Abstract
Diet1 modulates intestinal production of the hormone, fibroblast growth factor (FGF)15, which signals in liver to regulate bile acid synthesis. C57BL/6ByJ mice with a spontaneous Diet1-null mutation are resistant to hypercholesterolemia compared with wild-type C57BL/6J mice through enhanced cholesterol conversion to bile acids. To further characterize the role of Diet1 in metabolism, we generated Diet1−/− mice on the C57BL/6J genetic background. C57BL/6J Diet1−/− mice had elevated bile acid levels, reduced Fgf15 expression, and increased gastrointestinal motility and intestinal luminal water content, which are symptoms of bile acid diarrhea (BAD) in humans. Natural genetic variation in Diet1 mRNA expression levels across 76 inbred mouse strains correlated positively with Ffg15 mRNA and negatively with serum bile acid levels. This led us to investigate the role of DIET1 genetic variation in primary BAD patients. We identified a DIET1 coding variant (rs12256835) that had skewed prevalence between BAD cases and controls. This variant causes an H1721Q amino acid substitution that increases the levels of FGF19 protein secreted from cultured cells. We propose that genetic variation in DIET1 may be a determinant of FGF19 secretion levels, and may affect bile acid metabolism in both physiological and pathological conditions.
Keywords: intestine, fibroblast growth factor 15/19, Hybrid Mouse Diversity Panel
Cholesterol is eliminated from the body through the catabolism of cholesterol to bile acids, and subsequent loss of bile acids in the feces (1–3). The elimination of bile acids through the intestine is the basis for the reduction of serum cholesterol levels through the use of bile acid binding resins, which sequester bile acids in the intestine to form an insoluble complex (4–6). More recently, we identified a mutant mouse model that exhibits constitutive conversion of cholesterol to bile acids and elimination through feces and urine, leading to resistance to dietary cholesterol (7, 8). The underlying mutation occurs in the Diet1/Malrd1 gene, which encodes a 236 kDa protein that is expressed exclusively in epithelial cells lining the small intestinal villi and kidney proximal tubules (7). Because Diet1 is the original gene name and all studies published thus far use Diet1, we will continue to use this designation here. Diet1 influences enterohepatic signaling between the small intestine and liver to modulate hepatic bile acid synthesis. Specifically, Diet1 deficiency leads to reduced production and/or secretion by enterocytes of the hormone, fibroblast growth factor (FGF)15 (FGF19 in humans) (7). FGF15/19 is secreted by the ileum into the enterohepatic circulation, and downregulates bile acid synthesis by signaling through hepatic coreceptors (FGF receptor 4 and β klotho) (2, 9–11).
The augmentation of flux through the bile acid synthetic pathway has been successfully used therapeutically to control plasma cholesterol levels (2, 12, 13). However, the amount of bile acids lost through excretion must be carefully regulated, as increased bile acid levels in the intestine can trigger the flow of water into the colon, resulting in bile acid malabsorption, also known as bile acid diarrhea (BAD) (2, 14). One of the mechanisms by which elevated intestinal bile acids may influence the severity of BAD is by increasing colonic motility (15, 16). BAD may be classified into three types based on distinct etiologies (15). Type 1 BAD was first recognized in association with surgical resection or disease of the terminal ileum, which leads to impaired reabsorption of bile acids from the intestinal lumen (17). This results in elevated fecal bile acid levels and is treated with bile acid sequestrants (18). Type 2 BAD (referred to herein as “primary BAD”) was recognized in 1976 as a similar syndrome without obvious ileal disease (19). Primary BAD appears to be heterogeneous in etiology (20–22). Type 3 BAD encompasses chronic diarrheas caused by other conditions that can affect bile acid absorption, such as cholecystectomy or bacterial overgrowth in the small intestine (23).
Primary BAD is the most common of the bile acid malabsorption syndromes, estimated to affect about 1% of the European population, and accounting for about 30% of cases with chronic diarrhea and diarrhea-predominant irritable bowel syndrome (24). The definitive test for primary BAD is the measurement of fecal bile acid levels, but this is demanding and not usually performed clinically. Alternatively, BAD may be defined by administering a radiolabeled bile acid [selenium homocholic acid taurine (SeHCAT)] and determining the amount retained in the body after 7 days (25, 26). Patients with primary BAD eliminate the radiolabel more rapidly than controls, as determined by scanning with a γ camera (25). This test is available in Europe, but not in the United States. An additional screening method for BAD is the quantification of the bile acid synthetic intermediate, C4 (7α-hydroxy-cholest-4-ene-3-one), which is inversely correlated with SeHCAT retention (27).
A relationship between BAD and FGF19 levels has been identified by Walters and colleagues (28). BAD patients were diagnosed by reduced SeHCAT retention in the absence of bile acid sequestrants. In BAD patients, FGF19 levels were lower than normal and there was a positive correlation between FGF19 and percent SeHCAT retention (28). There was also an inverse relationship between FGF19 and C4 levels (16, 29).
Animal studies provide independent support for a key role of FGF15/19 in bile acid homeostasis. In monkeys, administration of anti-FGF19 antibodies produced severe bile acid loss and diarrhea (30). Furthermore, mice carrying a knockout mutation for FGF15 exhibited excess fecal bile acid secretion, as observed in humans (31). As described earlier, Diet1-deficient mice exhibit very low levels of FGF15 in the small intestine and constitutive upregulation of hepatic bile acid synthesis (7). Providing FGF15 to Diet1-deficient mice led to normalization of bile acid synthetic gene expression, demonstrating that Diet1 functions upstream of FGF15 to modulate bile acid levels (7).
In the current study, we characterize the effects of mouse Diet1-null mutation on an isogenic C57BL/6J background and find that these mice experience traits reminiscent of BAD in humans. We further assess whether natural genetic variation in Diet1 expression across more than six dozen inbred mouse strains is correlated with Fgf15 expression and bile acid traits. We expand our investigation to evaluate the presence of functional DIET1 variants in humans, and characterize a nonsynonymous coding variant that has a significantly different prevalence in control and BAD subjects, and which influences FGF19 levels secreted from cultured cells. These studies expand our knowledge of the role of variation in Diet1 levels and the sequence in traits associated with bile acid metabolism.
MATERIALS AND METHODS
Diet1−/− congenic mice
C57BL/6J and C57BL/6ByJ mice were obtained from the Jackson Laboratory (Bar Harbor, ME). Diet1−/− congenic mice were generated by backcrossing the Diet1 allele from C57BL/6ByJ mice onto the C57BL/6J background for eight generations. The Diet1 allele was genotyped using PCR primers specific for the mutant and wild-type alleles, as described previously (7). Heterozygous mice from the N8 backcross generation were intercrossed to produce wild-type (Diet1+/+) and Diet1−/− mice. Mice were placed on an atherogenic diet (TD88051; Harlan Laboratories, Madison, WI), as previously described (7), for 3 or 6 weeks before tissues were collected. All mouse studies were conducted in accordance with and approved by the Institutional Animal Research Committee of the University of California, Los Angeles.
