Abstract
Cholesterol 25-hydroxylase (CH25H) catalyzes the production of 25-hydroxycholesterol (25-HC), an oxysterol that can play an important role in different biological processes. However, the mechanisms regulating CH25H expression have not been fully elucidated. In this study, we determined that CH25H is highly expressed in mouse liver and peritoneal macrophages. We identified several liver X receptor (LXR) response elements (LXREs) in the human CH25H promoter. In HepG2 cells, activation of LXR by 25-HC or other oxysterols and synthetic ligands [T0901317 (T317) and GW3965] induced CH25H protein expression, which was associated with increased CH25H mRNA expression. 25-HC or T317 activated CH25H transcription in an LXRE-dependent manner. Thus, high-expressing LXRα or LXRβ activated CH25H expression, and the activation was further enhanced by LXR ligands. In contrast, inhibition of LXRα/β expression attenuated 25-HC or T317-induced CH25H expression. Deficiency of interferon γ expression reduced, but did not block, LXR ligand-induced hepatic CH25H expression. Activation of LXR also substantially induced macrophage CH25H expression. In vivo, administration of GW3965 to mice increased CH25H expression in both liver and peritoneal macrophages. Taken together, our study demonstrates that 25-HC can activate CH25H expression in an LXR-dependent manner, which may be an important mechanism to exert the biological actions of 25-HC.
Keywords: hepatocytes, 25-hydroxycholesterol, interferon-γ, liver X receptor
25-Hydroxycholesterol (25-HC) is produced from cholesterol. Although several cytochrome P450 enzymes may complete the conversion of cholesterol into 25-HC in vitro, cholesterol 25-hydroxylase (CH25H) has been demonstrated to play a critical role in the production of 25-HC, both in vitro and in vivo (1). 25-HC can play an important role in different biological processes, particularly in cholesterol metabolism. For instance, at the cellular level, 25-HC inhibits the activity of HMG-CoA reductase (HMGCR) to reduce cholesterol biosynthesis through inactivation of SREBP2 (2). However, mice with deficiency of CH25H expression have intact cholesterol metabolism (3), which challenges whether 25-HC physiologically functions as a negative regulator of SREBP2 activity and cholesterol biosynthesis. 25-HC also substantially enhances cholesterol esterification by activating ACAT (4).
Liver X receptor (LXR) α and β are ligand-activated transcription factors. LXR can play several important roles in the regulation of macrophage cholesterol metabolism and hepatic lipogenesis (5). For instance, LXR activation by synthetic ligands increases ABCA1 or ABCG1 to enhance excess cellular free cholesterol efflux to the extracellular cholesterol acceptor, apoA-I, or HDL, thereby inhibiting formation of lipid-laden macrophage/foam cells and the development of atherosclerosis (6). In addition to synthetic ligands such as T0901317 (T317) and GW3965, several oxysterols, including 25-HC, can function as endogenous LXR ligands. Thus, 25-HC can also induce macrophage ABCA1/ABCG1 expression in an LXR-dependent manner (7). However, the effect of 25-HC on foam-cell formation appears controversial. In vitro, treatment of macrophages with 25-HC increases cellular cholesterol accumulation and foam-cell formation. In vivo, a high 25-HC level is found in the atherosclerotic lesion areas of ApoE deficient (ApoE−/−) mice (8). Meanwhile, activation of LXR induces expression of SREBP-1 and its target genes in hepatocytes, such as FASN and acetyl-CoA carboxylase 1 (ACC1), which can result in severe lipid accumulation in the liver (9).
The effects of 25-HC on inflammation may need more investigation since the controversial results have also been reported. Activation of macrophages lacking CH25H expression leads to overproduction of inflammatory interleukin-1 (IL-1) family cytokines (3), which indicates the anti-inflammatory effects of 25-HC. 25-HC also reduces lipopolysaccharide (LPS)-induced macrophage TNF-α expression and secretion (10). In contrast, 25-HC enhances polyinosinic:polycytidylic acid-induced macrophage IL-6 production (11). In endothelial cells, 25-HC induces expression of adhesion molecules including intercellular adhesion molecule 1, vascular cell adhesion molecule 1, and E-selectin, in an LXR-independent manner (12). 25-HC is also involved in other immunological processes. For instance, 25-HC secreted from activated macrophages can suppress Toll-like receptor (TLR)-induced immunoglobulin A production (13).
