Abstract
Microglia are a major cellular component of gliomas, and abundant in the centre of the tumour and at the infiltrative margins. While glioma is a notoriously infiltrative disease, the dynamics of microglia and glioma migratory patterns have not been well characterized. To investigate the migratory behaviour of microglia and glioma cells at the infiltrative edge, we performed two-colour time-lapse fluorescence microscopy of brain slices generated from a platelet-derived growth factor-B (PDGFB)-driven rat model of glioma, in which glioma cells and microglia were each labelled with one of two different fluorescent markers. We used mathematical techniques to analyse glioma cells and microglia motility with both single cell tracking and particle image velocimetry (PIV). Our results show microglia motility is strongly correlated with the presence of glioma, while the correlation of the speeds of glioma cells and microglia was variable and weak. Additionally, we showed that microglia and glioma cells exhibit different types of diffusive migratory behaviour. Microglia movement fit a simple random walk, while glioma cell movement fits a super diffusion pattern. These results show that glioma cells stimulate microglia motility at the infiltrative margins, creating a correlation between the spatial distribution of glioma cells and the pattern of microglia motility.
Keywords: time lapse microscopy, PDGF rat model, anomalous diffusion, particle image velocimetry, glioblastoma
1. Introduction
Microglia function as part of the innate immune system within the central nervous system (CNS) and play a central role in the reaction of brain tissue to virtually all forms of injury and disease. Confocal time-lapse studies conducted in hippocampal slices in the normal brain, found that under normal conditions, the microglial cell body remains non-motile and has a ramified morphology with fine processes able to scan the microenvironment for signals that might indicate pathologies [1]. Activating pro-inflammatory signals such as TNF-α, lipopolysaccharides, and other cytokines elicit a series of morphological changes in which microglial fine processes are replaced by motile protrusions. These activated microglia acquire a motile phenotype able to translocate and phagocytose targets [1,2]. In the context of glioma, microglia comprise up to 30% of the primary tumour cellular composition [3–6] and their presence in glioma is positively correlated with histological grade [7].
Glioma is a diffuse, infiltrative disease; however, the dynamic behaviour of microglia and glial cell motility has not been well quantified. Characterizations of the cellular composition and molecular signatures of the glioma inner core versus the infiltrative margins reveal that the tumour edge is enriched in microglia [8]. Microglia abundance has been associated with increased tumour migration [3,8–13]. Previous dynamic studies of microglia in other pathologies have led to defining microglia activation states by examining morphology and migratory behaviour during brain trauma and pathologies [1,14,15]. These dynamic studies examine microglia activation when clearing away dead cells [16], their response to brain injury as a function of extracellular ATP concentration [17], and their surveillance behaviour in the brain under ‘resting’ conditions [18]. Describing the motility of cellular populations can therefore provide insight into activation states, and their interactions with other cellular populations. To date, the mathematical descriptions of microglial and glioma cell motility have not been well characterized. Here we use mathematical descriptions of anomalous diffusion and individual motility characteristics to examine the spatial and temporal patterns of migratory behaviour between glioma and microglia cells at the infiltrative edge. As a mathematical description, this work can serve to inform mathematical models of glioma dispersion, particularly models examining dispersion among two different populations. From a biological perspective, this work provides insight into how these cellular populations navigate within the same physical space, and how they may influence one another.
To study the dynamic behaviour of microglia and glioma cells within living brain tissue, we used a previously described rat glioma model, in which PDGFB-IRES-GFP expressing retrovirus is injected into the subcortical white matter, where it infects PDGFRα+ glial progenitors, and induces the formation of tumours that recapitulate histological features of human glioblastoma [19,20]. We generated acute slice cultures from this glioma model and performed two-colour time-lapse microscopy to visualize the motile behaviour of glioma cells and microglia at the infiltrative margins of glioma. We applied quantitative techniques on single cell tracking data and used particle image velocimetry to describe their migratory behavioural properties such as speed, directionality and movement type. Using the novel combination of these techniques, we were able to demonstrate that (i) the spatial patterns of microglial motility correlate with higher densities of tumour cells, and (ii) microglia and glioma cells exhibit different migratory behaviours, even when localized to the same region of brain tissue. These results show that glioma cells stimulate microglia to acquire a migratory phenotype at the infiltrative margins of glioma and provide new insight into the motile behaviour exhibited by glioma and microglia.
2. Materials and methods
2.1. Retroviral injections
The retrovirus pQ-PDGFB-IRES-GFP was generated as previously described [19,20] and concentrated to titres of 105 CFU ml−1. We anaesthetized neonatal Sprague Dawley rats (P2-P3) by submerging them in ice water for approximately 8 min. The heads were then placed in a stereotactic apparatus (Stoelting, Avondale, IL, USA) and virus was injected into the rostral subcortical white matter at stereotactic coordinates 2 mm lateral and 1 mm rostral to the bregma. A 33 gauge Hamilton microsyringe (Reno, NV, USA) was inserted to a depth of 1.5 mm, and 1–2 µl of virus was injected at a rate of 0.4 µl min−1. All animal experiments were performed according to the guidelines of the Institutional Animal Care and Use Committee at Columbia University.
