Abstract
The bacterial CRISPR-Cas system provides adaptive immunity against invading phages. Cas9, an RNA-guided endonuclease, specifically cleaves target DNA substrates and constitutes a well-established platform for genome editing. Recently, anti-CRISPR (Acr) proteins that inhibit Cas9 have been discovered, promising a useful off-switch for Cas9 to avoid undesirable off-target effects. Here, we report the solution structure and dynamics of Listeria monocytogenes AcrIIA4 that inhibits Streptococcus pyogenes Cas9 (SpyCas9). AcrIIA4 forms a compact monomeric αβββαα fold comprising three antiparallel β strands flanked by three α-helices and a short 310-helix. AcrIIA4 exhibits distinct backbone dynamics in fast and slow timescales at loop regions that form interaction surfaces for SpyCas9. In particular, the β1–β2 loop that binds to the RuvC domain of SpyCas9 is highly mobile, and the β1–β2 and α2–α3 loops that bind to the RuvC and C-terminal domains of SpyCas9, respectively, undergoes conformational exchanges in microsecond-to-millisecond time scales. AcrIIA4 binds to apo-SpyCas9 with KD ~4.8 μM, which compares to KD ~0.6 nM for AcrIIA4 binding to sgRNA-bound SpyCas9. Since the binary complex between AcrIIA4 and SpyCas9 does not compete with the target DNA binding, it can effectively disable the Cas9 nuclease activity by forming a tight ternary complex in the presence of sgRNA.
Introduction
The clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) proteins constitute a defense system in bacteria and archaea, providing a form of immunological memory against foreign genetic elements1,2. Upon infection, the bacterial host cell activates the CRISPR-Cas system to integrate short segments of the invading nucleic acids into the CRISPR loci. The integrated DNA sequences, also known as spacers, are transcribed into CRISPR RNAs that guide a single protein effector or a multi-protein effector complex to degrade the complementary sequences in foreign genes upon subsequent invasions. The CRISPR-Cas system is categorized into two classes, six types and thirty-three subtypes according to different organizations of signature cas genes3. Streptococcus pyogenes Cas9 (SpyCas9) belongs to the subtype II-A of the Class 2 CRISPR-Cas system. SpyCas9 has been extensively investigated and widely applied to genome editing due to its simplicity in generating double-strand DNA breaks at desired sites targeted by single-guide RNA (sgRNA)4–6.
Bacteria and phages have long been engaged in an arms race, developing a variety of arsenal to compete for invasion and defense7. Recently, anti-CRISPR (Acr) proteins that neutralize Cas9 activity have been discovered in phages targeting pathogenic bacterial strains8,9. Among them, AcrIIA2 and AcrIIA4 encoded by Listeria monocytogenes prophages have shown cross-strain inhibitory effects against SpyCas9, highlighting their potential as an off-switch for Cas9 for temporal and spatial control of genome editing. Recent structures of the AcrIIA4‒SpyCas9‒sgRNA complex obtained by X-ray crystallography and electron microscopy show that SpyCas9 binds to AcrIIA4 via the protospacer adjacent motif (PAM) interaction site and the RuvC domain10–12. Here we report the first solution structure and dynamics of AcrIIA4 using NMR spectroscopy. Backbone dynamics from 15N NMR relaxation data demonstrates that the binding interfaces for SpyCas9 exhibit fast internal motions and slow conformational exchanges in variable timescales. Further, we show that AcrIIA4 binds to SpyCas9 in the absence of sgRNA with an equilibrium dissociation constant (KD) of ~4.8 μM, which can associate with guide RNA to form a tight ternary complex, leading to the loss of Cas9 nuclease activity.
Results and Discussion
Structural description of AcrIIA4 determined by NMR spectroscopy
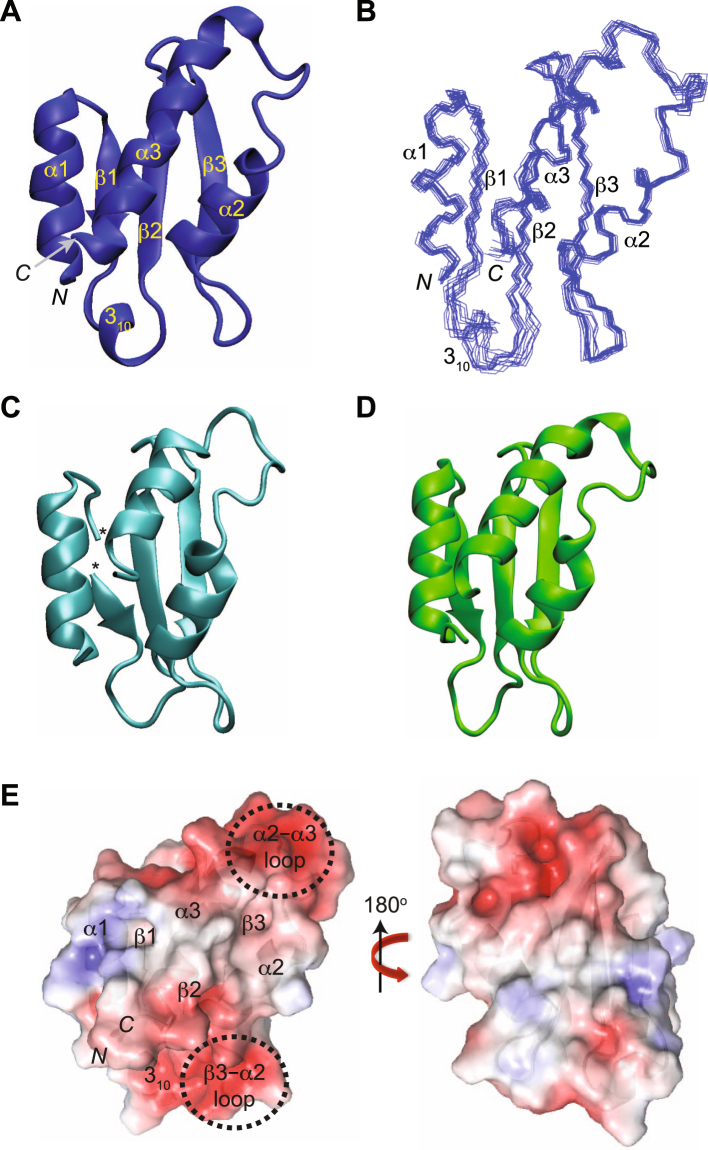
