Abstract
The remodeling of PUFAs by the Lands cycle is responsible for the diversity of phospholipid molecular species found in cells. There have not been detailed studies of the alteration of phospholipid molecular species as a result of serum starvation or depletion of PUFAs that typically occurs during tissue culture. The time-dependent effect of cell culture on phospholipid molecular species in RAW 264.7 cells cultured for 24, 48, or 72 h was examined by lipidomic strategies. These cells were then stimulated to produce arachidonate metabolites derived from the cyclooxygenase pathway, thromboxane B2, PGE2, and PGD2, and the 5-lipoxygenase pathway, leukotriene (LT)B4, LTC4, and 5-HETE, which decreased with increasing time in culture. However, the 5-lipoxygenase metabolites of a 20:3 fatty acid, LTB3, all trans-LTB3, LTC3, and 5-hydroxyeicosatrienoic acid, time-dependently increased. Molecular species of arachidonate containing phospholipids were drastically remodeled during cell culture, with a new 20:3 acyl group being populated into phospholipids to replace increasingly scarce arachidonate. In addition, the amount of TNFα induced by lipopolysaccharide stimulation was significantly increased in the cells cultured for 72 h compared with 24 h, suggesting that the remodeling of PUFAs enhanced inflammatory response. These studies supported the rapid operation of the Lands cycle to maintain cell growth and viability by populating PUFA species; however, without sufficient n-6 fatty acids, 20:3 n-9 accumulated, resulting in altered lipid mediator biosynthesis and inflammatory response.
Keywords: polyunsaturated fatty acid, Mead acid, volcano plot
Almost a century ago, the concept of essential fatty acid efficiency emerged (1), which led to the discovery that animal cells could not make PUFAs such as linoleic acid (LA; 18:2, n-6) or α-linolenic acid (ALA; 18:3, n-3). The absence of these fatty acids in the diet caused a diverse number of clinical signs (2) and altered levels of arachidonate (3). Cellular biochemistry responds to a lack of LA and ALA by chain elongation and desaturation of oleoyl-CoA (n-9) to eventually form an unusual PUFA, 5,8,11-eicosatrienoic acid (n-9), often called Mead acid after its discoverer (4). This PUFA retains biophysical properties critical for cellular membrane fluidity, but lacks the ability to be transformed into certain lipid mediators. The appearance of Mead acid esterified to cellular phospholipids is a result of the same biochemistry that normally takes place when PUFAs from the diet, such as ALA and arachidonic acid (AA), are incorporated into complex lipids. Once within the cell, these PUFAs are very quickly remodeled into cellular phospholipids. For example, the remodeling of AA by what has been termed the Lands cycle has been studied for a number of years (5) and is known to be responsible for the diversity of phospholipid molecular species seen in all cellular membranes. This phospholipid remodeling pathway is a result of CoA esters of PUFAs being synthesized by a family of long-chain acyl-CoA synthases (6) and the action of specific acyltransferases, including the lysophospholipid acyltransferases, LPCAT3/MBOAT5 (7) and LPIAT1/MBOAT7 (8), known to incorporate AA into phosphatidylcholine (PC) and phosphatidylinositol (PI), respectively.
Population of cellular membrane phospholipids with PUFAs, specifically AA, is a critical step in the biosynthesis of the eicosanoid lipid mediator family. The COX pathway leads to the formation of prostanoids by way of the prostaglandin H2 endoperoxide intermediate, that is metabolized to PGD2, PGE2, PGF2α, PGI2, and thromboxane (Tx)A2 by specific synthases (9). These prostanoids have a plethora of physiological and pathophysiological effects, including vasodilation and vascular leakage (PGE2), mast cell maturation, eosinophil recruitment and allergic responses (PGD2), vascular and respiratory smooth muscle contraction (PGF2α), and inhibition (PGI2) or activation (TxA2) of platelet aggregation (10).
The 5-lipoxygenase catalyzes oxygenation of AA to 5(S)-hydroperoxy-6-trans-8,11,14-cis-eicosatetraenoic acid, and further dehydration to the allylic epoxide, leukotriene (LT)A4. LTA4 is converted by LTA4 hydrolase to the dihydroxy acid, LTB4, and by LTC4 synthase to the glutathione conjugate, LTC4. LTB4 is a pro-inflammatory lipid mediator that activates and recruits neutrophils into inflammatory areas (11), whereas cysLTs (LTC4 and its metabolites, LTD4 and LTE4) induce bronchoconstriction and neutrophil extravasation, and also participate in vascular leakage (12).
Macrophages, which are a major source of various lipid mediators, respond to a variety of stimuli by activating phospholipase A2 and producing eicosanoids. The RAW 264.7 murine macrophage cell line has been used in numerous studies as a model of primary macrophages (13). RAW cells respond to lipopolysaccharide (LPS) and interferon-γ (IFN-γ) by producing nitric oxide and PGE2 (14). Following calcium ionophore A23187 stimulation, RAW cells also produced modest amounts of LTs and prostaglandins (15). The addition of LPS, phorbol 12-myristate 13-acetate, or A23187 resulted in an increase of lysophosphatidylinositol levels (16).
After oxidation of arachidonate and transformation of the free fatty acid into the bioactive chemical structure, the eicosanoid is transported out of the cell so that it can encounter one of numerous G protein-coupled receptors for prostaglandins (10) and LTs (12, 17) found on target cells. However, a large portion of the liberated arachidonate is not oxidized, but rather converted back into a CoA ester to complete the remodeling cycle or to undergo mitochondrial and peroxisomal β-oxidation.