2.2. Two-colour time-lapse microscopy of microglia and glioma cell migration in brain slice
Rat pups injected with PDGFB-IRES-GFP were killed by decapitation at 10–12 days post injection (dpi) as described previously [20,21]. Brains were isolated, and 300 μm coronal sections of the injected hemispheres were made using a McIlwain Tissue Chopper (Campden Instruments, Loughborough, UK). Sections that had a significant number of GFP+ cells around the level of the injection site indicating tumour formation were transferred onto a 0.4 μm culture plate insert (Millipore) and placed on top of a well with 1200 µl of serum-free medium of a 6-well glass bottom plate (MatTek, Ashland, MA, USA). The plate with the slices placed on top of the insert filter were transferred to 95% O2, 5% CO2 incubator for an hour of recovery. All slices were stained with the same concentrations of Isolectin-B4 (Cy5-IB4, Life Technology) as described previously [1]. Briefly, the slices were incubated in IB4 (5 µg ml−1) dissolved in serum free media for 30 min and rinsed three times also with serum-free media. Thus, all microglia were subjected to the same concentrations of lectin prior to imaging. The time-lapse experiments were performed in a stage-mounted incubator with CO2 and temperature control, using a Nikon TE2000 inverted fluorescence microscope (Melville, NY, USA) with the controls from Metamorph software, as previously described [21]. Images were acquired at 40× magnification at 3 min intervals for up to 24 h. The imaging analysis was performed in regions where the glioma cells were intermingled with the surrounding brain tissue, and thus represents the infiltrative margins of the tumour.
To verify that the Rhodamine-IB4+ slices labelled microglial cells, we subjected the Rhodamine-IB4 slices that were labelled and used for time-lapse microscopy experiments to double immunofluorescence staining with an antibody against Iba1, a well characterized marker of microglia [22]. The double immunofluorescent labelling of the slices that we tested with Rhodamine-IB4 and anti-Iba1 antibody conjugated to Cy5 convincingly revealed that the large majority of Iba1 cells are also IB4+ cells (electronic supplementary material, figure S1) as predicted from previous published studies [22,23].
2.3. Immunofluorescent staining of slices
After time-lapse microscopy was performed, some tumour slices labelled with Rhodamine-IB4 were fixed with 4% paraformaldehyde for a 15 min rinse with PBS and blocked with 5% horse serum with 1% triton X-100 for 1 h. Slices were rinsed three times with 0.1% Triton X-100 in PBS and incubated with anti-Iba1 antibody overnight at 4°C in blocking solution. Slices were then washed four times with 0.1% Triton X-100 in PBS for 15 min and incubated with anti-rabbit conjugated to Cy-5 for 2 h, washed four times for 15 min and mounted on slides. Confocal images of anti-Iba1 and IB4 were taken with a Zeiss and were colour combined in photoshop.
2.4. Single cell tracking
Using Metamorph software (Molecular Devices, CA), cells were manually tracked frame by frame over the course of 8–18 h, 15 min per frame and, in some cases, at 3 min per frame, in order to be able to track the fastest cells. Cells from each population were randomly chosen for tracking across three separate experiments, yielding 100 tumour cells, and 248 microglia in experiment 1, 190 tumour cells and 100 microglia in experiment 2, and 50 tumour cells and 55 microglia in experiment 3.
2.5. Assessment of general movement type
The single cell tracking data was used to generate mean squared displacement (MSD) versus time plots. For both the tumour and microglial cell populations, the MSD of cells were computed for overlapping time intervals equal to multiples of 15 min (e.g. 15, 30, 45 etc.) up until 18 h of observation. For example, overlapping time intervals of 15 min multiples indicates: MSD is calculated for 0–15 min, 15–30… 1065–1080. Then MSD is calculated for 0–30 min, 30–60… 1050–1080 up until 18 h of observation, and so on. The plot of average MSD versus time interval was then log transformed and a line fit to the data using the Matlab linear regression algorithm within the statistics toolbox. The fitted slope (λ) of the linear fit was then used for each cell type to determine the type of movement behaviour. A slope of l represents simple diffusion, and a slope not equal to 1 is considered anomalous with λ > 1 representative of super-diffusive and λ < 1 sub-diffusive behaviour. Super-diffusive behaviour is associated with persistence in directional movement, and sub-diffusive behaviour is associated with movement in restricted spaces [24].