AcrIIA4 (a.a. 1‒87) exists as a monomer in solution, and exhibits a well resolved 1H–15N HSQC NMR spectrum that is typically observed in compact folded proteins (Figures S1 and S2). We have determined the solution structure of AcrIIA4 using heteronuclear triple-resonance NMR spectroscopy. Backbone and side chain assignments were obtained using a suite of three-dimensional heteronuclear correlation NMR spectroscopy experiments. The backbone chemical shifts were completely assigned except for Thr28 that did not show backbone amide resonance in the 2D 1H–15N HSQC spectrum. Ser20, Ser24, Gln29, Glu47, and Gln65 did not show backbone amide resonances, but their aliphatic and carbonyl groups could be assigned from their sequential connectivities. The 1H assignment data covered 602 out of 646 1H atoms excluding those in exchangeable hydroxyl and amino groups, which amounts to the 93.2% completeness of 1H assignment. Three-dimensional 13C-separated NOE and 15N-separated NOE restraints were employed for the structure calculation using the Xplor-NIH program13. Residual dipolar couplings (RDCs) were measured in 10 mg/ml of pf1 phage alignment medium. The structure was determined using 1,782 NMR restraints including 1,504 experimental NOE-based distance restraints, 161 dihedral angle restraints, 79 backbone 1DNH RDC restraints, and 38 hydrogen bonding restraints (Table 1). The hydrogen bond restraints were introduced at the final stage of the structure calculation, based on the secondary structural information from NOE and chemical shift data. The hydrogen bond restraints were in perfect agreement with the short-range and medium range NOEs, and dihedral angle restraints. AcrIIA4 is comprised of three antiparallel β strands (β1, 16–19; β2, 29–33; β3, 40–44) and three flanking α helices (α1, 2–13; α2, 50–59; α3, 68–85) in an α1–β1–β2–β3–α2–α3 order (Fig. 1A). In addition, a short 310 helix (22–25) is formed in the loop connecting β1 and β2 strands. Superposition of the backbone atoms for the ensemble of the 20 lowest energy structures of AcrIIA4 demonstrates that overall secondary structures are well defined such that three helices are packed together upon a three-stranded β sheet (Fig. 1B). The connecting loop regions are less well-defined, and the β1‒β2 loop among others shows the largest backbone root-mean-square deviations (RMSD ~1.3 Å).
Table 1.
Restraints and structural statistics for AcrIIA4.
| Experimental restraints | <SA>* |
|---|---|
| Nonredundant NOEs | 1504 |
| Dihedral angles, ϕ/ψ/χ | 71/71/19 |
| Hydrogen bonds | 38 |
| Residual dipolar coupling, 1DNH | 79 |
| Total number of restraints | 1782 (20.5 per residue) |
| rms deviation from experimental restraints | |
| Distances (Å) (1504) | 0.029 ± 0.001 |
| Torsion angles (°) (161) | 0.90 ± 0.10 |
| Residual dipolar coupling R-factor (%)† | |
| 1DNH (%) (79) | 1.8 ± 0.2 |
| rms deviation from idealized covalent geometry | |
| Bonds (Å) | 0.002 ± 0 |
| Angles (°) | 0.47 ± 0.01 |
| Impropers (°) | 0.42 ± 0.01 |
| Coordinate precision (Å)*‡ | |
| Backbone | 0.61 ± 0.11 |
| Heavy atoms | 1.38 ± 0.09 |
| Ramachandran statistics (%)ठ| |
| Most favorable regions | 91.2 ± 1.1 |
| Allowed regions | 8.8 ± 1.1 |
*For the ensemble of the final 20 simulated annealing structures.
†The magnitudes of the axial and rhombic components of the alignment tensor were 7.3 Hz and 0.35, respectively.
‡Regions with secondary structures (residues 2–13, 16–19, 29–33, 40–44, 50–59, and 68–85).
§Calculated using the program PROCHECK39.
Figure 1.
Structures of AcrIIA4 as ribbon and surface representations. (A) Solution structure of AcrIIA4 in the free state. Secondary structures, as well as N- and C-termini are annotated. (B) Superposition of the backbone atoms of the final 20 simulated annealing structures of AcrIIA4. The structures are best-fit superposed against well-ordered secondary structures between residues 2–13, 16–19, 29–33, 40–44, 50–59, and 68–85. (C) AcrIIA4 from the crystal structure of the AcrIIA4–SpyCas9–sgRNA complex (PDB code 5XBL). (D) AcrIIA4 from the crystal structure of the AcrIIA4–SpyCas9–sgRNA complex (PDB code 5VW1). (E) Front and backside view of molecular surface representations of AcrIIA4 color-coded by electrostatic potential, ±5 kT. AcrIIA4 on the left panel is drawn in the same orientation as in (A).