The rapidity of cellular essential fatty acid deficiency, especially for cells carried in tissue culture, has not been widely appreciated. There have been a few studies with either supplementation of arachidonate or other PUFAs and the effect of serum starvation on cellular function (18). In such studies, a remarkable effect of depletion of AA has been noted (19). Current recommendations for cell culture suggest changing culture media (resupply of PUFAs) only as frequently as 3 days with no suggestion that this period of time could lead to important biochemical changes by depletion of PUFAs (20). There have not been detailed studies of the alteration of the lipidome, specifically those phospholipid molecular species altered as a result of serum starvation or depletion of PUFAs during tissue culture for times as short as 3 days. Reported here is the time-dependent effect of tissue culture on the production of eicosanoids, specifically prostaglandins and LTs, and time-dependent alterations in phospholipids in the membranes of cells carried in culture.
MATERIALS AND METHODS
Materials
Eicosanoid standards, including LTB3 and deuterium-labeled eicosanoids, were purchased from Cayman Chemical (Ann Arbor, MI). Phospholipid standards and deuterium-labeled phospholipids were purchased from Avanti Polar Lipids (Alabaster, AL). A23187 and LPS from Salmonella minnesota were purchased from Sigma-Aldrich (St. Louis, MO). DMEM was purchased from Corning (Manassas, VA) and Wako Pure Chemical (Osaka, Japan). All of the solvents were HPLC grade and purchased from Thermo Fisher Scientific (Waltham, MA). HBSS was purchased from Corning.
Cell culture
RAW 264.7 cells were purchased from American Type Culture Collection (Manassas, VA) and were cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum, 100 IU/ml penicillin, and 100 μg/ml streptomycin. The cells were grown in humidified air with 5% CO2 at 37°C. RAW cells (5 × 106 per 100 mm dish) were plated and cultured for 24 h (day 1), 48 h (day 2), and 72 h (day 3).
Quantification of eicosanoids produced in A23187-stimulated RAW cells
RAW 264.7 cells were cultured for 1, 2, and 3 days without medium change. The cells were washed with PBS, scraped, and centrifuged at 190 g for 5 min. Aliquots of cells (1 × 106/ml) suspended in HBSS containing Ca2+ and Mg+ were stimulated with 2 μM of A23187 for 30 min at 37°C. The reaction was stopped with 1 ml of ice-cold methanol and diluted with water containing 0.1% formic acid to yield a final methanol concentration of 20%. After centrifugation, deuterium-labeled internal standards were added and the supernatants loaded on Oasis HLB cartridges (Waters, Milford, MA). The column was sequentially washed with water containing 0.1% formic acid, 15% methanol containing 0.1% formic acid, and petroleum ether containing 0.1% formic acid. The samples were eluted with 200 μl of methanol containing 0.1% formic acid. For RP-HPLC-MS/MS, a Shimadzu LC system consisting of four LC-20AD pumps, a SIL-20AC autosampler, a CTO-20AC column oven, a FCV-12AH six-port switching valve, and a TSQ Quantum Ultra triple quadrupole mass spectrometer equipped with an ESI ion source (Thermo Fisher Scientific) were used (21). An aliquot of each sample (50 μl) was injected into the trap column, an Opti-Guard Mini C18, at a total flow rate of 500 μl/min. Three minutes after sample injection, the valve was switched to introduce the trapped sample to the analytical column, a Capcell Pak C18 MGS3 (Shiseido, Tokyo, Japan). Separation of lipids was achieved by a linear gradient using water and acetonitrile containing 0.1% formic acid. The total flow rate was 120 μl/min, the column temperature was set at 46°C, and the LC column eluent was introduced directly into a TSQ Quantum Ultra. All compounds were analyzed in a negative ion polarity mode. Eicosanoids were quantified by multiple reaction monitoring (MRM). The MRM transitions monitored were m/z 351 → 271 for PGE2 and PGD2, m/z 369 → 195 for TxB2, m/z 624 → 272 for LTC4, m/z 626 → 272 for LTC3, m/z 335 → 195 for LTB4, m/z 337 → 195 for LTB3, m/z 319 → 195 for 5-HETE, m/z 321 → 115 for 5-hydroxyeicosatrienoic acid, m/z 303 → 259 for AA, m/z 305 → 261 for MA, m/z 355 → 275 for [2H4]PGE2 and [2H4]PGD2, m/z 373 → 199 for [2H4]TxB2, m/z 629 → 272 for [2H5]LTC4, m/z 339 → 197 for [2H4]LTB4, m/z 327 → 116 for [2H8]5HETE, and m/z 311 → 267 for [2H8]AA. The identification of LTB3 and all trans-LTB3 came from the ion transition m/z 337 → 195 and at the retention times of 11.95 and 12.05 min, respectively, as established by analysis of commercial LTB3 separated under the same LC conditions. For accurate quantification, calibration curves were generated for each target eicosanoid using known reference standards and the same isotope-labeled internal standard. Automated peak detection, calibration, and calculation were carried out by the Xcalibur 1.2 software package.
Lipid extraction and NP-HPLC/MRM analysis of RAW 264.7 phospholipids
After addition of the deuterated internal standards, [2H31]16:0/18:1-phosphatidic acid (PA), [2H31]16:0/18:1-PC, [2H31]16:0/18:1-phosphatidylethanolamine (PE), [2H31]16:0/18:1-PG, [2H31]16:0/18:1-PI, and [2H31]16:0/18:1-phosphatidylserine (PS) (10 ng each), lipids from RAW 264.7 cells (3 × 106 cells) at each culture time point were extracted according to the Bligh-Dyer method (22). The organic phase was dried under a stream of nitrogen gas and resuspended in 100 μl of a mixture of 75% HPLC solvent A (hexane/isopropanol 30:40, v/v) and 25% solvent B (5 mM ammonium acetate in hexane/isopropanol/water 30:40:7, v/v/v). Samples were injected into an HPLC system connected to a triple quadrupole mass spectrometer (4000 QTRAP; SCIEX, Framingham, MA) and normal phase chromatography was performed using a silica HPLC column (Ascentis, 150 × 2.1 mm, 5 μm; Supelco, Bellefonte, PA) at a flow rate of 200 μl/min. Solvent B was maintained at 25% for 5 min, increased gradually to 60% in 10 min and then to 95% in 5 min, and was held for 20 min before reequilibration for 15 min. Mass spectrometric analysis was performed for approximately 100 molecular species of PA, PC, PE, PS, PG, PI, and bis(monoacylglycero)phosphate (BMP) that contained either 20:4 or 20:3 fatty acyl groups in the negative ion mode using MRM. The list of ion transitions is presented as supplemental Table S1. The ratios between the integrated area of each analyte and the integrated area of the corresponding internal standard for each class are normalized by cell number. This lipidomic analysis was repeated three times for each time point of tissue culture and the normalized abundance ratios subjected to statistical analysis using the R-based programs available at the Metabolomics Workbench (www.metabolomicsworkbench.org) sponsored by the Common Fund of the National Institutes of Health or an R-based program written using the Plotly R-package that provided information about the intensity of the specific phospholipid molecular species by size of indicating icon and color as well as being interactive. An HTML file of the interactive volcano plot shown in Fig. 4 is provided as supplemental material.