2.6. Localized movement analysis
To assess averaged regional behavioural properties of the cells we analysed the time-lapse microscopy images using the method of particle image velocimetry (PIV). PIV is a technique that determines velocity of particles over time, and has previously been used to determine motility and flow of fluorescently labelled cells [25]. In general, PIV analysis is done by dividing the region of interest into many smaller tile segments called interrogation windows. The cross-correlation of the pixel intensities between time frame i to frame i + 1 in each interrogation window is then calculated via a direct Fourier transform. The average movement of all the cells contained in that interrogation window is associated with the shift between frame_i to frame i + 1 corresponding to the highest correlation determined from the cross-correlation calculations. This average movement is then translated into a velocity by considering the time interval between frames. While the velocities calculated with PIV analysis are representative of an average velocity in the interrogation window, they are accounting for all of the cells in the field of view. This is of importance as single cell-tracking is limited by sampling since there are thousands of cells with a great deal of behavioural heterogeneity in our system of interest.
The field of view for our time-lapse microscopy images is 799 × 1042 µm for experiment 1 and 1392 × 1039 µm for experiments 2 and 3. To perform PIV analysis, we used PIVlab [26], a freely available Matlab package and considered interrogation windows of 102 × 102 µm (64 × 64 pixels). We were interested in the spatially resolved speed of the cells within each window, so PIV output velocity vectors were converted to all positive values, and then averaged into 64 by 64 pixel squares.
The background noise of time-lapse images was reduced using the band pass filter and background subtraction tools in ImageJ. After removal of the fluorescence background, the time-lapse images of glioma and microglia were separately converted into binary images so pixel noise was removed leaving only cell movement to be correlated. For correlations involving tumour speed (electronic supplementary material, figure S6) we weighted the linear fit by the density of tumour cells.
3. Results
3.1. Glioma cells induce microglial motility
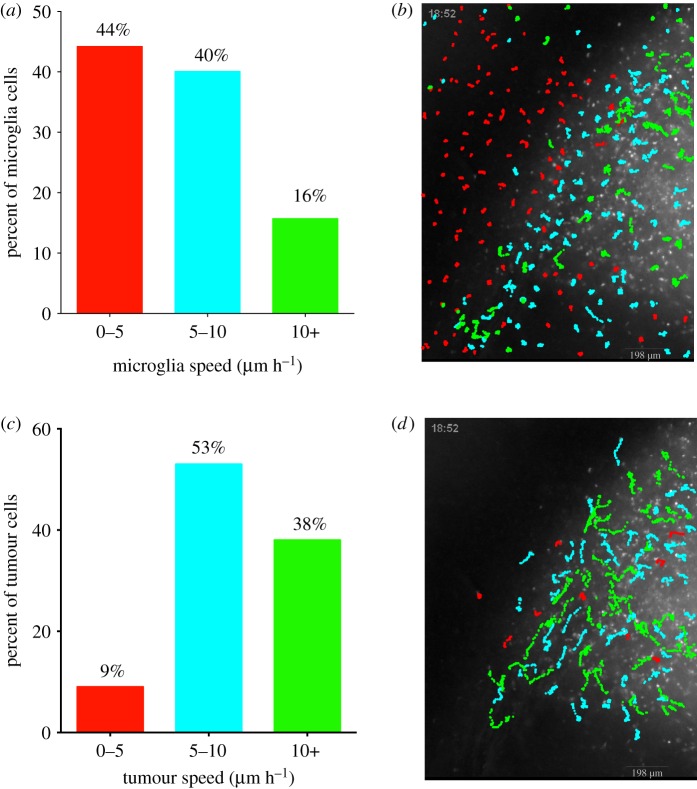
To investigate whether the migratory behaviour of microglia is influenced by the presence of glioma cells, two-colour fluorescence time-lapse microscopy from acute brain slices of a rat PDGFB-driven model was performed where the glioma cells were GFP+ [19,20] and microglia were labelled with isolectin IB4 conjugated to either Rhodamine or Cy5 [1]. At the glioma infiltrative edge, we observed that microglia exhibited heterogeneous migration speeds depending on their spatial proximity to the tumour. For example, in experiment 1, 44% of the tracked microglia moved between 0 and 5 µm h−1 (figure 1a), while 56% moved greater than 5 µm h−1. Interestingly, the majority of microglia that travelled between 0 and 5 µm h−1 were located in the peritumour area, beyond the margins of identifiable glioma cell infiltration, while microglia that moved greater than 5 µm h−1 were found in regions of glioma cell infiltration (figure 1b). Of particular note, 2% of microglia that were tracked travelled with speeds greater than 30 µm h−1 indicating large differences in migration speeds within the tumour infiltrative edge. Similar results were seen in experiments 2 and 3 (electronic supplementary material, table S1). The observation that microglia migrate significantly faster within regions of glioma infiltration provide dynamic evidence that glioma cells are inducing microglial activation. This analysis also showed that tracked glioma cells within the infiltrative margin of the tumour migrated significantly faster than the microglia. It was observed that 9% of tracked glioma cells travelled with speeds under 5 µm h−1, while 91% of glioma cells travelled greater than 5 µm h−1 (figure 1c).