The overall backbone fold of free AcrIIA4 in solution was similar to that of the crystal structures determined for the AcrIIA4‒SpyCas9‒sgRNA complexes, indicating that AcrIIA4 does not change the backbone conformation upon binding to SpyCas9‒sgRNA (Fig. 1C and D). The β1 strand was well-defined and the C-terminal α3 helix extended between Glu68 and Glu85 in the solution structure, which was consistent with the crystal structure with higher resolution (Fig. 1D). On the other hand, the β1‒β2 loop formed a 310 helix between Thr22 and Asn25 in the solution structure, but did not exhibit a regular secondary structure in complex with SpyCas9‒sgRNA.
AcrIIA4 features a cluster of aromatic residues that form a compact hydrophobic core via stacking and edge-to-face packing between the β3 strand, and α2 and α3 helices. The tightly packed aromatic side chains result in large ring current effects and unusual chemical shifts, such as Hβ of Glu71 (−0.1 ppm), Hα of Gln65 (0.4 ppm), and Hβ and Hγ of Met77 (0.17 ppm and −0.05 ppm). AcrIIA4 is an acidic protein (pI ~4.2), and the surface electrostatic potential indicates that negative charges are densely populated in the β3–α2 loop, the α2–α3 loop and the beginning of the α3 helix (Fig. 1E). It has been suggested that the negatively charged patches mimic the phosphate groups of nucleic acids that associate with the PAM interaction site of SpyCas910–12, which was also found in AcrF2 proteins inhibiting the subtype I-F effector complex14.
Backbone dynamics reveals characteristic motions at the binding interface
15N R1 and R2 relaxation rates and 1H–15N heteronuclear NOE were measured to characterize the backbone dynamics of AcrIIA4. The rotational correlation time (τc) was calculated as 6.2 ns from the ratio of 15N R2 and R1 relaxation rates in the ordered region, which is typical for the size of AcrIIA4 (~10.2 kDa). Secondary structural regions exhibited large 1H–15N heteronuclear NOE (>0.8), and uniform 15N R2 and R1 relaxation parameters indicating that the secondary structures were well structured and rigid without internal motions (Fig. 2). On the other hand, loop regions connecting the secondary structures manifested variable dynamic behavior in a wide range of timescales (Fig. 2). Reduced 1H–15N heteronuclear NOE (<0.8) and 15N R1 relaxation rates indicated that fast backbone dynamics in ps‒ns timescales was prevalent in the β1–β2 loop (Gly21–Ile27) region. In addition, increased 15N R2 relaxation rates indicated the presence of slower conformational exchanges in μs‒ms timescales in the β1–β2 loop (Thr22, Asn23, and Ser26), the β3–α2 loop (Asn48 and Ser50) and the α2–α3 loop (Gly62 and Glu68) regions. The conformational exchanges also resulted in complete line broadening of backbone amide resonances in the β1–β2 loop (Ser20, Ser24, Thr28, and Gln29), the β3–α2 loop (Glu47), and the α2–α3 loop (Gln65) regions.
Figure 2.

Plots of relaxation parameters of backbone amide groups of AcrIIA4. (A) 15N R1 relaxation, (B) 15N R2 relaxation, and (C) 1H–15N heteronuclear NOE data as a function of residue number. The secondary structures of AcrIIA4 are shown on top as a schematic representation. The relaxation parameters were obtained on an 800 MHz Bruker NMR spectrometer.
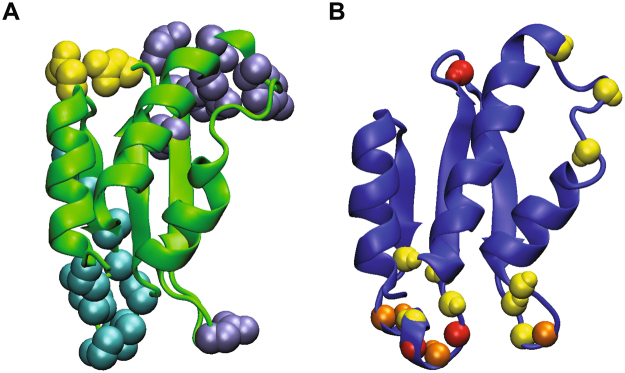
It is remarkable that the dynamic loop regions constitute the binding interfaces between AcrIIA4 and SpyCas9 in the crystal structure10–12. AcrIIA4 mainly interacts with the TOPO domain, the RuvC domain, and the C-terminal domain (CTD) of SpyCas9. Specifically, AcrIIA4 employs the α1–β1 and β2–β3 loops to interact with the TOPO domain, the β1 strand and the β1–β2 loop to interact with the RuvC domain, and the β2–β3, β3–α2, and α2−α3 loops to interact with the CTD (Fig. 3A). The interfacial loops exhibited significant backbone dynamics in a wide range of time scales. The β1–β2 loop that binds to the RuvC domain of SpyCas9 was highly mobile with fast internal motions in ps‒ns timescales as well as slower conformational exchanges in μs‒ms timescales (Fig. 3B). On the other hand, the β3–α2 loop and the α2–α3 loop that bind to the CTD of SpyCas9 exhibited extensive conformation exchanges in μs‒ms timescales. The binding interface for the TOPO domain appeared less dynamic compared to the other interfaces.