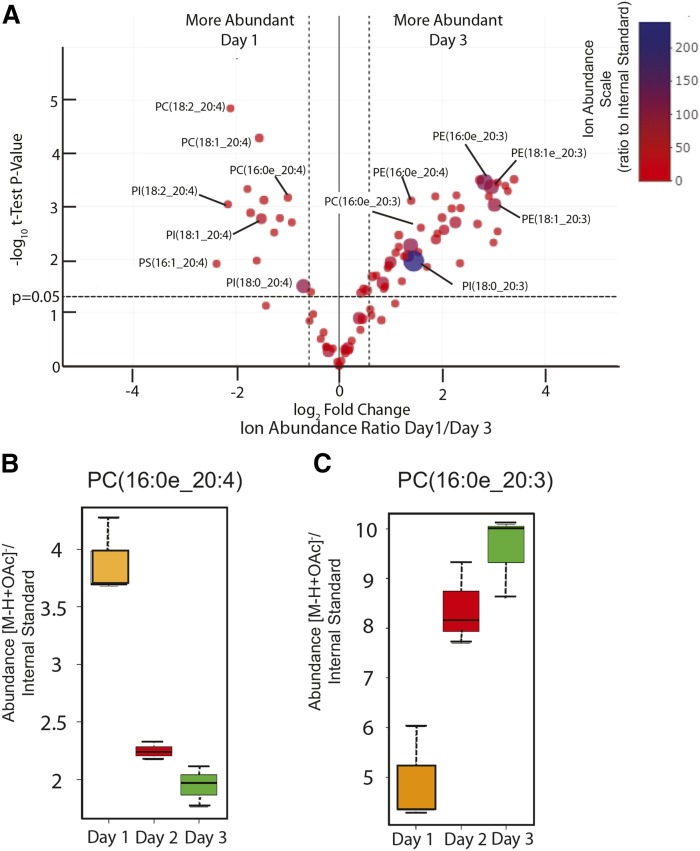
Fig. 4.
A: Volcano plot comparing the relative abundances of phospholipid molecular species in RAW 264.7 cells cultured for 1 or 3 days. Those species significantly more than 2-fold more abundant at day 1 are in the upper left sector with a P-value (t-test) greater than 0.05. Those species more abundant at day 3 are in the right upper sector. The size of the circle is related to abundance as well as the color (as per the color scale) in units of abundance of ion divided by the abundance of the internal standard for that class of phospholipid (abundance ratio). B, C: Box plots of the abundance ratio of PC(16e_20:4) and PC(16e_20:3) in RAW cells after 1, 2, or 3 days of culture. The bar in the quartile-indicating box is the median value, n = 3 for each day in culture.
Stimulated production of TNFα
RAW 264.7 cells were cultured for 1 and 3 days without medium change. The cells were washed with DMEM, scraped, and centrifuged at 190 g for 5 min. Aliquots of cells (1 × 106/ml) suspended in DMEM were cultured in a 24-well plate for 30 min, and the cells were stimulated with 100 ng/ml LPS for 1, 2, 4, 6, and 8 h. The concentration of TNFα in the medium was analyzed with the Mouse TNFα ELISA kits (eBioscience) according to the manufacturer’s instructions.
