik BR, Tong X, Cheong WF, Wenk MR, Monteiro A. 2018. Sex differences in 20-hydroxyecdysone hormone levels control sexual dimorphism in Bicyclus anynana butterfly wing patterns. Mol. Biol. Evol. 35(2), 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka K, Barmina O, Sanders LE, Arbeitman MN, Kopp A. 2011. Evolution of sex-specific traits through changes in HOX-dependent doublesex expression. PLoS Biol. 9, e1001131 ( 10.1371/journal.pbio.1001131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hobert O, Westphal H. 2000. Functions of LIM- homeobox genes. Trends Genet. 16, 75–83. ( 10.1016/S0168-9525(99)01883-1) [DOI] [PubMed] [Google Scholar]
- 24.Mann R, Carroll S. 2002. Molecular mechanisms of selector gene function and evolution. Curr. Opin. Genet. Dev. 12, 592–600. ( 10.1016/S0959-437X(02)00344-1) [DOI] [PubMed] [Google Scholar]
- 25.Whitney M, Tomoyasu Y, Wheeler SR, Denell RE. 2005. Ultrabithorax is required for membranous wing identity in the beetle Tribolium castaneum. Nature 433, 643–647. ( 10.1038/nature03272) [DOI] [PubMed] [Google Scholar]
- 26.Lewis EB. 1963. Genes and developmental pathways. Am. Zool. 3, 33–56. ( 10.1093/icb/3.1.33) [DOI] [Google Scholar]
- 27.Diaz-Benjumea FJ, Cohen SM. 1993. Interaction between dorsal and ventral cells in the imaginal disc directs wing development in Drosophila. Cell 75, 741–752. ( 10.1016/0092-8674(93)90494-B) [DOI] [PubMed] [Google Scholar]
- 28.Suzuki Y, Palopoli M. 2001. Evolution of insect abdominal appendages: are prolegs homologous or convergent traits? Dev. Genes Evol. 211, 486–492. ( 10.1007/s00427-001-0182-3) [DOI] [PubMed] [Google Scholar]
- 29.Shubin N, Tabin C, Carroll S. 1997. Fossils, genes and the evolution of animal limbs. Nature 388, 639–648. ( 10.1038/41710) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting this article are provided in the main text or as part of the electronic supplementary material.