Figure 3.
Binding interfaces of AcrIIA4 for SpyCas9‒sgRNA and mobile regions with backbone dynamics. (A) Binding interfaces of AcrIIA4 from the crystal structure in complex with SpyCas9‒sgRNA (PDB code 5VW1). AcrIIA4 is shown as a ribbon diagram, and interfacial residues in the complex are shown as a space-filling model. Interfaces for the TOPO domain, the RuvC domain, and the C-terminal domain of SpyCas9 are colored in yellow, cyan, and ice-blue, respectively. (B) Mobile regions with backbone dynamics of AcrIIA4 from the solution structure. AcrIIA4 is shown as a ribbon diagram, and backbone amide groups are shown as a space-filling model. Residues with fast internal motions (ps‒ns time scale) and slower conformational dynamics (μs‒ms time scale) are colored in red and yellow, respectively. Residues with both fast and slower dynamics are colored in orange.
It has been reported that the fast and slow motions characterized by NMR relaxation play an important role in the target search and recognition process of biomolecular interactions15–18. It is notable that the dynamic β1–β2 loop of AcrIIA4 directly interacts with the active site of the SpyCas9 nuclease, preventing the substrate DNA binding. SpyCas9 cleaves double-stranded DNA substrates using two nuclease domains, the HNH nuclease domain that processes the target DNA strand complementary to the guide RNA, and the RuvC nuclease domain that processes the non-target DNA strand. The β1–β2 loop of AcrIIA4 extends to the active site of RuvC, and participates in a hydrogen bonding network with catalytic residues, blocking the access of the non-target DNA strand. Meanwhile, the α2–α3 loop that recognizes the PAM interaction site for the target DNA strand manifests slower conformational exchanges. The α2–α3 loop does not directly block the active site of the nuclease, but occludes the binding site of the PAM sequence that is located immediately after the cleavage site of the target DNA strand. The loop dynamics of AcrIIA4 are thus linked to the target recognition and binding, with different time scales between the interaction surfaces.
AcrIIA4 binds to SpyCas9 in the absence of sgRNA
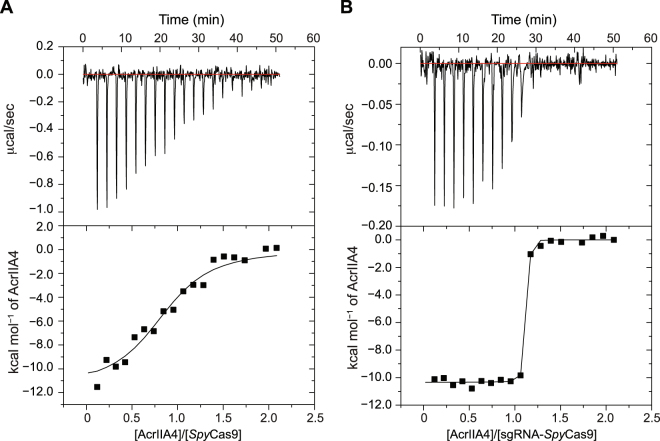
We quantitatively measured the binding thermodynamics between AcrIIA4 and apo-SpyCas9 using isothermal titration calorimetry (ITC). Our measurement revealed a large exothermic reaction between AcrIIA4 and SpyCas9 with KD = 4.8 ± 1.5 μM and a 1:1 stoichiometry (Fig. 4A). The interaction between AcrIIA4 and sgRNA bound-SpyCas9 showed a significantly higher affinity, with the KD value of 0.6 ± 0.1 nM (Fig. 4B). The binding affinity measured by ITC was similar to the previous report of KD = 4.1 ± 1.1 nM from the microscale thermophoresis data10. Thus, the presence of sgRNA promoted the interaction between AcrIIA4 and SpyCas9 by a factor of 8000 (Table 2). The weaker binding between AcrIIA4 and apo-SpyCas9 explains previous observations that AcrIIA4 did not co-elute with SpyCas9 in the absence of sgRNA from the size exclusion chromatography, though MBP pull-down assays showed a possible weak interaction between AcrIIA4 and apo-SpyCas910–12. We also note that AcrIIA4 did not exhibit measurable affinity to sgRNA alone (Figure S3). The binding of AcrIIA4 and SpyCas9 was largely an enthalpy-driven reaction, which can be explained by their binding surfaces comprising extensive electrostatic interactions and hydrogen bonding networks11. The entropic contribution to the binding was unfavorable, which reflects a decrease in conformational flexibility at the interfaces upon complex formation. Indeed, the increased affinity of AcrIIA4 to sgRNA-bound SpyCas9 largely originated from favorable entropic contribution, demonstrating that sgRNA binding to SpyCas9 relieved the entropic cost of SpyCas9 to accommodate AcrIIA4.
Figure 4.
ITC for the interaction (A) between AcrIIA4 and SpyCas9, and (B) between AcrIIA4 and sgRNA-bound SpyCas9. Raw ITC data (Top panel) and integrated heats of injection (bottom panel) are presented for the titration between AcrIIA4 and SpyCas9. In the bottom panel, the experimental data are shown as solid squares and the least squares best-fit curves derived from a simple one-site binding model are shown as a black line. The thermodynamic parameters for the complex formation are listed in Table 2.
Table 2.