Expression of CH25H protein in bone marrow-derived dendritic cells and macrophages can be upregulated by IFNs (14). This finding has led to identification of CH25H as one of the IFN-stimulated genes and 25-HC as a potent inhibitor of viral infection (15, 16).
Cellular 25-HC production is mainly determined by CH25H activity, indicating that CH25H expression can greatly influence 25-HC actions. For example, suppression of the IL-1 family cytokines and immunoglobulin A production by 25-HC is substantially impaired in mice lacking CH25H expression (3, 13). Macrophage CH25H expression can be activated by TLR4 ligands (e.g., LPS) and IFNs via an LXR-independent manner (16). However, it remains unknown whether CH25H expression can be regulated by other mechanisms. Interestingly, injection of mice with TLR ligand induces CH25H expression in multiple tissues, with the greatest effect in the liver. LPS treatment also elevates serum 25-HC levels, which solely depend on CH25H expression (13). Collectively, these findings indicate that both macrophages and hepatocytes may be an important source for 25-HC production. Indeed, we determined high CH25H expression in mouse liver and peritoneal macrophages in this study. By completing a sequence alignment analysis, we identified several LXR response elements (LXREs) in the human CH25H promoter. Combining the facts that 25-HC is an endogenous LXR ligand and that both LXR expression and oxysterol production can be detected in the cells with high cholesterol metabolism activity, such as hepatocytes and macrophages, we hypothesized that 25-HC can activate CH25H expression in an LXR-dependent manner, which may be an important mechanism to exert the biological actions of 25-HC.
MATERIALS AND METHODS
Reagents
25-HC, 22(R)-hydroxycholesterol and 22(S)-hydroxycholesterol were purchased from Sigma-Aldrich (St. Louis, MO). Synthetic LXR ligands, GW3965 and T317, were purchased from Cayman Chemical (Ann Arbor, MI). Rabbit anti-CH25H, anti-SREBP2, anti-FASN, anti-HMGCR, anti-β-actin and anti-GAPDH polyclonal antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX). Rabbit anti-ABCA1 polyclonal antibody was purchased from Novus Biologicals (Littleton, CO). Rabbit anti-LXRα, anti-LXRβ and anti-ABCG1 polyclonal antibodies were purchased from Proteintech (Chicago, IL). Dual-Luciferase assay kit was purchased from Promega (Madison, WI).
Cell lines
RAW264.7 and HepG2 cell lines were purchased from ATCC (Manassas, VA) and cultured in complete RPMI 1640 and DMEM medium containing 10% FBS, 50 μg/ml streptomycin, 100 units/ml penicillin, and 2 mM glutamine, respectively. Peritoneal macrophages and primary hepatocytes were isolated from male C57BL/6 mice and IFN-γ deficient (IFN-γ−/−) mice (C57BL/6 background), respectively, as described (17, 18). The protocols for experiments with mice were approved by the Ethics Committee of Nankai University and conformed to the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health. Cells received treatment in serum-free medium.
cDNA synthesis and quantitative real time PCR
Total RNA was extracted from cells followed by cDNA synthesis as described (19). Real-time PCR was conducted using SYBR Green PCR Master Mix (Bio-Rad) and the following primers: homo-CH25H forward, 5′-ATGTTGACCACGTGGAAGGT-3′, and homo-CH25H backward, 5′-TGGGAACTGTTTTCTTTGGG-3′; mus-CH25H forward, 5′-CCAGCTCCTAAGTCACGTC-3′, and mus-CH25H backward, 5′-CACGTCGAAGAAGGTCAG-3′; homo-GAPDH forward, 5′-ACAACTTTGGCATTGTGAA-3′, and homo-GAPDH backward, 5′-GATGCAGGGATGATGTTCTG-3′; mus-β-actin forward, 5′-ATGGAGGGGAATACAGCCC-3′, and mus-β-actin backward, 5′-TTCTTTGCAGCTCCTTCGTT-3′. CH25H mRNA expression was normalized by GAPDH or β-actin mRNA in the corresponding samples.