Figure 1.
Microglia and tumour cell tracks coloured by speed and plotted on top of tumour microscopy background. Data from 248 microglia tracks and 100 tumour cell tracks from 18 h of recording from one slice of tumour. (a) Per cent of microglia categorized by average speed of 0 to 5, 5 to 10 and greater than 10 µm h−1. (b) Microglia tracks plotted in their native position on top of tumour background, coloured by colour scheme of a based on average speed. (c) Per cent of tumour cells categorized by average speed as done in a. (d) Tumour cell tracks plotted in their native position on top of the tumour background as done in b.
3.2. Microglia demonstrate a unique migration pattern as compared to glioma cells at the infiltrative edge of glioma
In addition to individual cell speeds, we were interested in a more detailed characterization of the two cell types' movement patterns. We began by considering the question at the population level by assessing whether population movement patterns of all tracked cells resembled a simple, sub- or super-diffusive process. A simple diffusive process would imply the cells were moving in a fairly non-restricted environment in a pattern consistent with a random walk. A sub-diffusive process is associated with movement in a restricted environment, and a super-diffusive process is associated with persistent directionality in movement patterns [24].
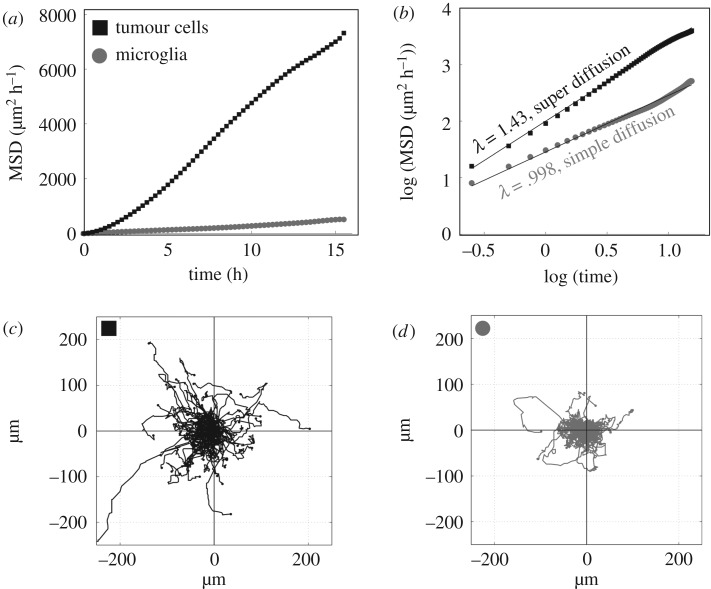
When all microglia were analysed as a population, microglia exhibited a movement pattern characteristic of a simple diffusive process across three separate experiments (λ = 0.98–1.0), whereas glioma cells exhibited super-diffusion (λ = 1.41–1.48) (figure 2b; electronic supplementary material, table S1). Figure 2a,b also highlights that the glioma cells, on average, travelled much farther distances than the microglia as the MSD of glioma cells is much greater over the same time span. These different migration patterns were particularly evident when examining a wind rose plot which show the individual trajectories of glioma and microglia cells initialized to the same starting point in space (figure 2c,d). The glioma cells exhibit much greater directionality (electronic supplementary material, figure S2), and cover more area than microglia.
Figure 2.
Movement behaviour of tumour cell and microglia populations. Data from experiment 1 includes 18 h of recording from one slice of tumour. (a) Mean square displacement over time plot show tumour cell populations move farther (black squares) over all time intervals compared with microglia (grey circles). (b) Log transformed plot of mean square displacement over time demonstrates that tumour cells exhibit super diffusion (λ = 1.43) in contrast to microglia (grey circle), which exhibit simple random walk (λ = 99). (c) One hundred tumour cell tracks starting from origin. (d) Two hundred and forty-eight microglia tracks starting from the origin. Tumour cell tracks cover much greater distance, and exhibit greater directionality compared to microglia tracks within the same region. These metrics are representative of the average or cumulative movement behaviour of the cell types, whereas the heterogeneity in individual cell movement patterns are notable in figure 1.