Thermodynamic parameters for the interaction between AcrIIA4 and SpyCas9 obtained by isothermal titration calorimetry at 25 °C.
| Description | KD (nM) | ΔG (kcal/mol) | ΔH (kcal/mol) | −TΔS (kcal/mol) |
|---|---|---|---|---|
| AcrIIA4:SpyCas9 | 4800 ± 150 | −7.3 ± 0.1 | −11.5 ± 0.7 | 4.2 ± 0.8 |
| AcrIIA4:SpyCas9‒sgRNA | 0.6 ± 0.1 | −12.6 ± 0.4 | −10.3 ± 0.1 | −2.3 ± 0.4 |
1H‒15N HSQC spectra of 15N-AcrIIA4 titrated with unlabeled SpyCas9 exhibited gradual line broadening without chemical shift changes. Due to the large molecular weight of SpyCas9 (m.w. ~158.4 kDa), backbone amide resonances of free AcrIIA4 gradually disappeared without any sign of new signals from the complex, such that 0.2 mM 15N-AcrIIA4 complexed with SpyCas9 at a 1:1 ratio resulted in complete line broadening, except for the C-terminal Asn87. Most amide resonances of free AcrIIA4 exhibited intensity reduction of >60%, when the complex formation reached 10% from the stoichiometric titration (Fig. 5). This indicates that the line broadening is caused not only by the large complex formation, but also by intermediate exchange from the interaction. It has been reported that the interaction surfaces in slow exchanging large macromolecular complexes can be identified by differential line broadening upon binding19. We examined the line broadening of individual residues of AcrIIA4 upon apo-SpyCas9 binding. The line broadening was less severe at the terminal and internal loop regions, and interfacial residues such as Tyr67 and Asp69 exhibited larger line broadening than others. The differential line broadening, however, did not unambiguously identify the binding interface of AcrIIA4. Comparison of amide resonance intensities of AcrIIA4 between free and 10% bound states indicated that the intensity reduction spanned most backbone amide groups. We examined the line broadening upon titration at 10 °C and 45 °C, but could not distinguish the binding interfaces based on the differential line broadening. We infer that the dynamics of AcrIIA4 and SpyCas9 in complex may have contributed to the intermediate exchange, obscuring the precise mapping of the binding interface.
Figure 5.

Intensity ratio of backbone amide resonances between AcrIIA4 in complex with SpyCas9 (10% bound) and free AcrIIA4. The intensity ratio of backbone amide groups of AcrIIA4 measured from 1H–15N HSQC spectra at 25 °C is shown as a function of residue number. The intensity of 0.2 mM AcrIIA4 in free state (Ifree), and in complex with 0.02 mM SpyCas9 (Ibound) are measured and the ratio is presented as Ibound/Ifree. The secondary structures of AcrIIA4 are shown on top as a schematic representation, and the binding interfaces of AcrIIA4 in the AcrIIA4‒SpyCas9‒sgRNA complex (PDB code 5VW1) are shown above the ratio as a visual guidance.
The crystal structure of the AcrIIA4–SpyCas9–sgRNA complex has shown that SpyCas9 employs the RuvC domain, the TOPO domain, and the CTD to interact with AcrIIA410–12. The interfaces at the TOPO domain and the CTD (red spheres in Figure S4) overlap with the PAM interaction site of SpyCas9 for the target strand of the DNA substrate. The interface at the RuvC domain (orange spheres in Figure S4) is the active site pocket that cleaves the non-target strand of the double-stranded DNA substrate. When the binding interfaces in the complex are mapped onto apo‒SpyCas9, interfaces from the RuvC domain and the CTD are relatively well maintained in apo‒SpyCas9 (dashed circle in red; Figure S4), but those from the TOPO domain are completely lost (dashed circle in black; Figure S4). Based on the structural alignment, we speculate that AcrIIA4 binds to apo-SpyCas9 in a similar manner to sgRNA-bound SpyCas9, and that the RuvC domain and the CTD provide the interaction surfaces for AcrIIA4 in the absence of sgRNA. Binding of SpyCas9 to sgRNA induces a major rearrangement of the helical REC domain, as well as ordering of the PAM interaction site at the TOPO domain and the CTD20,21. It is remarkable that the affinity of the AcrIIA4‒SpyCas9 interaction increases by a thousand-fold upon sgRNA binding10. Thus, the guide RNA binding and concomitant conformational rearrangement of SpyCas9 provide key interfaces not only for the DNA substrate but also for the inhibitor AcrIIA4.
It has been shown that AcrIIA4 competes with the double-strand DNA substrate for SpyCas9 binding with a ~17-fold higher affinity10. AcrIIA4 inhibited the substrate DNA binding to SpyCas9 in a concentration-dependent manner, but did not affect the dissociation of the DNA substrate from SpyCas9, indicating a slow dissociation kinetics12. In other words, AcrIIA4 effectively inhibits the nuclease activity of SpyCas9, but there is competition between the DNA substrate and AcrIIA4 targeting for the same interface on SpyCas9. We demonstrated that AcrIIA4 bound sequentially to SpyCas9 and then to guide RNA with increasing affinity. In the sequential binding mechanism, AcrIIA4 does not need to compete with target DNA for SpyCas9 binding, since target DNA recognition requires a pre-formed assembly of guide RNA and SpyCas9. The binary complex between AcrIIA4 and apo‒SpyCas9 has a moderate affinity, but it can rapidly turn into a tight ternary complex in the presence of mature guide RNA, effectively preventing the target DNA binding. The sequential binding mechanism would be advantageous for the anti-CRISPR function, especially when the anti-CRISPR genes are integrated as a prophage, and constitutively inactivates the host CRISPR-Cas system22.