Western blot analysis
Expression of CH25H, LXRα, LXRβ, SREBP1, SREBP2, FASN, HMGCR, ABCA1, ABCG1, and GAPDH protein was determined by Western blot with cellular proteins extracted from HepG2 cells, primary hepatocytes, RAW264.7 cells, peritoneal macrophages, or mouse liver as described (20). Briefly, after treatment, cells were washed with PBS and then lysed in an ice-cold lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 1 mM PMSF, 50 mM sodium fluoride, 1 mM sodium orthovanadate, 50 μg/ml aprotinin and leupeptin). A piece of mouse liver was removed and homogenized in the same lysis buffer above. Cellular lysate or liver homogenate was centrifuged for 10 min at 4°C at 14,000 g with a Microfuge. The supernatant was collected as the whole cellular extract or whole tissue protein. After determination of concentration, an equal amount of protein from each sample was loaded on a sodium dodecyl sulfate-polyacrylamide gel for electrophoresis. After electrophoresis, the proteins were transferred onto a nylon-enhanced nitrocellulose membrane. The membrane was blocked with a solution of 0.1% Tween 20/PBS (PBS-T) containing 5% dry fat-free milk for 1 h followed by incubation with primary antibody overnight at 4°C. After reblocking with PBS-T containing 5% milk, the blot was incubated with goat HRP-conjugated anti-rabbit secondary antibody or rabbit HRP-conjugated anti-mouse secondary antibody for 1 h at room temperature. After washing three times for 10 min each time with PBS-T, the membrane was incubated for 1 min in a mixture of equal volume of Western blot chemiluminescence reagent 1 and 2. The membrane was then exposed to X-ray film or subjected to C-DiGit Blot Scanner (Li-cor, Lincoln, NE).
Immunofluorescent staining
After treatment, expression of CH25H, HSPA5, or ATP1A1 protein in HepG2 cells was determined by immunofluorescent staining as described (21). Briefly, cells were fixed with 4% paraformaldehyde for 30 min, washed with PBS for 10 min, and blocked with 2% BSA for 2 h at room temperature. After incubation with primary antibody overnight at 4°C, cells were incubated with either Rhodamine- or FITC-conjugated secondary antibody for 2 h at room temperature. Cells were also stained with DAPI solution for the nucleus. Cells were observed under a fluorescence microscope (Leica, Wetzlar, Germany), and the images were photographed.
Construction of LXRα or LXRβ expression vector, normal or mutated CH25H promoter(s), and determination of promoter activity
The cDNAs encoding human LXRα and LXRβ cloned into pEGFP-C2 (C2) vector were constructed, and expression of exogenous LXRα and LXRβ protein was confirmed by Western blot as described (20).
The DNA for the human CH25H promoter (from −962 to +64) was generated by PCR with genomic DNA isolated from HepG2 cells and the following primers: forward, 5′-GGTACCTTGACGAACAACGCAGGTGG-3′ backward, 5′-GATATCGAGCAGTTGTGGCAGCTCAT-3′. The DNA was then ligated into pGL4.10 luciferase reporter vector, and the constructed normal CH25H promoter was named pCH25H. The promoters with LXRE mutation(s) as indicated in the inserted box in Fig. 3E were constructed with pCH25H and the primers containing mutated sequences using the Phusion Site-Directed Mutagenesis kit (New England Biolabs).
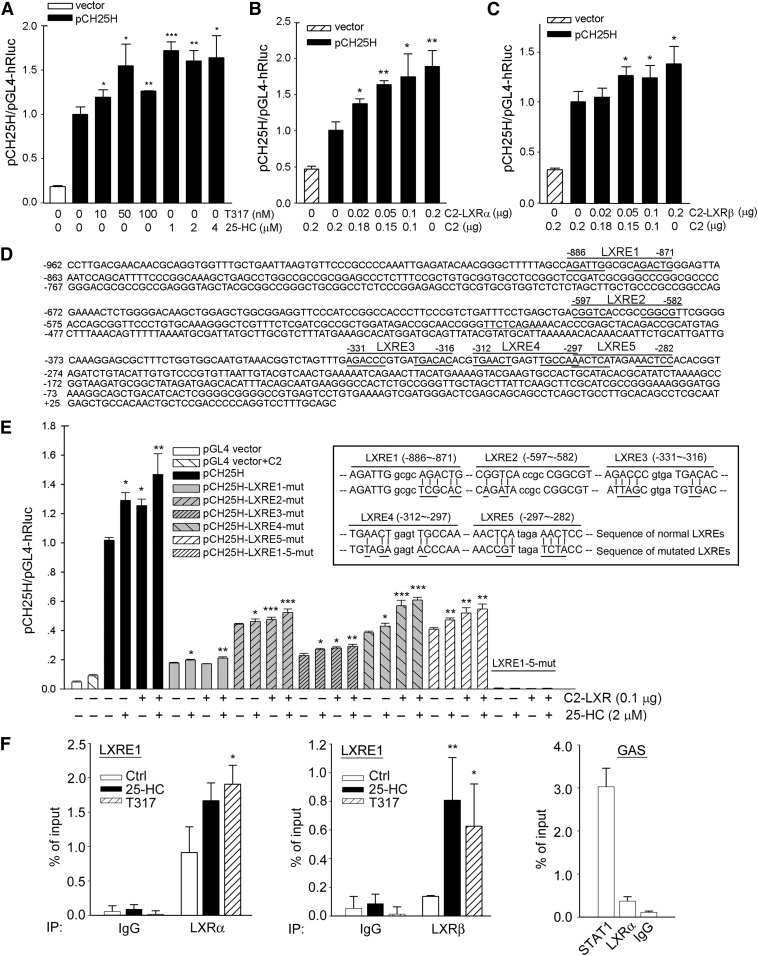
Fig. 3.