RESULTS
Eicosanoid profiling in A23187-stimulated RAW cells
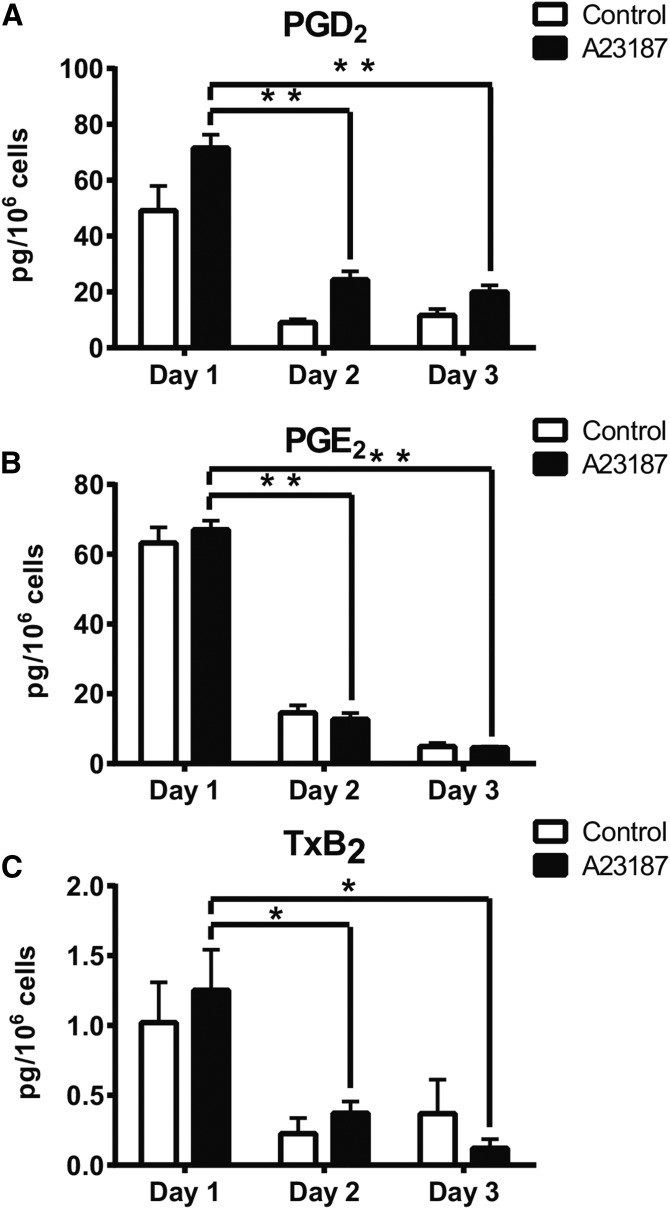
RAW 264.7 cells (5 × 106 cells in a 100 mm dish) were cultured for 24, 48, or 72 h without any medium changes. After incubation with or without calcium ionophore, A23187, eicosanoids were extracted and quantitated using a targeted RP-LC-MS/MS assay. The stimulated production of arachidonate metabolites derived from the cyclooxygenase pathway, TxB2, PGE2, and PGD2 (Fig. 1), was found to decrease for those cells cultured for increasing amounts of time.
Fig. 1.
Cyclooxygenase products of AA that appear in the supernatant of RAW 264.7 cells carried in culture for the indicated number of days and then stimulated (or not) with calcium ionophore A23187. A: Prostaglandin D2 production. B: Prostaglandin E2 production. C: TxB2 production. Data are expressed as average ± SEM, n = 3. Significance comparisons, *P < 0.01, **P < 0.001.
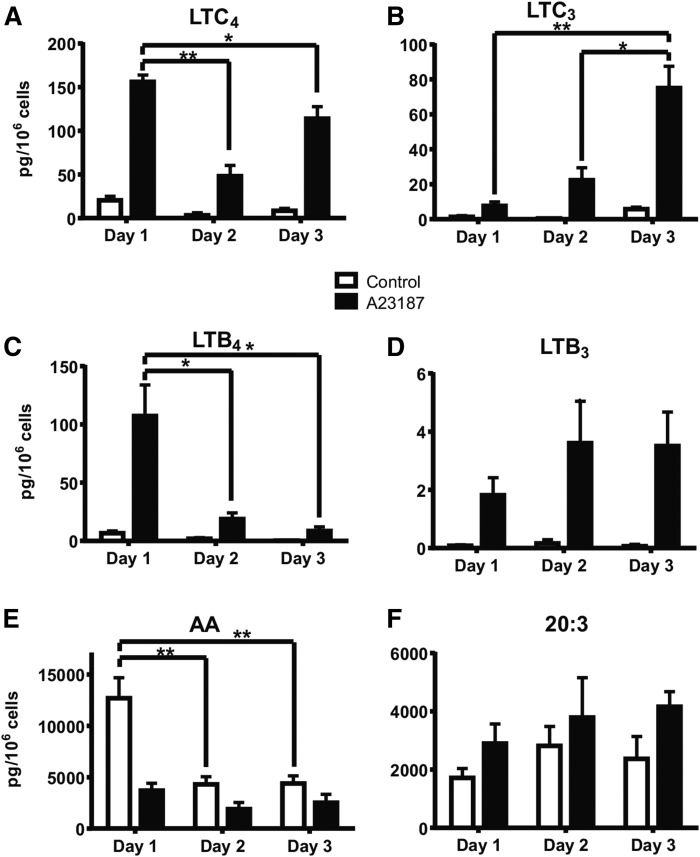
In a similar fashion, the absolute quantities of 5-lipoxygenase metabolites, LTC4 and LTB4, as well as free arachidonate, generated after calcium ionophore stimulation, were also found to decrease (Fig. 2A, C, E). However, there were new metabolites of the 5-lipoxygenase pathway that then increased with time after stimulation of RAW 264.7 cells in culture. The 5-lipoxygenase metabolites, LTC3 and LTB3, as well as a free 20:3 fatty acid, were found to time-dependently increase (Fig. 2B, D, F). These eicosanoid products were identified by their longer HPLC retention times, consistent with one double bond less, the observed m/z of the carboxylate anion, and the diagnostic product ion of each LT species (23). Analysis of the LTA3 nonenzymatic product, all trans-LTB3, was established by HPLC retention time and MS and this product was found to be more abundant than LTB3 (supplemental Fig. S2). The structure of the 20:3 fatty acid was determined by the Paternò-Büchi reaction as 5,8,11-eicosatrienoic acid (20:3 n-9), as described elsewhere (24). Free AA was found to decrease with increasing days in culture and a fatty acid with a reverse-phase HPLC retention time longer than arachidonate having a carboxylate anion [M-H]− at m/z 305 became quite abundant. This information was consistent with a 20:3 fatty acid that was released by a phospholipase activated by the addition of the calcium ionophore to those cells carried for more than 1 day in culture. The released arachidonate was time-dependently decreasing while the 20:3 fatty acid was increasing with days in culture. These results further suggested that AA-containing phospholipids were becoming depleted in those RAW 264.7 cells with time, while 20:3-containing phospholipids were increasing in response to the culture conditions (Fig. 3).
Fig. 2.
Levels of 20:4 and 20:3 fatty acids and their 5-lipoxygenase products that appear in the supernatant of RAW 264.7 cells carried in culture for the indicated number of days and then stimulated (or not) with calcium ionophore A23187. A: LTB4 production. B: LTB3 production. C: LTC4 production. D: LTC3 production. E: Free 20:4 accumulation. F: Free 20:3 accumulation. Data are expressed as average ± SEM, n = 3. Significance comparisons, *P < 0.01, **P < 0.001.