Mouse plasma lipid and bile acid measurements
Mice were fasted for 16 h and blood was collected from the retro-orbital sinus under isoflurane anesthesia. Enzymatic assays for total cholesterol and HDL cholesterol were performed as described (32). Total bile acid levels were assessed using Diazyme total bile acids assay kit (DZ042A-K; Diazyme Laboratories, Poway, CA).
Hybrid Mouse Diversity Panel analysis
The Hybrid Mouse Diversity Panel (HMDP) strains have been described previously (33). Briefly, 76 inbred and recombinant inbred mouse strains (three to five mice per strain) were assessed at 8 weeks for gene expression in the ileum for Diet1 and Fgf15. Biliary bile acid and cholesterol levels were determined by calorimetric assays from Diazyme and Thermo Fisher (A12216; Thermo Fisher Scientific, Grand Island, NY), respectively. Correlation analyses were performed in Prism (v7, GraphPad Software, La Jolla, CA).
Gene expression and quantitative PCR
Liver and small intestine were isolated from mice following a 16 h fast and RNA was prepared using Trizol (Invitrogen). Primer sequences for quantitative PCR of Diet1, Fgf15, Asbt, Cyp7a1, Cyp8b1, Cyp27, Fgfr4, and B2m (as a normalization gene) were reported previously (7). Rpl4 primer sequences were 5′gatgagctgtatggcacttgg3′ for forward primer and 5′cttgtgcatgggcaggtt3′ for reverse primer.
Determination of gastrointestinal transit time
Mice were fed an atherogenic diet for 2 weeks. After an overnight fast with ad libitum water, mice were gavaged with a semiliquid solution (0.2 ml) of 5% Evans blue and 1.5% methylcellulose (without additional food), as previously described (34). Mice were continuously monitored and the time to produce the initial blue fecal pellet was determined as a measure of gastrointestinal transit time.
Intestinal water content
Mice were fed an atherogenic diet for 2 weeks, then fasted for 18 h with ad libitum water, and refed for 3 h with atherogenic diet. The small intestine and colon were excised precisely and the contents collected by gentle extrusion. Contents were weighed, desiccated for 3 days, and then weighed again to determine the fluid and solid weights, as described previously (35–37). Weights of the water and dry intestinal contents were expressed as absolute amounts, and after normalization to intestinal lengths.
Study subjects
Genomic DNA was studied in an initial cohort of 44 subjects that has been described before (38) (Table 1). Twenty-two of the subjects had chronic diarrhea due to primary BAD, with SeHCAT 7 day retention between 1% and 7%. The control group of 22 subjects had normal bowel habits and no evidence of malabsorption. Both groups were of predominantly Caucasian origin with no subjects of African descent. The DNA samples were obtained under the approval of the local Research Ethics Committees (Institutional Review Boards) of Hammersmith, Queen Charlotte’s and Chelsea, Acton, and Wrightington, Wigan, and Leigh Hospitals. Informed consent was obtained from all subjects. The DNA was sequenced under a protocol approved by the University of California, Los Angeles Institutional Review Board.
TABLE 1.
Clinical characteristics of BAD case/control cohorts
| Group 1 | Group 2 | All Subjects | |
| Number | 44 | 78 | 122 |
| Age range (years) | 27–80 | 17–90 | 17–90 |
| Male:female | 13:31 | 34:43 | 47:74 |
| SeHCAT retention range (%) | 0.5–7.3a | 0.4–90 | 0.4–90 |
Non-BAD subjects were not assessed for percent SeHCAT retention.
Genomic DNA was also studied in an additional cohort of patients, who have also been previously described (28) (Table 1). This cohort consisted of a prospectively recruited set of 78 patients with chronic diarrhea who underwent SeHCAT testing and FGF19 measurements. This cohort was also predominantly Caucasian in origin.
DIET1 sequencing
The exons and flanking intron sequences of DIET1 were amplified by PCR using the oligonucleotide primer sequences listed in supplemental Table S1. PCR products were treated with recombinant exonuclease I and shrimp alkaline phosphatase (Agilent Technologies, Santa Clara, CA), and sequenced on a Biosystems 3730 capillary DNA analyzer using BigDye terminator cycle sequencing reagents. Variants were confirmed by amplification of an independent PCR product and sequencing both strands.
The rs12256835 genotyping
Genomic DNA was extracted from patient blood using QiAmp DNA mini kit (Qiagen, Manchester, UK) according to the manufacturer’s protocols. Samples were genotyped for the rs12256835 variant using a TaqMan allelic discrimination assay (Life Sciences, Paisley, UK). PCR was performed on 96-well reaction plates and analyzed using a StepOnePlus RT-PCR machine (Life Sciences) to obtain an allelic discrimination plot, including a negative water control. Reaction volume was 15 μl per well, including 7.5 μl TaqMan genotyping Master Mix (2×), 0.375 μl SNP assay mix (40×), 6.125 nuclease-free water, and 1 μl genomic DNA. Standard PCR conditions were: 95°C for 10 min, followed by 40 cycles of amplification at 95°C for 15 s, and 60°C for 1 min, followed by 60°C for 30 s.
Serum FGF19 measurements
Serum FGF19 levels were measured by commercial ELISA assay (FGF19 Quantikine ELISA; R&D Systems, Minneapolis, MN), as previously described. Aliquots (100 μl) of serum were assayed in duplicate according to the manufacturer’s protocols.
Expression constructs and site-directed mutagenesis
Expression plasmids for human Diet1-V5 and FGF19-Myc were generated previously (7). Site-directed mutagenesis was performed on the plasmid expressing Diet1-1721H to generate Diet1-1721Q using the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies) with primers gaagctcactgtgcacagtatacaagcacaacag (hDIET1_H1721Q-F) and ctgttgtgcttgtatactgtgcacagtgagcttc (hDIET1_H1721Q-R). Site-directed mutagenesis was performed on the plasmid expressing Diet1-1712D to generate Diet1-1721G using primers cataagccagattgctctggtaggtctgatgaagc (hDIET1_H1712G-F) and gcttcatcagacctaccagagcaatctggcttatag (hDIET1_H1712G-R).