LXR activation induces CH25H transcription in an LXRE-dependent manner. A: HepG2 cells were transfected with the normal human CH25H promoter (pCH25H) for 4 h followed by treatment with T317 or 25-HC at the indicated concentrations overnight. B, C: HepG2 cells in 48-well plates were cotransfected with pCH25H plus LXRα or LXRβ expression vector for 20 h. D: The sequence of human CH25H promoter (from −962 to +64) with indication of positions for LXRE1, 2, 3, 4, and 5, and the 6 nucleotides on each side of these LXREs were underlined. E: Cells were transfected with DNA for normal pCH25H promoter or mutated pCH25H promoter(s) (the mutated nucleotides in each LXRE were underlined in the sequences in the inserted box) plus LXRα/β expression vectors for 4 h followed by the indicated treatment for 16 h. Cells were also transfected with Renilla luciferase for internal control. After treatment, cellular proteins were extracted and used to determine activity of firefly and Renilla luciferase with the Dual-Luciferase Reporter Assay System. The promoter activity of each sample was normalized to the promoter activity of the control sample [normal pCH25H alone (A, E) or normal pCH25H plus C2 empty vector (B, C)]. The activity of the control sample was defined as 1. *P < 0.05; **P < 0.01; ***P < 0.001 vs. normal pCH25H alone (A, E) or normal pCH25H plus C2 empty vector (B, C) in the corresponding group (n = 4). F: After treatment with T317 (100 nM) or 25-HC (2 μM) for 12 h, chromatin was isolated from HepG2 cells followed by immunoprecipitation with normal IgG (negative control), anti-LXRα, anti-LXRβ, or anti-STAT1 antibody (positive control). The qPCR was conducted with the primers for LXRE1 and GAS. The primers for LXRE1 (from −886 to −871) are, forward (from −988 to −967), 5′-AGGCTTCAGGAACTCACATCTC-3′, and reverse (from −884 to −867), 5′-CTCCCAGTCTGCGCCAAT-3′; for GAS (from −513 to −505, the conserved sequence for GAS is TTCNNNGAA and central complementary) are, forward (from −623 to −606), 5′-CCTTCCCGTCTGATTTCC-3′, and reverse (from −358 to −341), 5′-CCGTTTACATTGCCACCA-3′. *P < 0.05; **P < 0.01 vs. control in the corresponding group (n = 3).
To determine normal or mutated pCH25H promoter activity, HepG2 cells were transfected with DNA for normal or mutated pCH25H promoter plus DNA for Renilla (for normalization of transfection efficiency) using LipofectamineTM 2000 (Invitrogen). After 20 h of transfection plus treatment, cells were lysed with the lysis buffer supplied in the Dual-Luciferase Reporter Assay System (Promega, Madison, WI). The cellular lysate was then used to determine firefly and Renilla luciferase activity as described (22). The promoter activity in each sample was initially defined as the ratio of firefly luciferase activity to Renilla luciferase activity in the same sample, and then the promoter activity of each sample was further normalized to the promoter activity of the control group (normal pCH25H promoter or normal pCH25H plus the C2 empty vector), and the promoter activity in the control group was defined as 1.