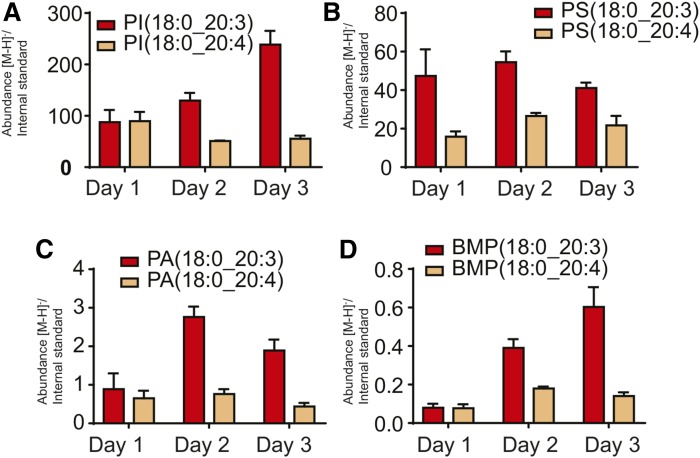
Fig. 3.
Phospholipid molecular species containing either 20:4 (orange bars) or 20:3 (red bars) esterified to PI (A), PS (B), PA (C), or BMP (D). Data are expressed as average ± SEM, n = 3.
Phospholipid remodeling during cell culture without medium changes
The molecular species of phospholipids present in RAW 264.7 cells at various times in cell culture were determined by a targeted NP-HPLC/MRM assay, which analyzed the samples for specific phospholipid molecular species that contained either 20:4 or 20:3 fatty acyl groups and the corresponding deuterated internal standards (supplemental Table S1). This negative ion-based ESI assay was based on scheduled ion transitions for molecular ions of phospholipids yielding either the specific carboxylate anion at m/z 303 (20:4) or the anion at m/z 305 (20:3) in a tandem quadrupole mass spectrometer. In this way, information as to class of phospholipid (PA, PS, PI, PE, PC, PG, or BMP) came from the HPLC retention time, which also defined those ion transitions used during the elution of this class of phospholipid. The mass of the target ions [M − H]− or [M + OAc]− (in the case of PC) along with information of the specific PUFA fatty acyl group could be used to precisely assign the phospholipid to the molecular species level (25). For these studies, only the fatty acids 20:3 and 20:4 were included in the list of ion transitions. The measured abundance of each molecular species was normalized to the abundance of the deuterated internal standard for that phospholipid class and the number of cells extracted.
When the 20:4- and 20:3-containing molecular species of each of the phospholipid classes were examined, the abundance of many 20:3 phospholipid species increased with time in culture relative to the corresponding 20:4 molecular species (Fig. 3). This was quite obvious for the abundant PI species, PI(18:0_20:4) and PI(18:0_20:3) (Fig. 3A), PA(18:0_20:4) and PA(18:0_20:3) (Fig. 3C), and even many phospholipids of minor abundance, such as BMP(18:0_20:4) and BMP(18:0_20:3) (Fig. 3D). However, some phospholipid species changed very little even though they were fairly abundant, such as PS(18:0_20:4) and PS(18:0_20:3) (Fig. 3B). Other major phospholipid species containing esterified arachidonate decreased at day 2, but then increased on day 3 in the same trend as the LTC4 production, but not other eicosanoids (supplemental Fig. S3). The measurement of the precise abundance of molecular species at one particular time does not reveal the dynamic nature of the Lands remodeling pathway, the specificity of the various lysophospholipid acyltransferases (26), or the phospholipases that remodel phospholipid molecular species in cells. These complications are superimposed on the dramatic increase in Mead acid production. The results confirmed that many phospholipid molecular species were being populated with the 20:3 fatty acyl group during culture without media change. A complete list of the identified phospholipid species is available in supplementary Table S2.
An unbiased evaluation of the changes in phospholipid molecular species that occurred with time in culture was carried out by statistical comparisons of phospholipid abundances on day 1 to day 2 and on day 1 to day 3. A volcano plot that compared fold change of ion intensity (log 2-fold change) to significance (log t-test) (Fig. 4) revealed a striking number of phospholipid molecular species whose relative abundances differed by factors of 2- to 8-fold. Many PC, PI, and PS molecules containing 20:4 fatty acids were significantly more abundant on day 1 compared with day 3 (right side of Fig. 4), and several phospholipids containing esterified 20:3 significantly increased on day 3 compared with day 1 (left upper sector of Fig. 4). In general, the most significant positive fold abundances on day 1 relative to day 3 corresponded to the arachidonate-containing molecular species of PC and PI, while the majority of the negative fold abundances were PE and BMP phospholipid species that contained esterified 20:3. As an example, a box plot of the data on PC(16:0e_20:4) revealed a 2-fold decrease from day 1 to day 3 (Fig. 4B), while PC(16e:0_20:3) had over a 2-fold increase in abundance from day 1 to day 3 (Fig. 4C). The PC molecular species, PC(18:1e_20:4) and PC(18:0e_20:4), had just the opposite relative abundance changes from day 1 to day 3 in culture (supplemental Fig. S3). This pattern of arachidonate-content in these ether PC species was similar to the trend for the stimulated production of LTC4 from these cells at the corresponding day of culture. Overall, these results revealed that the molecular species of arachidonate phospholipids were drastically remodeled during cell culture without medium change and that a new fatty acyl group was being populated into the phospholipid classes to replace the increasingly scarce arachidonate. That new fatty acid was 5,8,11-eicosatrienoic acid, also called Mead acid (n-9).