FGF19 secretion assay
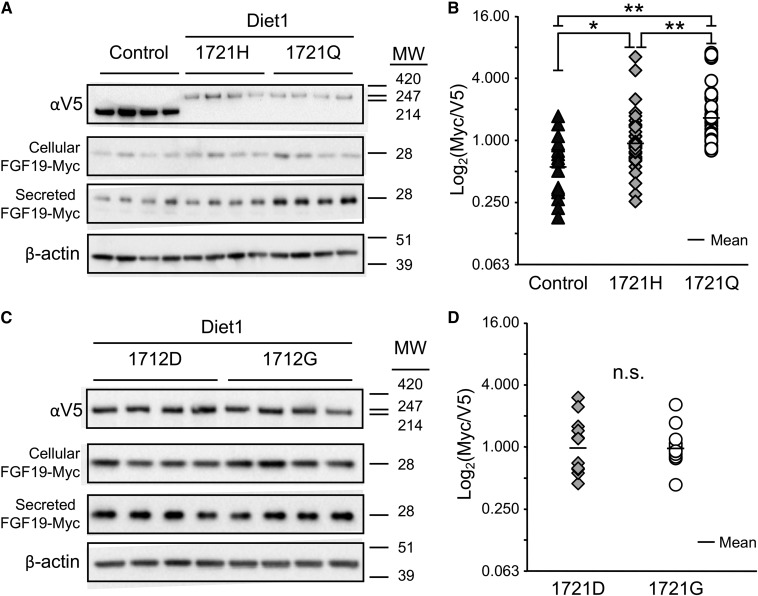
The effect of Diet1 on FGF19 secretion was assessed as previously described (7). Briefly, HEK293T cells (American Type Culture Collection, Manassas, VA) were transfected (in triplicate or quadruplicate) with pcDNA6 expression plasmids for FGF19-Myc in combination with a V5-tagged control gene construct (Kdm5c), Diet1-1721H, Diet1-1721Q, Diet1-1712D, or Diet1-1712G (BioT transfection reagent; Bioland, Paramount, CA). After 24 h, an equal amount of fresh medium was added to each well. The following day, culture medium was collected to quantify the levels of FGF19 secreted in 24 h, and cells were collected to quantify cellular FGF19 and Diet1 protein levels by Western blot. Diet1 was detected with rabbit anti-V5 (1:5,000; Bethyl Laboratories, Montgomery, TX); FGF19 was detected with rabbit anti-Myc (1:10,000; Bethyl Laboratories); and β-actin was detected with a mouse monoclonal antibody (1:10,000; Sigma, St. Louis, MO). After secondary antibody application, protein bands were detected with ECL2 chemiluminescent reagent (ThermoScientific, Rockford, IL) and the Bio-Rad ChemiDoc XRS+ system with a CCD camera and ImageLab software (Bio-Rad, Hercules, CA). The protein band intensity from the resulting image was quantified using the ImageLab software. The amount of FGF19 was normalized to Diet1 protein levels. Western blot images shown in Fig. 6A were adjusted and cropped in Adobe Photoshop.
Fig. 6.
DIET1 variant rs12256835 influences secretion of FGF19 in a cell-based assay. A: Representative results of an FGF19 secretion assay comparing Diet1-1721H and -1721Q. FGF19-Myc was expressed in HEK293T cells in combination with either Diet1-1721H-V5, Diet1-1721Q-V5, or an irrelevant control protein (Kdm5c-V5). Cellular Diet1 protein levels and secreted FGF19 levels were visualized by Western blot. MW, molecular mass in kD. B: Quantification of FGF19 secretion from nine experiments of the type shown in A. Each point is one independent transfection (done in triplicate or quadruplicate), representing the amount of FGF19 secreted per unit of cellular V5 expression for control protein (Kdm5c), Diet1-1721H, or Diet1-1721Q. The overall P-value for all three conditions is P = 3.70 × 10−7 (one-way ANOVA). Individual comparisons were made using Tukey’s HSD post hoc test on log2-normalized values. Small horizontal lines represent geometric mean of samples for each protein isoform (*P < 0.05, **P < 0.01). C: Representative results of an FGF19 secretion assay comparing Diet1-1721D and -1721G. Details are as in A. D: Quantification of FGF19 secretion from five experiments of the type shown in C. No statistically significant difference (n.s.) in the amount of extracellular FGF19 was detected between Diet1-1712D and -1712G (P = 0.945; one-way ANOVA).
Diet1/FGF19 coimmunoprecipitation
Protein coimmunoprecipitation was performed as previously described (7). HEK293T cells were transfected as described for the FGF19 secretion assay above. After 48 h, cells were lysed in PBS containing 1% NP-40 (Sigma), 1× Complete Mini protease inhibitor cocktail (Roche, Indianapolis, IN), and 0.1% phosphatase inhibitors 2 and 3 (Sigma). Cell lysates were incubated overnight at 4°C with 1 μg of mouse anti-V5 antibody (Life Technologies, Grand Island, NY), 4 μg of mouse anti-Myc antibody (Millipore, Billerica, MA), or 4 μg nonspecific mouse IgG (Jackson ImmunoResearch, West Grove, PA). Protein A/G PLUS agarose beads (Santa Cruz Biotechnologies, Paso Robles, CA) were added for 3 h at 4°C. After three washes with 1× PBS + 0.1% NP-40, protein was eluted from the beads with 1× loading buffer and 1% β-mercaptoethanol, boiled for 5 min, and analyzed by Western blot, as described in the preceding section.
Statistical analysis
Fisher’s exact tests and one-way ANOVA were performed using VasserStats (http://www.vassarstats.net/). Two-way ANOVA was performed using NCSS or GraphPad Prism. Linear regressions were performed with WinStat for Excel. Pair-wise comparisons were made using Student’s t-test.
RESULTS
Generation of Diet1−/− C57BL/6J mice
We previously identified the Diet1 gene in the C57BL/6ByJ mouse strain, which carries a naturally occurring mutation leading to a null allele for Diet1 (7). Because the C57BL/6ByJ mouse strain is >99% genetically identical to C57BL/6J mice, the latter strain was previously used as the wild-type control in studies of Diet1 (7, 8, 39, 40). However, to ensure that all effects of Diet1 deficiency could be attributed to the Diet1-null mutation (and not influenced by C57BL/6ByJ and C57BL/6J variation at loci other than Diet1), we generated a congenic strain by breeding the Diet1-null allele onto the C57BL/6J background for eight generations. As expected, the resulting C57BL/6J Diet1−/− mice had undetectable Diet1 mRNA expression in the ileum (Fig. 1A).
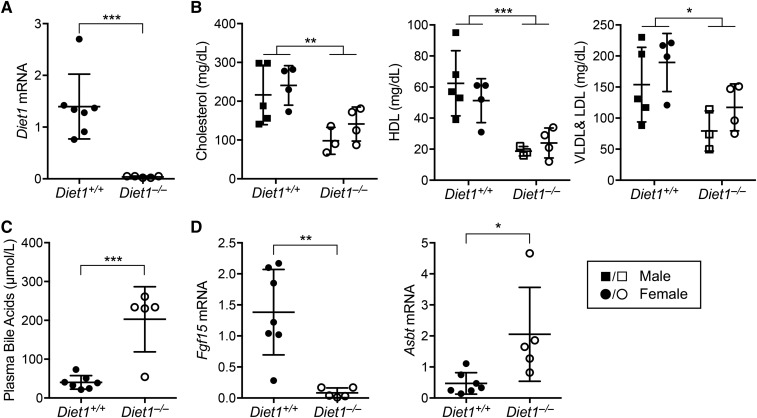
Fig. 1.