In summary, we have determined the solution structure of the anti-CRISPR protein AcrIIA4, and investigated the backbone dynamics using NMR spectroscopy. AcrIIA4 adopts a small and compact backbone fold, but exhibits significant dynamics in the loop region that serves as the binding interface for SpyCas9. Notably, AcrIIA4 can bind to SpyCas9 in the absence of sgRNA, and the binary complex can associate with sgRNA to form a tight ternary complex, avoiding competition with the DNA substrate for SpyCas9 binding.
Materials and Methods
Cloning, protein expression, and purification
The synthetic AcrIIA4 (a.a. 1–87) gene was cloned into a modified pBT7-N-His vector (ATCC). The plasmid was introduced into Escherichia coli strain BL21star(DE3) cells (Invitrogen), and grown in LB or minimal media with 15NH4Cl and/or 13C6-glucose as sole nitrogen or carbon sources, respectively. Cells were grown at 37 °C to OD600 ~0.8, induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 16 °C, and harvested by centrifugation after 16 hours of induction. Cell pellets were resuspended in 20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 20 mM imidazole, and 1 mM phenylmethylsulfonylfluoride, lysed using Emulsiflex C3 (AVESTIN) and centrifuged at 20,000 g for 30 min. The clear supernatant was loaded onto a HisTrap column (GE Healthcare) equilibrated with 20 mM Tris-HCl, pH 7.4, 200 mM NaCl, and 20 mM imidazole, and eluted with 500 mM imidazole. The His6-tag was removed using TEV protease in 20 mM Tris-HCl, pH 8.0, 5 mM β-mercaptoethanol (BME), and 200 mM NaCl, and the digestion reaction was loaded onto the HisTrap column. The flow-through was collected and loaded onto a Superdex 75 column (GE Healthcare) equilibrated with 20 mM Tris-HCl, pH 7.4, 200 mM NaCl. Lastly, AcrIIA4 was loaded onto a MonoQ column (GE Healthcare) and eluted with a gradient of 0.1‒1 M NaCl.
The synthetic S. pyogenes Cas9 (a.a. 1–1368) gene was cloned into a pET28b vector with a C-terminal His6-tag, and transformed into E. coli strain BL21(DE3) cells. The cells were cultured in LB medium at 37 °C to OD600 ~0.7. Protein expression was induced by the addition of 0.5 mM IPTG, followed by incubation at 17 °C for 16 hours. Cells were harvested by centrifugation and resuspended in 20 mM Tris, pH 7.5, 300 mM NaCl, 5 mM BME, and 10% (w/v) glycerol. After sonication and centrifugation, the supernatant was loaded onto a 5 mL HisTrap HP column (GE Healthcare) pre-equilibrated with 20 mM Tris, pH 7.5, 300 mM NaCl, 5 mM BME, 10% (w/v) glycerol, and 30 mM imidazole. SpyCas9 was eluted with a linear gradient of 450 mM imidazole and then dialyzed against 20 mM Tris-HCl, pH 7.5, 175 mM NaCl, 10% (w/v) glycerol, and 5 mM BME. The SpyCas9 fraction was loaded onto a 5 mL HiTrap SP column (GE Healthcare) pre-equilibrated with the dialysis buffer. SpyCas9 was eluted with a linear gradient of 1 M NaCl. The protein was further purified using a HiLoad 16/60 Superdex200 column (GE Healthcare) equilibrated with 20 mM Tris-HCl pH 8.0, 200 mM NaCl, 5% (w/v) glycerol, and 2 mM dithiothreitol.
Preparation of sgRNA
The DNA template for RNA transcription was prepared by using the Giga-prep kit (Epigentics). The sgRNA was prepared in vitro by mixing rNTPs (rATP, rGTP, rCTP, and cUTP), MgCl2, T7 RNA polymerase (P266L mutant)23, inorganic pyrophosphatase, and the DNA template in the transcription buffer. After 6 hour transcription at 37 °C, the synthesized RNA was precipitated by ethanol treatment overnight, purified using 12% denaturing PAGE (19:1 cross-linking ratio), and electro-eluted (Elutrap, Whatman). The purified RNA was desalted and exchanged into water using Amicon (Millipore).