We then examined whether the microglia movement pattern was modified due to an apparent activation state defined by speed. By aggregating microglia tracks based on average speeds as defined in figure 1, we found that microglia moving at speeds greater than 5 µm h−1 (located primarily in higher tumour densities) travelled larger net distances in terms of their MSD, compared to those travelling less than 5 µm h−1 (located primarily in lower tumour densities). However, microglia often followed a behaviour that was characterized at most by a simple diffusive process throughout all three speed subdivisions (electronic supplementary material, table S1). Together, figures 2 and electronic supplementary material, table S1 show that as a population, microglia move according to a simple diffusive process, and that this tends to holds true for microglia located in regions of glioma infiltration, and for microglia that are located beyond the margins of glioma infiltration.
To look at the individual migratory properties of microglia and tumour cells, we used the single cell trajectories to assess the efficiency of direction of each cell by computing a metric of directionality called confinement ratio [24]. Linear movement is considered more efficient (i.e. confinement ratio closer to 1). The confinement ratio is defined as the displacement (net distance from start to finish) divided by the total cumulative distance travelled by a cell. Interestingly, when grouping microglia by three average speed categories (0–5, 5–10, 10+ μm h−1) and assessing average microglia directionality per category, we found that microglia directionality was increased among lower speed categories compared to microglia grouped in higher speed categories. Glioma cell movement exhibited the opposite behaviour. Glioma cell directionality was increased among higher speed categories compared to glioma cells grouped in lower speed categories (electronic supplementary material, figure S3). Not surprisingly, on average glioma cells were more directional than microglia across all three speed categories, highlighting the stark differences in movement behaviour between microglia and glioma. We found that the relationships between speed and directionality were evident among microglia cells, which tended to display lower confinement ratios among areas of high glioma cell density. However, we did not see any relationship between the directionality of glioma cell migration and microglia cell density (electronic supplementary material, figure S4).
3.3. Glioma cell motility is weakly correlated with microglia motility
To explore the possibility that microglia and glioma motility were spatially correlated, we used PIV to generate a spatial map of average speeds for each population, as described in Materials and methods. This provided the opportunity to assess cell movement without being confined to using only a sample of cells that were tracked. The output of this method is a coarsened grid, with a PIV-generated average velocity of cell movement assigned to each grid square. After converting the velocities to speeds, the correlation of the co-localized speed of the glioma and microglia populations can be assessed by associating the average speed for each grid square of the glioma cells to the average speed of its corresponding grid square from the microglia cells. On the same coarsened grid, we also created an average density map of glioma cells by calculating the average intensity within each grid square and considered the correlation between microglia speed and glioma density.
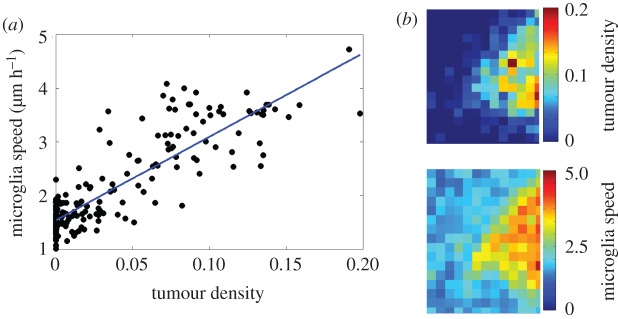
Using this approach, we found that microglia speed was strongly correlated with the local density of glioma cells, which is consistent with our results using single cell tracking data (figure 3) and was found across three separate experiments (electronic supplementary material, figure S5a–c). We then assessed the correlation of the speeds of the microglia and glioma cell populations. We observed that in regions further away from the tumour infiltrative edge that both glioma cells and microglia exhibited little motility. However, when the speed of microglia and glioma cells was compared across the entire microscopic field of three separate experiments, the correlation was found to be weak and variable (electronic supplementary material, figure S5d–f). Together, these results show that glioma cells stimulate microglial motility at the infiltrative margins of the tumour, creating a strong correlation between the spatial distribution of glioma cells and the pattern of microglial motility. However, glioma cells and microglia showed very different migratory behaviour, even when crawling through the same microenvironment. This suggests that glioma cells and microglia are either responding to different migratory cues, or are responding to same cues, but in different ways.
Figure 3.
Relationship between microglia and tumour density from experiment 1. Analysis conducted over 18 h of movie from one tumour. (a) Correlation between microglia speed and tumour density (R2 = 0.75, p < 001). (b) Heat maps of tumour density and microglia speed. Red regions are of high tumour densities, or high microglia speed. Correlation was performed by correlating each square of the heat-maps of tumour density and microglia speed to one another.
4. Discussion
We have applied a unique quantitative analysis that brings together individual cell tracking data and particle image velocimetry to infer both individual and population movement behaviour for glioma cells and microglia. To our knowledge, this is the first study to provide evidence of the dynamic activation of microglia due to infiltrating glioma and to investigate motility behaviour of glioma and microglia within the brain through slice culture. We have shown that microglia exhibit increased motility at the infiltrative margins of glioma and that microglia and glioma movement behaviour are categorically different. Microglia exhibited a diffuse process, moving as a simple random walk, whereas glioma cells exhibited a super-diffusive migratory behaviour with significantly increased directionality compared to microglia. These differences in migratory behaviour were seen even when microglia and glioma cells were migrating through the same regions of brain tissue. These findings highlight the nature by which microglia are activated due to glioma, and the movement behaviour they exhibit in response to glioma.