Altered bile acid, cholesterol, and ileal gene expression levels in C57BL/6J Diet1−/− mice. A: Confirmation of Diet1 deficiency in C57BL/6J Diet1−/− mouse ileum. B: Reduced plasma total, HDL, and VLDL cholesterol levels in C57BL/6J Diet1−/− mice compared with isogenic Diet1+/+ mice fed an atherogenic diet for 3 weeks (**P < 0.01 and ***P < 0.001 on two-way ANOVA). C: Elevated plasma bile acid levels in C57BL/6J Diet1−/− mice fed an atherogenic diet for 3 weeks. D: Reduced ileal Fgf15 and elevated Asbt expression in C57BL/6J Diet1−/− mice fed an atherogenic diet 3 weeks. For all panels, n = 5–7 mice per genotype, as indicated. For panels A, C, and D, statistical analyses via Student’s t-test; *P < 0.05, **P < 0.01, ***P < 0.001.
We assessed the C57BL/6J Diet1−/− mice for traits originally observed in the C57BL/6ByJ mice (7) and found that the effects on bile acid metabolism were recapitulated. Data shown are for mice fed a short-term (3 week) atherogenic diet, which enhanced the phenotypic differences between wild-type and Diet1-deficient mice (8). As observed previously for C57BL/6ByJ mice (7, 8, 39), C57BL/6J Diet1−/− mice exhibited approximately 2-fold lower plasma cholesterol and lipoprotein levels and >4-fold higher plasma bile acid levels than Diet1+/+ mice (Fig. 1B, C). Intestinal gene expression was altered in the congenic strain similarly to C57BL/6ByJ mice, with dramatically reduced Fgf15 mRNA and increased Asbt mRNA (Fig. 1D). Diet1 deficiency in C57BL/6ByJ mice did not influence hepatic Fgfr4 gene expression and increased the expression of cholesterol synthetic genes (Cyp27, Cyp7a1, and Cyp8b1) (8). C57BL/6J Diet1−/− mice similarly exhibited wild-type Fgfr4 and elevated Cyp27 levels, but effects on Cyp7a1 and Cyp8b1 exhibited biological variability (supplemental Fig. S1A). Thus, the plasma and intestinal phenotypes originally observed in C57BL/6ByJ mice were present in Diet1-deficient C57BL/6J mice.
Diet1 deficiency causes intestinal diarrhea in the mouse
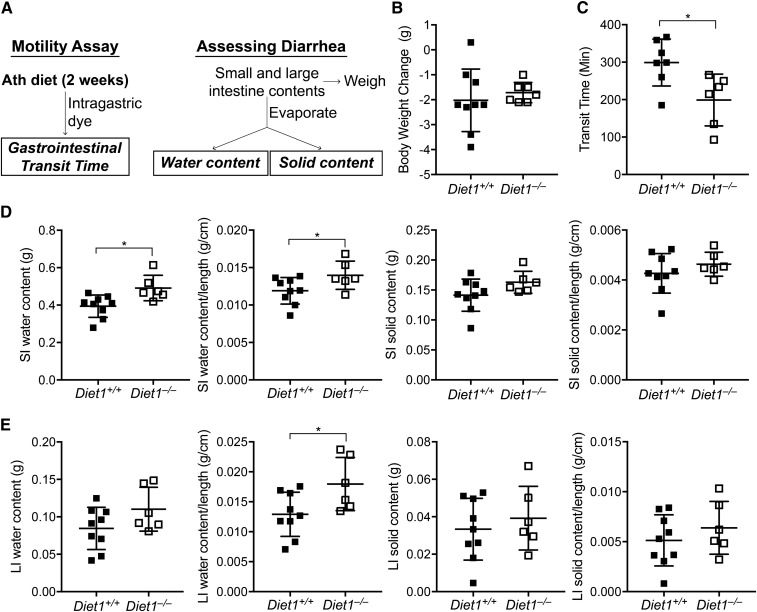
We hypothesized that Diet1 is required for normal gastrointestinal function, and assessed intestinal motility and the presence of diarrhea (Fig. 2A). Diet1−/− mice exhibited normal food and water intake and body weight while eating an atherogenic diet (Fig. 2B; supplemental Fig. S1B). However, Diet1−/− mice had reduced transit time of an intragastric bolus through the intestinal tract (Fig. 2C), indicating increased intestinal motility (16, 21, 30). The increase in intestinal motility in conjunction with increased bile acid levels in feces (8) raised the possibility that these animals may experience BAD. We assessed the intestinal content of water and solids after acute feeding of the atherogenic diet for 3 h (see the scheme in Fig. 2A). Small and large intestine were collected and intestinal contents extruded and weighed. Following evaporation, the weight difference in intestinal contents was used to calculate the content of water and solids. Diet1−/− mice had increased water content of both small and large intestine per unit of intestinal length, indicating diarrhea (Fig. 2D, E). Increased water content was also observed in the small intestine of Diet1−/− mice when determined in fasted mice (supplemental Fig. S1C). By contrast, the intestinal solids content was not affected by Diet1 (Fig. 2D, E), suggesting a secretory diarrhea in Diet1−/− mice. The elevated bile acid levels, increased gastrointestinal motility, and increased intestinal water content in the absence of alterations in dry content suggested parallels between Diet1−/− mice and patients with BAD.
Fig. 2.
Diet1−/− mice exhibit reduced gastrointestinal transit time and diarrhea. A: Study design to measure gastrointestinal transit time and diarrhea. Mice were fed an atherogenic diet for 2 weeks and gastrointestinal transit time was measured after gastric administration of a bolus containing dye. The time to first fecal pellet containing dye was determined. Diarrhea was assessed in fed animals by quantifying small and large intestine water and solid contents (described further in the Materials and Methods). B: Diet1−/− and Diet1+/+ mice have similar body weight after 2 weeks on atherogenic diet. C: Diet1−/− mice exhibit reduced total gastrointestinal transit time, indicating increased intestinal motility. D: Small intestinal (SI) water content is increased in Diet1−/− mice when expressed as absolute amount and when normalized to intestinal length. There were no differences in SI solids content between genotypes. E: Large intestine (LI) water content normalized to length is increased in Diet1−/− mice. There were no differences in LI solids content between genotypes. Statistical analyses via Student’s t-test; *P < 0.05; n = 6–9.
Genetic variation in mouse Diet1 expression positively correlates with Fgf15 mRNA levels and negatively correlates with biliary bile acid and cholesterol levels
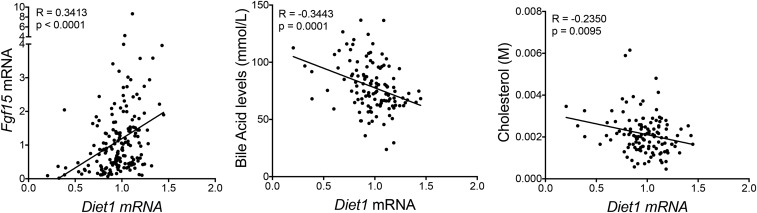
Our data, thus far, indicated that complete deficiency of Diet1 influences Fgf15 mRNA and plasma bile acid and cholesterol levels (Fig. 1) (7, 8). We wondered whether more modest naturally occurring variations in Diet1 expression levels across different genetic backgrounds would also correlate with these traits. To assess this, we analyzed Diet1 and Fgf15 mRNA levels in ileum, biliary cholesterol levels, and bile acid levels across 76 inbred mouse strains, which comprise the HDMP (33). We observed a positive correlation between Diet1 and Fgf15 expression levels and a negative correlation between Diet1 mRNA and both bile acid and cholesterol levels in gallbladder (Fig. 3). These results strengthen the relationship between Diet1 expression levels and these parameters, and indicate that small variations in Diet1 expression levels are associated with differences in Fgf15 mRNA, bile acid, and cholesterol levels.