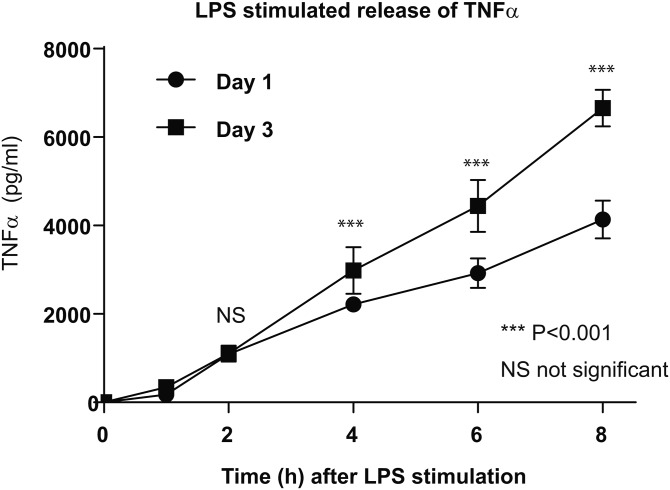
TNFα stimulated response of RAW in culture without medium changes
The fatty acid composition of membrane phospholipids in macrophages can influence inflammatory responses (27, 28). While lauric acid (C12:0) enhanced the recruitment of Toll-like receptor 4 (TLR4) and its adaptor proteins, TIR domain-containing adaptor inducing interferon-β and myeloid differentiation factor 88, into lipid rafts and induced TLR4 activation, DHA inhibited the recruitment to lipid rafts and the activation of TLR4 (29). The amount of TNFα induced after 4 h LPS stimulation was significantly higher in those cells cultured for 72 h compared with 24 h without medium change (Fig. 5), suggesting that the remodeling of phospholipid molecular species enriched with esterified arachidonate to esterified Mead acid enhanced the inflammatory response of macrophages.
Fig. 5.
Production of TNFα by LPS (100 ng/ml) stimulation for 0, 1, 2, 4, 6, and 8 h in RAW cells cultured for 24 h (day 1) and 72 h (day 3) without medium change. Data are expressed as average ± SD, n = 6. ***P < 0.001 (paired t-test).
DISCUSSION
The ability to carry cells in tissue culture has had an enormous impact on revealing specific biochemical events as well as cell biology. While artifacts that emerge by using such in vitro methods are known, there has been little attention paid to the alterations of lipid composition introduced by such protocols. In addition to this, it has been an established protocol to use serum starvation in many studies, including studies of T and B cell immunology, as a practical means to induce specific biochemical events (18, 30). This is particularly relevant for the essential biochemical molecules that cannot be synthesized de novo. A striking class of such molecules is the PUFAs of the n-6 and n-3 families that require exogenous LA or ALA to populate a large family of unsaturated fatty acids found in cellular membranes esterified to phospholipids. This family includes AA that is the precursor of a family of bioactive lipid mediators, such as prostanoids and LTs. With long-term culture without supplementation of these essential fatty acids, the n-6 and n-3 fatty acids containing phospholipids can be depleted. This was readily demonstrated in this study by the depletion of AA in phospholipids of all classes, even after just 2 or 3 days of continuous culture.
The depletion of AA stores is not without consequences, as observed for the generation of prostaglandins and LTs following stimulation of cells carried in long-term culture. Prostaglandins require a sequence of three double bonds interrupted by methylene groups in a specific structural position along the 20 carbon chain so that a hydrogen atom can be abstracted by PGH synthase at carbon-13 to form an oxygen radical, 11-hydrogenperoxy, as the initial step of the cyclooxygenase mechanism (31). This three double bond structural requirement for cyclooxygenase is observed in arachidonate (5,8,11,14-eicosatetraenoic acid) as well as in dihomo-γ-linolenic acid (8,11,14-eicosatrienoic acid), but not in the isomeric Mead acid (5,8,11-eicosatetraenoic acid). Thus, the accumulation of Mead acid in cell membrane phospholipids would be expected to have a measurable effect on prostaglandin biosynthesis. This is exactly what was found when RAW 264.7 cells carried in culture for 2 to 3 days were stimulated with a calcium ionophore to release AA for PGH synthase.
On the other hand, the biosynthesis of LTs involves 5-lipoxygenase abstracting a hydrogen atom from carbon-7 from a fatty acid substrate that also has homo-conjugated three double bond structure. This structural requirement leads to formation of the 5-hydroperoxy radical that can be further converted by 5-lipoxygenase to form a triene-conjugated epoxide. Arachidonate has this structural prerequisite as well as Mead acid, but not dihomo-γ-linolenic acid. In this case of 5-lipoxygenase, LT products of the 3 series (LTC3 and LTB3) can be formed, but LTA4 hydrolase is not very efficient for the production of LTB3 (32). LTA3 was previously found to inhibit LTB4 production by blocking the binding of LTA4 to LTA4 hydrolase (33). Thus, while the LTC3 time-dependently increased, the all trans-LTB3 and LTB3, which can also be formed from LTA3, were not abundantly found. Nevertheless the all trans-LTB3 product was found to be more abundant, consistent with the nonenzymatic hydrolysis of LTA3 derived from Mead acid. These products were defined several years ago (32–35) and have similar, but somewhat less potent, actions through the LTB-R and cysLTR receptors (35).
Cells respond to depletion of essential PUFAs by converting oleic acid (n-9), which can be made from acetate, into a PUFA. These steps involve a Δ6-desaturase (FADS1) to form 6,9-octadecadienoic acid. This fatty acid can then be elongated by two carbons (after making the CoA ester) by an elongase, such as ELOVL4, to form 8,11-eicosadienoic acid (n-9). Subsequent oxidation of this diene fatty acyl-CoA ester by the Δ5-disaturase (FADS1) results in the synthesis of Mead acid (20:3, n-9). This biochemical response by the cells is critical in maintaining viability and growth of the cell, perhaps by sustaining membrane fluidity (4).