The migratory behaviour of microglia could be indicative of the variety of functions that microglia may be performing. Previous dynamic studies of microglia motility have been in the context of the normal brain in hippocampal slices and suggest that the low directionality exhibited by the majority of microglia may enable more contact with cells in a short period of time, resembling a surveillance function [1]. It has also been observed by others that surveying microglia are territorial, meaning they mutually repel one another upon contact [18]. Previous work characterizing microglia movement during spreading depression showed that microglia move in a Lévy flight type pattern, a movement process where microglia were observed to translocate large distances depending on the state of synaptic activity [27].
We found individual microglia motility was widely heterogeneous in terms of speed and directionality. At a population level this was characterized by examining the slope of the log-transformed MSD over time. Our analysis revealed that microglia move according to a simple diffusive process (slope of 1), throughout different regions of the field. While we have quantified population movement behaviour of microglia as a simple random walk, we noted a large degree of intercellular heterogeneity in their movement behaviour. While the majority of microglia moved between 0 and 10 µm h−1, 2% of the tracked microglia cells moved between 30 and 70 µm h−1, which was much faster than the average speed of the tracked tumour cells. These ultra-fast microglia were actually moving characteristically faster than the tumour cells, but they exhibited much less directionality in their movement. It is possible that these ultra-fast microglia may have been in an increased activation state, or had been stimulated by some other means as their migration path moved around and through many other microglia. This was in contrast to the majority of microglia behaviour, which we observed to not overlap migration paths with other microglia (see supplementary video).
The mechanisms by which microglia may be affecting glioma invasion are not well understood. We found that glial cell migratory behaviour was super-diffusive, and characterized by an overall average increased speed and directionality compared to microglia. One possibility for the differences between glial cells and microglia is that activated microglia may condition the local microenvironment in ways that facilitate motility, for example, by modifying extracellular space to reduce the impedance to migration. If this were case, then one may expect microglial and glioma cell motility to be spatially correlated. Yet, there only existed a weak and variable correlation between tumour and microglia motility implying that microglia and glioma cells did not exhibit similar motility patterns (electronic supplementary material, figure S6). However, it is possible that the conditioning of the microenvironment by microglia may happen on a different time scale, e.g. earlier than the tumour cell invasion.
Previous work in PDGF-driven tumours have shown that PDGF stimulates both the migration and proliferation of glioma cells [20]. Migrating tumour cells were seen to proliferate along their route, stopping to undergo mitosis for a short time (minimum 1 h) before daughter cells resumed migration. Moreover, PDGFRa+ progenitor cells were seen to co-opt surrounding vasculature early in tumour development, and time-lapse microscopy confirmed that a subset of GFP+ cells migrate along the abluminal surface of blood vessels [20]. This perivascular migration likely contributed to the highly directional behaviour seen among the glioma cell population compared to microglia. Similarly, in a more recent study, oligodendrocyte progenitor cells (OPCs) were found to migrate along the vasculature in the developing nervous system and required physical interactions with the vascular endothelium [28]. Furthermore, the authors showed that Wnt signalling regulates interactions between OPCs and blood vessels via a Cxcr4 dependent mechanism. In light of these findings, future studies should examine a possible role of Wnt and Cxcr4 in the perivascular migration of glioma cells.
While previous work has found evidence of an association between microglia and tumour dispersion, our dynamic study at the tumour infiltrative edge demonstrated that higher tumour densities was associated with microglia dispersion, however the relationship between microglia motility and tumour motility was weakly correlated. One explanation for this is that other intrinsic cellular mechanisms might be responsible for their differing migratory behaviours that are not explained by the differences in physical properties of the extracellular space alone. Another possibility is that other elements of microglia activation could influence glioma dispersion that would not be revealed in a motility analysis alone.
Our experimental system allowed us to recapitulate elements of human glioblastoma using a PDGF-driven rat model of GBM. Tumours were induced by stereotactic injections of PDGFB-expressing retrovirus into the subcortical white matter. It is possible that tissue damage at the injection site would cause a transient stimulation of microglial motility; however, the slices were generated at a time when tumours had grown extensively, and the margins of the tumour were many hundreds of microns from the injection site. Furthermore, our analysis shows that the speed of microglial migration was spatially correlated with tumour cell density (figure 3). Therefore, our results show that glioma cells are stimulating microglial motility.