Fig. 3.
Diet1 mRNA levels correlate positively with Fgf15 mRNA, and negatively with biliary bile acid and cholesterol levels across 76 mouse strains. Diet1 mRNA levels in ileum vary across 76 inbred mouse strains, and correlate positively with Fgf15 mRNA levels. Diet1 mRNA levels correlate negatively with bile acid and cholesterol levels in bile collected from the gallbladder. Each point on the graphs represents a single mouse strain (three to five mice per strain) from the HDMP (see the Materials and Methods). The mice were fed a standard chow diet.
Sequence variation in DIET1
As a recently annotated gene, DIET1 probes were not available in existing exome sequencing platforms when we began our study, and DIET1 has not been resequenced in any disease cohort. To evaluate the potential role of rare or common DIET1 variants in primary BAD, we sequenced the complete DIET1 coding region and exon/intron junctions in a case-control cohort (22 cases, 22 controls). The basic characteristics of the cohort are summarized in Table 1. Primary BAD cases were defined as individuals with very low SeHCAT retention levels (<7.3%), and controls were randomly selected from patients with normal bowel habits (38).
We observed a total of 34 variants distributed along the length of DIET1 (Fig. 4). Of these, 25 produce nonsynonymous amino acid substitutions, including two novel single nucleotide variants (SNVs) observed in single individuals from the Case group, one novel SNV observed in a single individual from the Control group, and a frame-shift mutation at position 1221 detected in a single control subject (supplemental Fig. S2; supplemental Tables S2, S3). Because these variants occurred at low frequency, there was no statistical evidence of an enrichment of rare variants in the primary BAD cases, and we did not pursue these variants further.
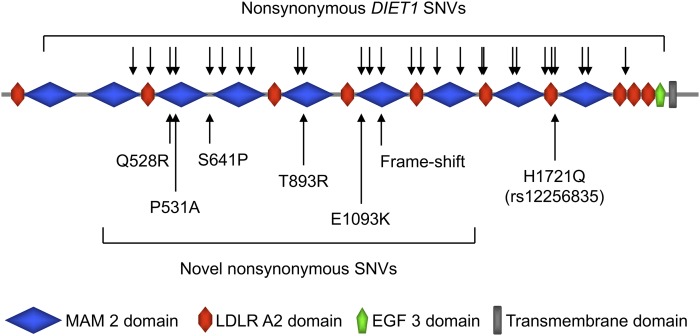
Fig. 4.
Location of nonsynonymous variants in human Diet1 protein. Schematic of Diet1 protein domain structure. Arrows above the diagram indicate the 25 nonsynonymous variants and one single-nucleotide deletion detected in cases and controls. Arrows below the diagram indicate the novel nonsynonymous variants, the frame-shift mutation, and the position of rs12256835.
A common DIET1 variant with skewed frequency distribution between primary BAD case and control subjects
Among the nonsynonymous variants identified in DIET1, one previously recorded common variant [rs12256835, 5163 T>G; minor allele frequency (MAF) 0.125] was present at statistically different levels in BAD cases and controls (Fisher’s exact test: nominal P = 0.0005; adjusted for multiple testing, P = 0.0175) (supplemental Table S3). The rs12256835 major allele encodes a histidine residue at position 1721 of the Diet1 protein (Fig. 4), and the minor allele encodes a glutamine residue (H1721Q). The minor (Q) allele was detected exclusively in control samples (11 of 44 alleles). This polymorphism has a frequency of 0.3181 in dbSNP (build 150), suggesting that it is a common source of structural variation in the Diet1 protein in the general population. Based on existing database information from dbSNP, the allele frequency for rs12256835 is similar in European and East Asian populations (MAF 0.17–0.25, respectively), but deviates substantially in African populations (MAF 0.63). However, individuals of African descent were not present in our study sample, and cannot account for the difference in rs12256835 allele frequency that we observed between BAD cases and controls.
We investigated the association of the rs12256835 genotype with BAD in an additional 78 individuals, for an extended sample of 122 individuals (see Table 1 for the characteristics of the initial group, the extended group, and the total cohort). The individuals with primary BAD (defined as SeHCAT ≤10%) were more likely to have the T allele than the G allele (Fisher’s exact test: P = 0.035; Table 2). The reduced frequency of the minor allele (G) in those with primary BAD in the extended sample group was consistent with our observations in the initial cohort.
TABLE 2.
Allele frequencies for rs12256835 in all subjects
| T Allele | G Allele | P | Odds Ratio (95% CI) | |
| SeHCAT ≤10% | 83 | 13 | 0.035 | 0.470 (0.235, 0.940) |
| SeHCAT >10% | 111 | 37 |
The number of T and G alleles genotyped in 122 subjects are shown, segregated as BAD cases (SeHCAT ≤10%) and controls (SeHCAT >10%). The third column lists the P-value of association as calculated by Fisher’s exact test, and the odds ratio (with 95% CIs indicated) is given in the final column.
Walters and colleagues previously established a positive relationship between percent SeHCAT retention and FGF19 levels (28). To extend our analysis of the rs12256835 genotype association with BAD, we determined fasting FGF19 levels in the group of 78 patients described above. The cohort was separated into BAD cases (SeHCAT ≤10%) and controls (SeHCAT >10%) and classified according to the genotype at rs12256835 (TT vs. TG and G/G). FGF19 values were significantly different between BAD cases and controls (P = 0.0088; two-way ANOVA), and there was a significant interaction between rs12256835 genotype and BAD case/control status (P = 0.016; Table 3). Post hoc Student’s t-tests showed that, in BAD cases, FGF19 levels were significantly lower in those with the T/T genotype compared with those with one or more G alleles (P = 0.0488, Table 3).
TABLE 3.
The rs12256835 genotype and FGF19 levels
| FGF19 (pg/ml) | |||
| T/T | T/G and G/G | ||
| SeHCAT ≤10% | Mean ± SEM | 119 ± 19a | 253 ± 56 |
| Number | 14 | 12 | |
| SeHCAT >10% | Mean ± SEM | 246 ± 24 | 215 ± 28 |
| Number | 30 | 22 | |
Fasting FGF19 levels for a cohort of 78 patients are shown, segregated as BAD cases (SeHCAT ≤10%) and controls (SeHCAT >10%) and classified according to the genotype at rs12256835. FGF19 values were log-normalized before statistical analysis. FGF19 values were significantly different between BAD cases and controls [P = 0.0088 (two-way ANOVA)], and there was a significant interaction between rs12256835 genotype and BAD case/control status (P = 0.016).