RAW cells are murine macrophage cell lines and can produce the inflammatory cytokines and mediators, such as TNFα, IL1β, and IL6. Previous studies have shown that fatty acid exposure and the fatty acid composition of inflammatory cells influence the functions of these cells, and the contents of AA, EPA, and DHA appear to be especially important. The depletion of these PUFAs modulates the TLR4-mediated signaling pathway and consequent inflammatory responses. Mead acid biosynthesis from oleic acid was found to be rapidly induced in RAW cells during cell culture without medium change and may compensate the depletion of PUFAs of the n-3 and n-6 families, as well as enhance the inflammatory cytokine response.
The use of statistical tools to provide an overview of the significant changes in arachidonoyl- and eicosatrienoyl-containing phospholipids readily revealed the significant depletion of arachidonate from most classes of phospholipids. While some molecular species were not significantly altered during the 2 and 3 days of unsupplemented culture, as indicated by those unlabeled species in the volcano plots, there were many more phospholipids that did change. In addition, a number of new species became quite apparent, while the most abundant arachidonate species at day 1 decreased with the appearance of the corresponding 20:3 species of that class. This clearly indicated the rapid operation of the Lands cycle to maintain membrane fluidity by populating PUFA species that were being depleted during the normal remodeling of phospholipids and release of AA by various phospholipases. The most abundant phospholipid species containing AA included PC and PI on day 1, while on day 2, the PE and PS species that contain 20:3 were becoming much more significant. Even the difference between day 3 and day 2 indicated remodeling of PS, PC, phosphatidylglycerol, and PE molecular species. The overall effect of these changes in PUFA molecular species was revealed by a reduced ability to generate PGE2 and PGD2 as time for these cells in culture progressed, as well as the significant diminution of the ability to make LTC4 and LTB4. In part, this was made up by an increased production of LTB3 and LTC3 with days in culture as the altered inflammatory cytokine response to LPS.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Duncan Claypool who wrote the interactive Volcano Plot program in the R-language.
Footnotes
Abbreviations:
- AA
- arachidonic acid
- ALA
- α-linolenic acid
- BMP
- bis(monoacylglycero)phosphate
- LA
- linoleic acid
- LPS
- lipopolysaccharide
- LT
- leukotriene
- MRM
- multiple reaction monitoring
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PGD2
- prostaglandin D2
- PGE2
- prostaglandin E2
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
- TLR4
- Toll-like receptor 4
- Tx
- thromboxane
This work was supported, in part, by Japan Society for the Promotion of Science Grants 16K08596, 15KK0320, 15H05904, and 15H04708 (T.O., T.Y.); Takeda Science Foundation (T.O.); and National Institutes of Health Grant HL117798 (R.C.M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Burr G. O., and Burr M. M.. 1929. A new deficiency disease produced by the rigid exclusion of fat from the diet. J. Biol. Chem. 82: 345–367. [DOI] [PubMed] [Google Scholar]
- 2.Farrell P. M., Gutcher G. R., Palta M., and DeMets D.. 1988. Essential fatty acid deficiency in premature infants. Am. J. Clin. Nutr. 48: 220–229. [DOI] [PubMed] [Google Scholar]
- 3.Holman R. T. 1960. The ratio of trienoic: tetraenoic acids in tissue lipids as a measure of essential fatty acid requirement. J. Nutr. 70: 405–410. [DOI] [PubMed] [Google Scholar]
- 4.Fulco A. J., and Mead J. F.. 1959. Metabolism of essential fatty acids. VIII. Origin of 5,8,11-eicosatrienoic acid in the fat-deficient rat. J. Biol. Chem. 234: 1411–1416. [PubMed] [Google Scholar]
- 5.Yamashita A., Hayashi Y., Nemoto-Sasaki Y., Ito M., Oka S., Tanikawa T., Waku K., and Sugiura T.. 2014. Acyltransferases and transacylases that determine the fatty acid composition of glycerolipids and the metabolism of bioactive lipid mediators in mammalian cells and model organisms. Prog. Lipid Res. 53: 18–81. [DOI] [PubMed] [Google Scholar]
- 6.Ellis J. M., Frahm J. L., Li L. O., and Coleman R. A.. 2010. Acyl-coenzyme A synthetases in metabolic control. Curr. Opin. Lipidol. 21: 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shindou H., and Shimizu T.. 2009. Acyl-CoA:lysophospholipid acyltransferases. J. Biol. Chem. 284: 1–5. [DOI] [PubMed] [Google Scholar]
- 8.Lee H. C., Inoue T., Sasaki J., Kubo T., Matsuda S., Nakasaki Y., Hattori M., Tanaka F., Udagawa O., Kono N., et al. . 2012. LPIAT1 regulates arachidonic acid content in phosphatidylinositol and is required for cortical lamination in mice. Mol. Biol. Cell. 23: 4689–4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith W. L., and Murphy R. C.. 2015. The eicosanoids: cyclooxygenase, lipoxygenase, and epoxygenase pathways. In Biochemistry of Lipids, Lipoproteins and Membranes. N. Ridgeway and R. McLeod, editors. Elsevier Science, Oxford, UK. 260–295. [Google Scholar]
- 10.Hirata T., and Narumiya S.. 2012. Prostanoids as regulators of innate and adaptive immunity. Adv. Immunol. 116: 143–174. [DOI] [PubMed] [Google Scholar]
- 11.Okuno T., Yokomizo T., Hori T., Miyano M., and Shimizu T.. 2005. Leukotriene B4 receptor and the function of its helix 8. J. Biol. Chem. 280: 32049–32052. [DOI] [PubMed] [Google Scholar]