NMR Spectroscopy
The NMR sample contained 2 mM 13C,15N-AcrIIA4 in 20 mM sodium phosphate, pH 7.4, 100 mM NaCl, 0.01% NaN3. NMR spectra were recorded at 25 °C on Bruker 600 MHz and 800 MHz spectrometers equipped with a z-shielded gradient triple resonance probe. Sequential and side chain assignments of 1H, 15N and 13C resonances were achieved by three-dimensional triple resonance through-bond scalar correlation experiments CBCA(CO)NH, HNCACB, HNCO, HN(CA)CO, HBHA(CO)NH, HCCH-TOCSY, and 15N-TOCSY-HSQC. 3D 13C-separated NOESY and 15N-separated NOESY experiments were obtained using a mixing time of 120 ms. The acquisition parameters for the 3D experiments are described in detail as Supplementary Information. The χ1 angle restraints were obtained based on the quantification of intraresidue NOEs between backbone amide proton, α proton, and β methylene protons24. Residual 1DNH dipolar couplings were obtained by taking the difference in the J splitting values measured in oriented (10 mg/ml of pf1 phage alignment media) and isotropic (water) AcrIIA4 using 2D in–phase/antiphase 1H‒15N HSQC spectra25. 15N-R1 and 15N-R1ρ relaxation rates, and heteronuclear 1H‒15N NOE were measured using pulse schemes described previously26,27 on the Bruker 800 MHz spectrometer. Delays of 10, 50, 100, 400, 800, 1200, 1500 ms were used for the R1 relaxation measurement, and 0, 4, 12, 24, 40, 60, 80, 96 ms used for R1ρ experiment. 15N-R2 relaxation rates were obtained from the 15N-R1 and 15N-R1ρ relaxation rates using the equation, R2 = (R1ρ − R1 sin2 θ)/cos2 θ, where θ = arctan(ΩN/γNB1), ΩN is the resonance offset, and γNB1 is the spin-lock field strength28. For the heteronuclear 1H‒15N NOE measurement, 1H saturation was achieved using 3 s of 120° 1H pulses separated by 5 ms intervals. 2 s of relaxation delays were used for 15N-R1 and 15N-R1ρ relaxation measurements, and 4 s of the relaxation delay was used for the heteronuclear 1H‒15N NOE measurement. NMR spectra were processed using the NMRPipe29, and analyzed using PIPP30, and NMRView31 programs.
Structure calculation
Interproton distance restraints were derived from the NOE spectra and classified into distance ranges according to the peak intensity. ϕ/ψ torsion angle restraints were derived from backbone chemical shifts using the program TALOS+32. The solution structure of AcrIIA4 was calculated by simulated annealing in torsion angle space using the Xplor-NIH program13. The target function of simulated annealing included a covalent geometry, a quadratic van der Waals repulsion potential33, square-well potentials for interproton distance and torsion angle restraints, hydrogen bonding, RDC restraints34, harmonic potentials for 13Cα/13Cβ chemical shift restraints35, a multidimensional torsion angle database potential of mean force36, and a radius of gyration term37. The radius of gyration represents a weak overall packing potential, and residues 1‒86 of AcrIIA4 were selected for the term. Structures were displayed using VMD-XPLOR software38. The violations from the structure calculation are described in detail as Supplementary Information.
Isothermal Titration Calorimetry
The ITC experiment was performed at 25 °C using an iTC200 calorimeter (Malvern). To characterize the affinity of AcrIIA4 to SpyCas9, 50 µM SpyCas9 in the cell was titrated with 500 µM of the AcrIIA4 in 20 mM sodium phosphate, pH 7.4, 100 mM NaCl, 5 mM BME, and 0.01% NaN3. Twenty consecutive 2 μL aliquots of AcrIIA4 were titrated into the cell and the duration of each injection was 4 sec, with injections occurring at intervals of 150 sec. The heats associated with dilution of the substrates were subtracted from the measured heats of binding. ITC titration data were analyzed using the Origin version 7.0 program provided with the instrument.
Data availability
Atomic coordinates and NMR restraints of the reported solution structure have been deposited in the Protein Data Bank under accession code 5XN4 and in the Biological Magnetic Resonance Bank under accession code 36085, respectively.
Electronic supplementary material
Acknowledgements
This work was supported by the Cooperative Research Program for Agriculture Science & Technology Development, Rural Development Administration (PJ011112), and the Creative-Pioneering Researchers Program through Seoul National University (500-20170161). We thank the Research Institute of Agriculture and Life Sciences, the high-field NMR facility at the Korea Institute of Science and Technology, the Korea Basic Science Institute, and the National Center for Inter-University Research Facilities. We thank Ms. So Young An and Ms. Ji Yeon Shin for the help with sgRNA preparation.
Author Contributions
I.K. and J.Y.S. designed the study; I.K., D.K., M.H., N.K.K., and J.Y.S. performed the experiments; I.K., M.J., E.B., and J.Y.S. analyzed the data; I.K., M.J., E.B., and J.Y.S. wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-22177-0.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koonin EV, Makarava KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang W, Marraffini LA. CRISPR-Cas: new tools for genetic manipulations from bacterial immunity systems. Annu. Rev. Microbiol. 2015;69:209–228. doi: 10.1146/annurev-micro-091014-104441. [DOI] [PubMed] [Google Scholar]
- 5.Sternberg SH, Doudna JA. Expanding the biologist’s toolkit with CRISPR-Cas9. Mol. Cell. 2015;568:568–574. doi: 10.1016/j.molcel.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 6.Wright AV, Nunez JK, Doudna JA. Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 7.Samson JE, Magadán AH, Sabri M, Moineau S. Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 2013;11:675–687. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- 8.Pawluk A, et al. Naturally occurring off-switches for CRISPR-Cas9. Cell. 2016;167:1829–1838. doi: 10.1016/j.cell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rauch BJ, et al. Inhibition of CRISPR-Cas9 with bacteriophage proteins. Cell. 2017;168:150–158. doi: 10.1016/j.cell.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong D, et al. Structural basis of CRISPR–SpyCas9 inhibition by an anti-CRISPR protein. Nature. 2017;546:436–439. doi: 10.1038/nature22377. [DOI] [PubMed] [Google Scholar]