Post hoc Student’s t-tests confirmed lower FGF19 levels in those with primary BAD carrying the T/T genotype (P < 0.05).
The Diet1 H1721Q variant influences FGF19 production
The data described above suggested that the Diet1 protein variants encoded by the two alleles at rs12256835 may have functional differences. The Diet1 protein has a predicted modular structure, with repeating interspersed LDL receptor and meprin/A5-protein/PTPmu (MAM) domains (7). The H1721Q amino acid change occurs at the transition between the seventh LDL receptor domain and the adjacent MAM domain (Fig. 4). We previously demonstrated that Diet1 influences bile acid levels by modulating FGF15 or FGF19 production in mouse intestine and in a human intestinal cell line, respectively (7). We also observed partial colocalization of Diet1 and FGF19 in cultured enterocytes, and demonstrated that the proteins coimmunoprecipitate (7). We therefore assessed whether the Diet1 H and Q variants differ in the ability to bind FGF19 and/or to promote FGF19 secretion.
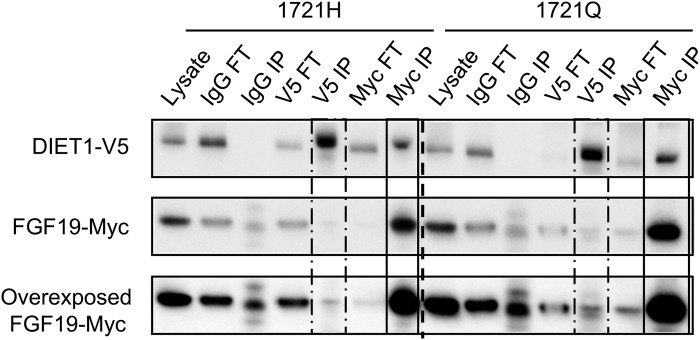
To assess whether the H-to-Q substitution alters Diet1/FGF19 protein interaction, we coexpressed epitope-tagged Diet1 and FGF19 in cultured cells, immunoprecipitated the protein complex, and quantified the amount of each Diet1 isoform pulled down by FGF19. Expression of the 1721H and 1721Q Diet1 variants at similar levels (see the “Lysate” lanes in Fig. 5) led to the pull-down of a similar proportion of Diet1 by immunoprecipitation of FGF19 (black boxes in Fig. 5). Thus, the two Diet1 isoforms interacted with FGF19 similarly.
Fig. 5.
Diet1-1721H and -1721Q isoforms do not differentially affect Diet1-FGF19 interaction. Coimmunoprecipitation of Diet1-1721H and Diet1-1721Q with FGF19. FGF19-Myc was coexpressed in HEK293T cells with Diet1-1721H-V5 or Diet1-1721Q-V5. Input lanes show similar levels of expression of the two Diet1 isoforms. Bands corresponding to precipitation of FGF19 with Diet1-1721H are on the left of the dashed line, and bands corresponding to precipitation of FGF19 with Diet1-1721Q are on the right of the dashed line. Dashed boxes indicate immunoprecipitation of Diet1 (V5 IP); solid boxes indicate immunoprecipitation of FGF19 (Myc IP). Both Diet1 isoforms interact with FGF19 with similar efficiency. A representative blot from eight experiments is shown.
To assess the ability of the two Diet1 protein variants to promote FGF19 protein secretion, we coexpressed V5-tagged Diet1-1721H or -1721Q with Myc-tagged FGF19 in HEK293T cells (Fig. 6A, B). Cells were collected 48 h after transfection to quantify intracellular FGF19, and medium was collected during the final 24 h to quantify secreted FGF19. FGF19 was quantified by Western blot analysis and normalized to the amount of Diet1 protein expressed (Fig. 6B), as well as to actin levels (supplemental Fig. S3). As a control for specificity of the Diet1 effect, we analyzed the levels of FGF19 secretion in response to expression of an irrelevant protein with a similar molecular weight to Diet1 (Kdm5c-V5). A representative experiment (from a total of nine individual experiments) is shown in Fig. 6A. The presence of Diet1-1721H or -1721Q protein increased FGF19 production compared with the control protein. The amount of intracellular FGF19 was similar for the 1721H or 1721Q Diet1 variants, but the amount of FGF19 secreted into the culture medium in 24 h was greater for Diet1-1721Q than Diet1-1721H (Fig. 6A, B; supplemental Fig. S3). This relationship was confirmed in the meta-analysis of nine independent experiments (each performed in triplicate or quadruplicate). The Diet1-1721Q variant enhanced FGF19 secretion by approximately 70% compared with Diet1-1721H (Fig. 6B; P < 0.01). By contrast, another Diet1 variant identified in our study and located in the same protein domain (D1712G) did not affect FGF19 secretion (Fig. 6C, D). Thus, it appears that the differential effect of the 1721H and 1721Q Diet1 proteins on FGF19 secretion is related to the specific amino acid substitution.
DISCUSSION
The current study expands our understanding of Diet1 deficiency on intestinal function and assesses the effects of Diet1/DIET1 genetic variation on traits related to FGF15/19 and bile acid metabolism. We evaluated a new Diet1−/− mouse strain on a C57BL/6J background in comparison to isogenic C57BL/6J wild-type mice (in contrast to the previous studies comparing C57BL/6ByJ mice, which carry the natural Diet1-null allele, to C57BL/6J mice). The Diet1−/− C57BL/6J mice recapitulated previous phenotypes in intestinal gene expression, as well as plasma lipid and bile acid levels (7, 8). We also found that Diet1 deficiency increases gastrointestinal transit time and intestinal water content, which, together with elevated bile acid levels and reduced FGF15 levels, parallel primary BAD in humans. The increased intestinal water content was not apparent when Diet1−/− mice were fed a standard chow diet (unpublished observations), suggesting that elevated circulating bile acid levels that are stimulated by short-term atherogenic diet contributed to the diarrheal phenotype, as is seen in patients with BAD.
In addition to effects of the extreme genetic condition of Diet1 deficiency, we assessed whether modest naturally occurring variations in Diet1 influence traits that are affected in Diet1−/− mice. We found that genetic variation in Diet1 intestinal mRNA expression across a panel of 76 inbred mouse strains correlated positively with Fgf15 mRNA levels, and negatively with bile acid and cholesterol levels. These results underscore that small changes in Diet1 mRNA levels (such as might occur among individuals in the human population) influence Fgf15 expression levels, as well as bile acid and cholesterol levels. In the future, it will be interesting to determine whether the genetic variations in Diet1 expression levels are associated with cis-acting regulatory sequence variants that differ across mouse strains.