- 12.Haeggström J. Z., and Funk C. D.. 2011. Lipoxygenase and leukotriene pathways: biochemistry, biology, and roles in disease. Chem. Rev. 111: 5866–5898. [DOI] [PubMed] [Google Scholar]
- 13.Maurya M. R., Gupta S., Li X., Fahy E., Dinasarapu A. R., Sud M., Brown H. A., Glass C. K., Murphy R. C., Russell D. W., et al. . 2013. Analysis of inflammatory and lipid metabolic networks across RAW264.7 and thioglycolate-elicited macrophages. J. Lipid Res. 54: 2525–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hulkower K. I., Pollock J. S., Walsh R. E., Huang R., Otis E. R., Brooks C. D., and Bell R. L.. 1996. Leukotrienes do not regulate nitric oxide production in RAW 264.7 macrophages. Prostaglandins Leukot. Essent. Fatty Acids. 55: 145–149. [DOI] [PubMed] [Google Scholar]
- 15.Nieves D., and Moreno J. J.. 2006. Role of 5-lipoxygenase pathway in the regulation of RAW 264.7 macrophage proliferation. Biochem. Pharmacol. 72: 1022–1030. [DOI] [PubMed] [Google Scholar]
- 16.Zoeller R. A., Wightman P. D., Anderson M. S., and Raetz C. R.. 1987. Accumulation of lysophosphatidylinositol in RAW 264.7 macrophage tumor cells stimulated by lipid a precursors. J. Biol. Chem. 262: 17212–17220. [PubMed] [Google Scholar]
- 17.Yokomizo T. 2015. Two distinct leukotrienes B4 receptors, BLT1 and BLT2. J. Biochem. 157: 65–71. [DOI] [PubMed] [Google Scholar]
- 18.Jain S., Suklabaidya S., Das B., Raghav S. K., Batra S. K., and Senapati S.. 2015. TLR4 activation by lipopolysaccharide confers survival advantage to growth factor deprived prostate cancer cells. Prostate. 75: 1020–1033. [DOI] [PubMed] [Google Scholar]
- 19.Traynor-Kaplan A., Kruse M., Dickson E. J., Dai G., Vivas O., Yu H., Whittington D., and Hille B.. 2017. Fatty-acyl chain profiles of cellular phosphoinositides. Biochim. Biophys. Acta. 1862: 513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masters J. R., and Stacey G. N.. 2007. Changing medium and passaging cell lines. Nat. Protoc. 2: 2276–2284. [DOI] [PubMed] [Google Scholar]
- 21.Matsunobu T., Okuno T., Yokoyama C., and Yokomizo T.. 2013. Thromboxane A synthase-independent production of 12- hydroxyheptadecatrienoic acid, a BLT2 ligand. J. Lipid Res. 54: 2979–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bligh E. G., and Dyer W. J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 23.Murphy R. C. 2015. Eicosanoids and bioactive lipid mediators. In Tandem Mass Spectrometry of Lipids: Molecular Analysis of Complex Lipids. S. Gaskell, editor. Royal Society of Chemistry, London. 40–74. [Google Scholar]
- 24.Murphy R. C., Okuno T., Johnson C. A., and Barkley R. M.. 2017. Analysis of double bond positions in polyunsaturated fatty acids. Anal. Chem. 89: 8545–8553. [DOI] [PubMed] [Google Scholar]
- 25.Liebisch G., Vizcaíno J. A., Köfeler H., Trötzmuller M., Griffiths W. J., Schmitz G., Schmitz G., Spener F., and Wakelam M. J.. 2013. Shorthand notation for lipid structures derived from mass spectrometry. J. Lipid Res. 54: 1523–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin S. A., Gijon M. A., Voelker D. R., and Murphy R. C.. 2014. Measurement of lysophospholipid acyltransferase activities using substrate competition. J. Lipid Res. 55: 782–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raphael W., and Sordillo L. M.. 2013. Dietary polyunsaturated fatty acids and inflammation: The role of phospholipid biosynthesis. Int. J. Mol. Sci. 14: 21167–21188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calder P. C. 2017. Omega-3 fatty acids and inflammatory processes: from molecules to man. Biochem. Soc. Trans., 45: 1105–1115. [DOI] [PubMed] [Google Scholar]
- 29.Wong S. W., Kwon M-J., Choi A. M. K., Kim H-P., Nakahira K., and Hwang D. H.. 2009. Fatty acids modulate Toll-like receptor 4 activation through regulation of receptor dimerization and recruitment into lipid rafts in a reactive oxygen species-dependent manner. J. Biol. Chem. 284: 27384–27392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka M., Suganami T., Kim-Saijo M., Toda C., Tsuiji M., Ochi K., Kamei Y., Minokoshi Y., and Ogawa Y.. 2011. Role of central leptin signaling in the starvation-induced alteration of B-cell development. J. Neurosci. 31: 8373–8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith W. L., Urade Y., and Jakobsson P. J.. 2011. Enzymes of the cyclooxygenase pathways of prostanoid biosynthesis. Chem. Rev. 111: 5821–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakschik B. A., Morrison A. R., and Sprecher H.. 1983. Products derived from 5,8,11-eicosatrienoic acid by the 5-lipoxygenase-leukotriene pathway. J. Biol. Chem. 258: 12797–12800. [PubMed] [Google Scholar]
- 33.Evans J. F., Nathaniel D. J., Zamboni R. J., and Ford-Hutchinson A. W.. 1985. Leukotriene A3. A poor substrate but a potent inhibitor of rat and human neutrophil leukotriene A4 hydrolase. J. Biol. Chem. 260: 10966–10970. [PubMed] [Google Scholar]
- 34.Lefkowith J. B., Jakschik B. A., Stahl P., and Needleman P.. 1987. Metabolic and functional alterations in macrophages induced by essential fatty acid deficiency. J. Biol. Chem. 262: 6668–6675. [PubMed] [Google Scholar]
- 35.Hammarström S. 1981. Conversion of 5,8,11-eicosatrienoic acid to leukotrienes C3 and D3. J. Biol. Chem. 256: 2275–2279. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.