- 11.Yang H, Patel DJ. Inhibition mechanism of an anti-CRISPR suppressor AcrIIA4 targeting SpyCas9. Mol. Cell. 2017;67:117–127. doi: 10.1016/j.molcel.2017.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin J, et al. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci. Adv. 2017;3:e1701620. doi: 10.1126/sciadv.1701620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwieters CD, Kuszewski J, Clore GM. Using Xplor–NIH for NMR molecular structure determination. Prog. Nucl. Magn. Reson. Spectrosc. 2006;48:47–62. doi: 10.1016/j.pnmrs.2005.10.001. [DOI] [Google Scholar]
- 14.Chowdhury S, et al. Structure reveals mechanisms of viral suppressors that intercept a CRISPR RNA-guided surveillance complex. Cell. 2017;169:47–57. doi: 10.1016/j.cell.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalodimos CG, et al. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 16.Lee GM, et al. The structural and dynamic basis of Ets-1 DNA binding autoinhibition. J. Biol. Chem. 2005;280:7088–7099. doi: 10.1074/jbc.M410722200. [DOI] [PubMed] [Google Scholar]
- 17.Bhabha G, et al. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science. 2011;332:234–238. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovermann M, Rogne P, Wolf-Watz M. Protein dynamics and function from solution state NMR spectroscopy. Q. Rev. Biophys. 2016;49:e6. doi: 10.1017/S0033583516000019. [DOI] [PubMed] [Google Scholar]
- 19.Matsuo H, et al. Identification by NMR spectroscopy of residues at contact surfaces in large, slowly exchanging macromolecular complexes. J. Am. Chem. Soc. 1999;121:9903–9904. doi: 10.1021/ja991881g. [DOI] [Google Scholar]
- 20.Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. A Cas9–guide RNA complex preorganized for target DNA recognition. Science. 2015;348:1477–1482. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- 21.Jinek M, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guillerez J, Lopez PJ, Proux F, Launay H, Dreyfus M. A mutation in T7 RNA polymerase that facilitates promoter clearance. Proc. Natl. Acad. Sci. USA. 2005;102:5958–5963. doi: 10.1073/pnas.0407141102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner G, et al. Protein structures in solution by nuclear magnetic resonance and distance geometry. The polypeptide fold of the basic pancreatic trypsin inhibitor determined using two different algorithms, DISGEO and DISMAN. J. Mol. Biol. 1987;196:611–639. doi: 10.1016/0022-2836(87)90037-4. [DOI] [PubMed] [Google Scholar]
- 25.Bax A, Kontaxis G, Tjandra N. Dipolar couplings in macromolecular structure determination. Methods. Enzymol. 2001;339:127–174. doi: 10.1016/S0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]
- 26.Farrow NA, Zhang OW, Forman-Kay JD, Kay LE. A heteronuclear correlation experiments for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J. Biomol. NMR. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 27.Tjandra N, Wingfield P, Stahl S, Bax A. Anisotropic rotational diffusion of perdeuterated HIV protease from 15N NMR relaxation measurements at two magnetic fields. J. Biomol. NMR. 1996;8:273–284. doi: 10.1007/BF00410326. [DOI] [PubMed] [Google Scholar]
- 28.Davis DG, Perlman ME, London RE. Direct measurements of the dissociation-rate constant for inhibitor-enzyme complexes via the T1 rho and T2 (CPMG)methods. J. Magn. Reson. Ser. B. 1994;104:266–275. doi: 10.1006/jmrb.1994.1084. [DOI] [PubMed] [Google Scholar]
- 29.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 30.Garrett DS, Powers R, Gronenborn AM, Clore GM. A common sense approach to peak picking in two-, three-, and four-dimensional spectra using automatic computer analysis of contour diagrams. J. Magn. Reson. 1991;95:214–220. doi: 10.1016/j.jmr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Johnson BA, Blevins RA. NMRView: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 32.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nilges M, Gronenborn AM, Brunger AT, Clore GM. Determination of three-dimensional structures of proteins by simulated annealing with interproton distance restraints. Application to crambin, potato carboxypeptidase inhibitor and barley serine proteinase inhibitor 2. Protein Eng. 1988;2:27–38. doi: 10.1093/protein/2.1.27. [DOI] [PubMed] [Google Scholar]
- 34.Clore GM, Gronenborn AM, Tjandra N. Direct refinement against residual dipolar couplings in the presence of rhombicity of unknown magnitude. J. Magn. Reson. 1998;131:159–162. doi: 10.1006/jmre.1997.1345. [DOI] [PubMed] [Google Scholar]
- 35.Kuszewski J, Gronenborn AM, Clore GM. The impact of direct refinement against 13Cα and 13Cβ chemical shifts on protein structure determination by NMR. J. Magn. Reson. Ser. B. 1995;106:92–96. doi: 10.1006/jmrb.1995.1017. [DOI] [PubMed] [Google Scholar]
- 36.Clore GM, et al. The three-dimensional structure of α1-purothionin in solution: combined use of nuclear magnetic resonance, distance geometry and restrained molecular dynamics. EMBO J. 1986;5:2729–2735. doi: 10.1002/j.1460-2075.1986.tb04557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuszewski J, Gronenborn AM, Clore GM. Improving the packing and accuracy of NMR structures with a pseudopotential for the radius of gyration. J. Am. Chem. Soc. 1999;121:2337–2338. doi: 10.1021/ja9843730. [DOI] [Google Scholar]
- 38.Schwieters CD, Clore GM. The VMD-XPLOR visualization package for NMR structure refinement. J. Magn. Reson. 2001;149:239–244. doi: 10.1006/jmre.2001.2300. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and NMR restraints of the reported solution structure have been deposited in the Protein Data Bank under accession code 5XN4 and in the Biological Magnetic Resonance Bank under accession code 36085, respectively.