The phenotype of Diet1−/− mice led us to investigate whether DIET1 genetic variation influences the severity of BAD in humans. We identified a prevalent (MAF of approximately 30% across populations) nonsynonymous variant in the DIET1 coding region that is associated with SeHCAT retention levels in BAD patients and which alters levels of FGF19 secretion in cultured cells. Previous genetic studies of primary BAD have focused on a few candidate genes with established roles in bile acid metabolism. A rare mutation in SLC10A2/ASBT was identified in one family with congenital BAD, but variations in SLC10A2 appear not to be associated with adult-onset primary BAD (41, 42). Because DIET1 was not annotated until recently (7), this gene is not represented on existing expression microarrays or exome sequencing platforms, and little gene expression or sequence data are available for this gene in human samples. Our DIET1 resequencing identified several rare nonsynonymous amino acid substitutions and a frame-shift mutation. One common DIET1 variant (rs12256835), which alters amino acid composition (H1721Q), was significantly associated with BAD status.
We found that Diet1-1721Q protein promotes increased FGF19 levels in the medium in a cultured cell assay system compared with Diet1-1721H. This effect is likely due to the specific position and amino acid alteration, as a D1712G nonconservative substitution that is located only nine amino acids away had no effect on FGF19 secretion. A full understanding of the role of residue 1721 in Diet1 protein function will require additional protein structure-function studies. An attractive hypothesis is that H1721Q amino acid change affects Diet1 protein conformation and/or interactions with other cellular proteins, potentially affecting FGF19 trafficking and secretion.
Our findings raise the possibility that the Diet1 H1721Q variant could contribute to the 7-fold variation in plasma FGF19 levels that have been observed in healthy individuals (11, 43). Resequencing DIET1 in additional subjects will be required to reveal whether DIET1 rare mutations or additional common variants contribute to variations in bile acid metabolism.
Supplementary Material
Acknowledgments
The authors thank Drs. Lucy Abbott, Kieran Moriarty, Sanjeev Pattni, and Ian Johnston for collecting BAD patient DNA samples, Jenny Link for the Kdm5c expression construct, Jake Lusis for the use of mouse metabolic cages, and Todd G. Kirchgessner, Petia Sipkova, and Xiaoquin Liu for measurement of HMDP strain bile acid and cholesterol levels.
Footnotes
Abbreviations:
- BAD
- bile acid diarrhea
- FGF
- fibroblast growth factor
- HMDP
- Hybrid Mouse Diversity Panel
- MAF
- minor allele frequency
- MAM
- meprin/A5-protein/PTPmu
- SeHCAT
- selenium homocholic acid taurine
- SNV
- single nucleotide variant
This work was supported by National Heart, Lung, and Blood Institute Grants HL102661 (K.R.) and HL28481 (K.R., T.Q.d.A.V.), the Bardhan Research and Education Trust (J.R.F.W.), and the Wellcome Trust (J.R.F.W.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare no conflicts of interest.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Russell D. W. 2009. Fifty years of advances in bile acid synthesis and metabolism. J. Lipid Res. 50: S120–S125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Aguiar Vallim T. Q., Tarling E. J., and Edwards P. A.. 2013. Pleiotropic roles of bile acids in metabolism. Cell Metab. 17: 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawson P. A., and Karpen S. J.. 2015. Intestinal lipid metabolism: new developments and current insights intestinal transport and metabolism of bile acids. J. Lipid Res. 56: 1085–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Staels B., Handelsman Y., and Fonseca V.. 2010. Bile acid sequestrants for lipid and glucose control. Curr. Diab. Rep. 10: 70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.1984. The Lipid Research Clinics Coronary Primary Prevention Trial results. I. Reduction in incidence of coronary heart disease. JAMA. 251: 351–364. [DOI] [PubMed] [Google Scholar]
- 6.1984. The Lipid Research Clinics Coronary Primary Prevention Trial results. II. The relationship of reduction in incidence of coronary heart disease to cholesterol lowering. JAMA. 251: 365–374. [PubMed] [Google Scholar]
- 7.Vergnes L., Lee J. M., Chin R. G., Auwerx J., and Reue K.. 2013. Diet1 functions in the FGF15/19 enterohepatic signaling axis to modulate bile acid and lipid levels. Cell Metab. 17: 916–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phan J., Pesaran T., Davis R. C., and Reue K.. 2002. The Diet1 locus confers protection against hypercholesterolemia through enhanced bile acid metabolism. J. Biol. Chem. 277: 469–477. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann A. F. 2009. Chronic diarrhea caused by idiopathic bile acid malabsorption: an explanation at last. Expert Rev. Gastroenterol. Hepatol. 3: 461–464. [DOI] [PubMed] [Google Scholar]
- 10.Hofmann A. F., Mangelsdorf D. J., and Kliewer S. A.. 2009. Chronic diarrhea due to excessive bile acid synthesis and not defective ileal transport: A new syndrome of defective FGF19 release. Clin. Gastroenterol. Hepatol. 7: 1151–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angelin B., Larsson T. E., and Rudling M.. 2012. Circulating fibroblast growth factors as metabolic regulators–a critical appraisal. Cell Metab. 16: 693–705. [DOI] [PubMed] [Google Scholar]
- 12.Gälman C., Östlund-Lindqvist A-M., Björquist A., Schreyer S., Svensson L., Angelin B., and Rudling M.. 2003. Pharmacological interference with intestinal bile acid transport reduces plasma cholesterol in LDL receptor/apoE deficiency. FASEB J. 17: 265–267. [DOI] [PubMed] [Google Scholar]
- 13.Insull W. 2006. Clinical utility of bile acid sequestrants in the treatment of dyslipidemia: a scientific review. South. Med. J. 99: 257–273. [DOI] [PubMed] [Google Scholar]
- 14.Mekhjian H. S., Phillips S. F., and Hofmann A. F.. 1979. Colonic absorption of unconjugated bile acids. Dig. Dis. Sci. 24: 545–550. [DOI] [PubMed] [Google Scholar]
- 15.Pattni S., and Walters J. R.. 2009. Recent advances in the understanding of bile acid malabsorption. Br. Med. Bull. 92: 79–93. [DOI] [PubMed] [Google Scholar]
- 16.Pattni S. S., Brydon W. G., Dew T., and Walters J. R.. 2012. Fibroblast growth factor 19 and 7α-hydroxy-4-cholesten-3-one in the diagnosis of patients with possible bile acid diarrhea. Clin. Transl. Gastroenterol. 3: e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann A. F. 1967. The syndrome of ileal disease and the broken enterohepatic circulation: cholerheic enteropathy. Gastroenterology. 52: 752–757. [PubMed] [Google Scholar]