This slice culture system may provide limitations to a motility analysis. Cell disperse in a 3D plane but our cell tracking measurements were measured in 2D. In some cases, cells might have exhibited high motility along the z-axis, but appear to have a slower motility when tracked on the x-y plane. Another limitation might be that there exists a temporal relationship between these two cell populations that was separate from a spatial relationship; and was unable to be unravelled by correlating their spatial-temporal relationship in the time interval under study. However, these particular possible limitations did not prevent us from detecting a relationship between microglia speed and tumour density. Further work is therefore warranted to further understand the complicated relationship between microglia and glioma cells, and particularly how these two cellular populations may influence tumour growth and dispersion.
5. Conclusion
The spatio-temporal behaviour of microglia and glioma cells in the intact brain tissue was examined here by using techniques to characterize movement behaviours. We found that the increased densities of glioma cells is correlated with increased activation of microglia, microglia exhibited a diffusive phenotype consistent with a simple random walk, and glioma cells moved with super-diffusive phenotype even within the same space. This study has implications for the functional interpretation of the differences of their movement strategy performed by glioma cells and microglia as these two cell types are responding to different migratory cytokines, or responding to the same ones in different ways. Further work remains to characterize the cellular mechanism by which glioma stimulates microglial motility, and the potential effects that microglia may have on glioma cell migration.
Supplementary Material
Data accessibility
All data, code, and tracking movies used to generate the results of this manuscript can be found in the data suppository Dryad, located here doi:10.5061/dryad.s4d28.
Authors' contributions
J.J. performed the data analysis, and drafted the manuscript; O.G. carried out the PDGF experiments, participated in data analysis and drafting of manuscript; A.H.-D. participated in coordination of the study, drafting of manuscript, conception of analysis techniques; S.N. carried out cellular tracking; R.C.R. participated in coordination of study and drafting of manuscript; J.G. participated in conception of analysis and drafting of manuscript; S.C.M. participated in drafting of manuscript and data analysis; P.A.S. participated in PIV analysis technique; A.R.A.A. participated in coordination of study, conception of analysis techniques, and drafting of manuscript; K.R.S. participated in coordination of study, drafting of manuscript, conception of analysis techniques, design of study; P.C. conceived of experiments, design and coordination of study, and drafting of manuscript. All authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
We gratefully acknowledge the support of the James S. McDonnell Foundation, the Ivy Foundation, the Mayo Clinic and the NIH (R01 NS060752, R01 CA164371, U54 CA210180, U54 CA143970, U54 CA193489)
References
- 1.Stence N, Waite M, Dailey ME. 2001. Dynamics of microglial activation: a confocal time-lapse analysis in hippocampal slices. Glia 33, 256–266. ( 10.1002/1098-1136(200103)33:3%3C256::AID-GLIA1024%3E3.0.CO;2-J) [DOI] [PubMed] [Google Scholar]
- 2.Kettenmann H, Hanisch U, Noda M, Verkhratsky A. 2011. Physiology of microglia. Physiol. Rev. 91, 461–553. ( 10.1152/physrev.00011.2010.) [DOI] [PubMed] [Google Scholar]
- 3.Markovic DS, et al. 2009. Gliomas induce and exploit microglial MT1-MMP expression for tumor expansion. Proc. Natl Acad. Sci. USA 106, 12 530–12 535. ( 10.1073/pnas.0804273106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kennedy BC, Showers CR, Anderson DE, Anderson L, Canoll P, Bruce JN, Anderson RCE. 2013. Tumor-associated macrophages in glioma: friend or foe? J. Oncol. 2013, 486912 ( 10.1155/2013/486912) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graeber MB, Scheithauer BW, Kreutzberg GW. 2002. Microglia in brain tumors. Glia 40, 252–259. ( 10.1002/glia.10147) [DOI] [PubMed] [Google Scholar]
- 6.Badie B, Schartner J. 2001. Role of microglia in glioma biology. Microsc. Res. Tech. 54, 106–113. ( 10.1002/jemt.1125) [DOI] [PubMed] [Google Scholar]
- 7.Komohara Y, Ohnishi K, Kuratsu J, Takeya M. 2008. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J. Pathol. 216, 15–24. ( 10.1002/path.2370) [DOI] [PubMed] [Google Scholar]