- 18.Hofmann A. F., and Poley J. R.. 1969. Cholestyramine treatment of diarrhea associated with ileal resection. N. Engl. J. Med. 281: 397–402. [DOI] [PubMed] [Google Scholar]
- 19.Thaysen E. H., and Pedersen L.. 1976. Idiopathic bile acid catharsis. Gut. 17: 965–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oduyebo I., and Camilleri M.. 2017. Bile acid disease: the emerging epidemic. Curr. Opin. Gastroenterol. 33: 189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walters J. R. F. 2014. Bile acid diarrhoea and FGF19: new views on diagnosis, pathogenesis and therapy. Nat. Rev. Gastroenterol. Hepatol. 11: 426–434. [DOI] [PubMed] [Google Scholar]
- 22.Bajor A., Kilander A., Fae A., Gälman C., Jonsson O., Öhman L., Rudling M., Sjövall H., Stotzer P-O., and Ung K-A.. 2006. Normal or increased bile acid uptake in isolated mucosa from patients with bile acid malabsorption. Eur. J. Gastroenterol. Hepatol. 18: 397–403. [DOI] [PubMed] [Google Scholar]
- 23.Fromm H., and Malavolti M.. 1986. Bile acid-induced diarrhoea. Clin. Gastroenterol. 15: 567–582. [PubMed] [Google Scholar]
- 24.Wedlake L., A’hern R., Russell D., Thomas K., Walters J. R. F., and Andreyev H. J. N.. 2009. Systematic review: the prevalence of idiopathic bile acid malabsorption as diagnosed by SeHCAT scanning in patients with diarrhoea-predominant irritable bowel syndrome. Aliment. Pharmacol. Ther. 30: 707–717. [DOI] [PubMed] [Google Scholar]
- 25.Merrick M. V., Eastwood M. A., and Ford M. J.. 1985. Is bile acid malabsorption underdiagnosed? An evaluation of accuracy of diagnosis by measurement of SeHCAT retention. Br. Med. J. (Clin. Res. Ed.). 290: 665–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vijayvargiya P., Camilleri M., Shin A., and Saenger A.. 2013. Methods for diagnosis of bile acid malabsorption in clinical practice. Clin. Gastroenterol. Hepatol. 11: 1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eusufzai S., Axelson M., Angelin B., and Einarsson K.. 1993. Serum 7-alpha-hydroxy-4-cholesten-3-one concentrations in the evaluation of bile-acid malabsorption in patients with diarrhea: correlation to SeHCAT test. Gut. 34: 698–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pattni S. S., Brydon W. G., Dew T., Johnston I. M., Nolan J. D., Srinivas M., Basumani P., Bardhan K. D., and Walters J. R. F.. 2013. Fibroblast growth factor 19 in patients with bile acid diarrhoea: a prospective comparison of FGF19 serum assay and SeHCAT retention. Aliment. Pharmacol. Ther. 38: 967–976. [DOI] [PubMed] [Google Scholar]
- 29.Walters J. R. F., Tasleem A. M., Omer O. S., Brydon W. G., Dew T., and le Roux C. W.. 2009. A new mechanism for bile acid diarrhea: defective feedback inhibition of bile acid biosynthesis. Clin. Gastroenterol. Hepatol. 7: 1189–1194. [DOI] [PubMed] [Google Scholar]
- 30.Pai R., French D., Ma N., Hotzel K., Plise E., Salphati L., Setchell K. D. R., Ware J., Lauriault V., Schutt L., et al. 2012. Antibody-mediated inhibition of fibroblast growth factor 19 results in increased bile acids synthesis and ileal malabsorption of bile acids in cynomolgus monkeys. Toxicol. Sci. 126: 446–456. [DOI] [PubMed] [Google Scholar]
- 31.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., et al. 2005. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2: 217–225. [DOI] [PubMed] [Google Scholar]
- 32.Cohen R. D., Castellani L. W., Qiao J. H., VanLenten B. J., Lusis A. J., and Reue K.. 1997. Reduced aortic lesions and elevated high density lipoprotein levels in transgenic mice overexpressing mouse apolipoprotein A-IV. J. Clin. Invest. 99: 1906–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghazalpour A., Rau C. D., Farber C. R., Bennett B. J., Orozco L. D., van Nas A., Pan C., Allayee H., Beaven S. W., Civelek M., et al. 2012. Hybrid mouse diversity panel: a panel of inbred mouse strains suitable for analysis of complex genetic traits. Mamm. Genome. 23: 680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ge X., Ding C., Zhao W., Xu L., Tian H., Gong J., Zhu M., Li J., and Li N.. 2017. Antibiotics-induced depletion of mice microbiota induces changes in host serotonin biosynthesis and intestinal motility. J. Transl. Med. 15: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang K. S., Ma T., Filiz F., Verkman A. S., and Bastidas J. A.. 2000. Colon water transport in transgenic mice lacking aquaporin-4 water channels. Am. J. Physiol. Gastrointest. Liver Physiol. 279: G463–G470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y., Li Z., Yang Y., Lin L., and Zhang H.. 2013. Role of glucagon-like peptide-1 in the pathogenesis of experimental irritable bowel syndrome rat models. Int. J. Mol. Med. 31: 607–613. [DOI] [PubMed] [Google Scholar]
- 37.Lee D. K., Jang S., Baek E. H., Kim M. J., Lee K. S., Shin H. S., Chung M. J., Kim J. E., Lee K. O., and Ha N. J.. 2009. Lactic acid bacteria affect serum cholesterol levels, harmful fecal enzyme activity, and fecal water content. Lipids Health Dis. 8: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balesaria S., Pell R. J., Abbott L. J., Tasleem A., Chavele K-M., Barley N. F., Khair U., Simon A., Moriarty K. J., Brydon W. G., et al. 2008. Exploring possible mechanisms for primary bile acid malabsorption: evidence for different regulation of Ileal bile acid transporter transcripts in chronic diarrhoea. Eur. J. Gastroenterol. Hepatol. 20: 413–422. [DOI] [PubMed] [Google Scholar]
- 39.Mouzeyan A., Choi J., Allayee H., Wang X., Sinsheimer J., Phan J., Castellani L. W., Reue K., Lusis A. J., and Davis R. C.. 2000. A locus conferring resistance to diet-induced hypercholesterolemia and atherosclerosis on mouse chromosome 2. J. Lipid Res. 41: 573–582. [PubMed] [Google Scholar]
- 40.Vergnes L., Phan J., Strauss M., Tafuri S., and Reue K.. 2003. Cholesterol and cholate components of an atherogenic diet induce distinct stages of hepatic inflammatory gene expression. J. Biol. Chem. 278: 42774–42784. [DOI] [PubMed] [Google Scholar]
- 41.Oelkers P., Kirby L. C., Heubi J. E., and Dawson P. A.. 1997. Primary bile acid malabsorption caused by mutations in the ileal sodium-dependent bile acid transporter gene (SLC10A2). J. Clin. Invest. 99: 1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montagnani M., Love M. W., Rossel P., Dawson P. A., and Qvist P.. 2001. Absence of dysfunctional ileal sodium-bile acid cotransporter gene mutations in patients with adult-onset idiopathic bile acid malabsorption. Scand. J. Gastroenterol. 36: 1077–1080. [DOI] [PubMed] [Google Scholar]
- 43.Gälman C., Angelin B., and Rudling M.. 2011. Pronounced variation in bile acid synthesis in humans is related to gender, hypertriglyceridaemia and circulating levels of fibroblast growth factor 19. J. Intern. Med. 270: 580–588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.