- 8.Bettinger I, Thanos S, Paulus W. 2002. Microglia promote glioma migration. Acta Neuropathol. 103, 351–355. ( 10.1007/s00401-001-0472-x) [DOI] [PubMed] [Google Scholar]
- 9.Coniglio SJ, Eugenin E, Dobrenis K, Stanley ER, West BL, Symons MH, Segall JE. 2012. Microglial stimulation of glioblastoma invasion involves epidermal growth factor receptor (EGFR) and colony stimulating factor 1 receptor (CSF-1R) signaling. Mol. Med. 18, 519–527. ( 10.2119/molmed.2011.00217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Põlajeva J, et al. 2014. Glioma-derived macrophage migration inhibitory factor (MIF) promotes mast cell recruitment in a STAT5-dependent manner. Mol. Oncol. 8, 50–58. ( 10.1016/j.molonc.2013.09.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhai H, Heppner FL, Tsirka SE. 2011. Microglia/macrophages promote glioma progression. Glia 59, 472–485. ( 10.1002/glia.21117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu F, et al. 2014. Glioma-derived versican promotes tumor expansion via glioma-associated microglial/macrophages Toll-like receptor 2 signaling. Neuro. Oncol. 17, 200–210. ( 10.1093/neuonc/nou324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coniglio SJ, Seagall JE. 2013. Review: molecular mechanism of microglia stimulated glioblastoma invasion. Matrix. Biol. 32, 372–380. ( 10.1016/j.matbio.2013.07.008) [DOI] [PubMed] [Google Scholar]
- 14.Boche D, Perry VH, Nicoll JAR. 2013. Review: activation patterns of microglia and their identification in the human brain. Neuropathol. Appl. Neurobiol. 39, 3–18. ( 10.1111/nan.12011) [DOI] [PubMed] [Google Scholar]
- 15.Hanisch U-K, Kettenmann H. 2007. Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat. Neurosci. 10, 1387–1394. ( 10.1038/nn1997) [DOI] [PubMed] [Google Scholar]
- 16.Petersen MA, Dailey ME. 2004. Diverse microglial motility behaviors during clearance of dead cells in hippocampal slices. Glia 206, 195–206. ( 10.1002/glia.10362) [DOI] [PubMed] [Google Scholar]
- 17.Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, Gan W. 2005. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 8, 752–758. ( 10.1038/nn1472) [DOI] [PubMed] [Google Scholar]
- 18.Nimmerjahn A, Kirchhoff F, Helmchen F. 2005. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318. ( 10.1126/science.1110647) [DOI] [PubMed] [Google Scholar]
- 19.Assanah M, Lochhead R, Ogden A, Bruce J, Goldman J, Canoll P. 2006. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J. Neurosci. 26, 6781–6790. ( 10.1523/JNEUROSCI.0514-06.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Assanah MC, Bruce JN, Suzuki SO, Chen A, Goldman JE, Canoll P. 2009. PDGF stimulates the massive expansion of glial progenitors in the neonatal forebrain. Glia 57, 1835–1847. ( 10.1002/glia.20895) [DOI] [PubMed] [Google Scholar]
- 21.Beadle C, Assanah MC, Monzo P, Vallee R, Rosenfeld SS, Canoll P. 2008. The role of myosin II in glioma invasion of the brain. Mol. Biol. Cell 19, 3357–3368. ( 10.1091/mbc.E08-03-0319) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito D, Imai Y, Ohsawa K, Nakajima K, Fukuuchi Y, Kohsaka S. 1998. Microglia-specific localisation of a novel calcium binding protein, Iba1. Mol. Brain Res. 57, 1–9. ( 10.1016/S0169-328X(98)00040-0) [DOI] [PubMed] [Google Scholar]
- 23.Markovic DS, Glass R, Synowitz M, van Rooijen N, Kettenmann H. 2005. Microglia stimulate the invasiveness of glioma cells by increasing the activity of metalloprotease-2. J. Neuropathol. Exp. Neurol. 64, 754–762. ( 10.1097/01.jnen.0000178445.33972.a9) [DOI] [PubMed] [Google Scholar]
- 24.Beltman JB, Marée AFM, de Boer RJ. 2009. Analysing immune cell migration. Nat. Rev. Immunol. 9, 789–798. ( 10.1038/nri2638) [DOI] [PubMed] [Google Scholar]
- 25.Petitjean L, Reffay M, Grasland-Mongrain E, Poujade M, Ladoux B, Buguin A, Silberzan P. 2010. Velocity fields in a collectively migrating epithelium. Biophys. J. 98, 1790–1800. ( 10.1016/j.bpj.2010.01.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thielicke W, Stamhuis EJ. 2014. PIVlab – towards user-friendly, affordable and accurate digital particle image velocimetry in MATLAB. J. Open Res. Softw. 2, e30 ( 10.5334/jors.bl) [DOI] [Google Scholar]
- 27.Grinberg YY, Milton JG, Kraig RP. 2011. Spreading depression sends microglia on levy flights. PLoS ONE 6, e19294 ( 10.1371/journal.pone.0019294.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai H-H, et al. 2016. Oligodendrocyte precursors migrate along vasculature in the developing nervous system. Science 351, 379–384. ( 10.1126/science.aad3839) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data, code, and tracking movies used to generate the results of this manuscript can be found in the data suppository Dryad, located here doi:10.5061/dryad.s4d28.