CRISPR-Cas9-mediated deletion of LXRα or LXRβ expression
HepG2 cells lacking LXRα or LXRβ expression were established using clustered regulatory interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) technology (23). The guide RNA was designed by using the online CRISPR Design Tool (http://tools.genome-engineering.org). The sequences of the guide oligos for LXRα and LXRβ are 5′-TCGGCTTCGCAAATGCCGTC-3′ and 5′-ACCCCGGCAGGCATAGCGCC-3′, respectively. After ligation, HepG2 cells were transfected with plasmid of pSpCas9 (BB)-2A-Puro vector or vector ligated with guide oligos. The selection of mutated clonal cell lines was completed using the standard protocol (23), and the deletion of target gene expression was confirmed by Western blot. The cells lacking LXRα and LXRβ expression were defined as CRISPR-LXRα and CRISPR-LXRβ cells, respectively, and the corresponding control cells were defined as CRISPR-Ctrl cells. To further determine the role of LXRs in the CH25H-induction, CRISPR-LXRα cells were transfected with LXRβ siRNA followed by T317 treatment. The siRNA sequence against LXRβ was purchased from Santa Cruz Biotechnology (catalog no. sc-45316).
ChIP-qPCR analysis
The binding of LXREs in the human CH25H promoter with LXRα or LXRβ protein was determined by chromatin immunoprecipitation (ChIP)-quatitative (q)PCR methods. Briefly, 5x107 HepG2 cells were treated with 25-HC or T317 overnight followed by isolation of chromatin (22). The input PCR was conducted with DNA extracted from the sonicated chromatin after reversal of the cross-linking. Immunoprecipitation was conducted with the same amount of chromatin from each sample based on the input and anti-LXRα, anti-LXRβ or anti-STAT1 (as a positive control antibody) polyclonal antibody, or normal IgG (as a negative control antibody). The qPCR was conducted with the detailed protocol as described (24) and the corresponding primers are listed in the legend of Fig. 3.
Determination of cellular total cholesterol and triglyceride levels
HepG2 cells in serum-free medium were treated with T317 or 25-HC for 16 h. After treatment, total cellular proteins and total lipids were extracted, followed by determination of total cholesterol and triglyceride levels using assay kits, and normalized by cellular protein levels (25).
Isolation of primary hepatocytes and in vivo studies
C57BL/6 wild-type mice and IFN-γ deficient (IFN-γ−/−, C57BL/6 background) mice were purchased from the Animal Center of Nanjing University and the protocol for the animal study was approved by the Animal Ethics Committee of Nankai University.
Mouse primary hepatocytes were isolated from C57BL/6 or IFN-γ−/− mice by a collagenase perfusion method. Briefly, after the animals were anesthetized, the midline laparotomy was performed, and the inferior vena cava was cannulated with an angiocatheter. The liver was then perfused with 1 ml heparin (320 U/ml), 40 ml solution I (Kreb’s solution containing 0.1 mM EGTA) and 30 ml solution II (Kreb’s solution containing 2.74 mM CaCl2 and 0.05% collagenase I) at 37°C, sequentially. The perfused liver was then passed through a 400 μm screening size filter by flushing with cold DMEM medium. The isolated hepatocytes were collected after centrifuge for 5 min at 50 g, resuspended with DMEM medium, and plated in 6-well plates (the cell density is ∼1x106 cells/well). The viability of the isolated hepatocytes was ∼90%, which was determined by the trypan blue exclusion method.
To determine whether LXR activation can induce CH25H expression in mouse liver and peritoneal macrophages, C57BL/6 mice (males, ∼8-week old) were randomly divided into two groups (5/group), and they then received the following treatment for one week: control group mice were fed normal chow; GW3965 group mice were fed normal chow containing GW3965, a synthetic LXR ligand, at a dose of 20 mg/day/kg bodyweight. At the end of treatment, mice were anesthetized and euthanized. Mouse blood, peritoneal macrophage, and liver samples were collected. Blood was used to prepare serum. A piece of liver was used to prepare frozen sections or extract total lipids. The hepatic lipid content was determined by Oil Red O staining of frozen liver sections and triglyceride (TG) quantitative assay with the liver total lipid extract as described (26). Total cellular protein and RNA were extracted from a piece of liver or peritoneal macrophages followed by determination of CH25H protein and mRNA expression by Western blot and quantitative real-time PCR, respectively.
Data analysis
All experiments were repeated at least three times, and the representative results are presented. Data were presented as mean ± SE and analyzed by a Student’s t-test using Prism 5 (GraphPad Software, Inc.). The significant difference was considered if P < 0.05 (n ≥ 3).
RESULTS
Activation of LXR induces CH25H protein expression in HepG2 cells
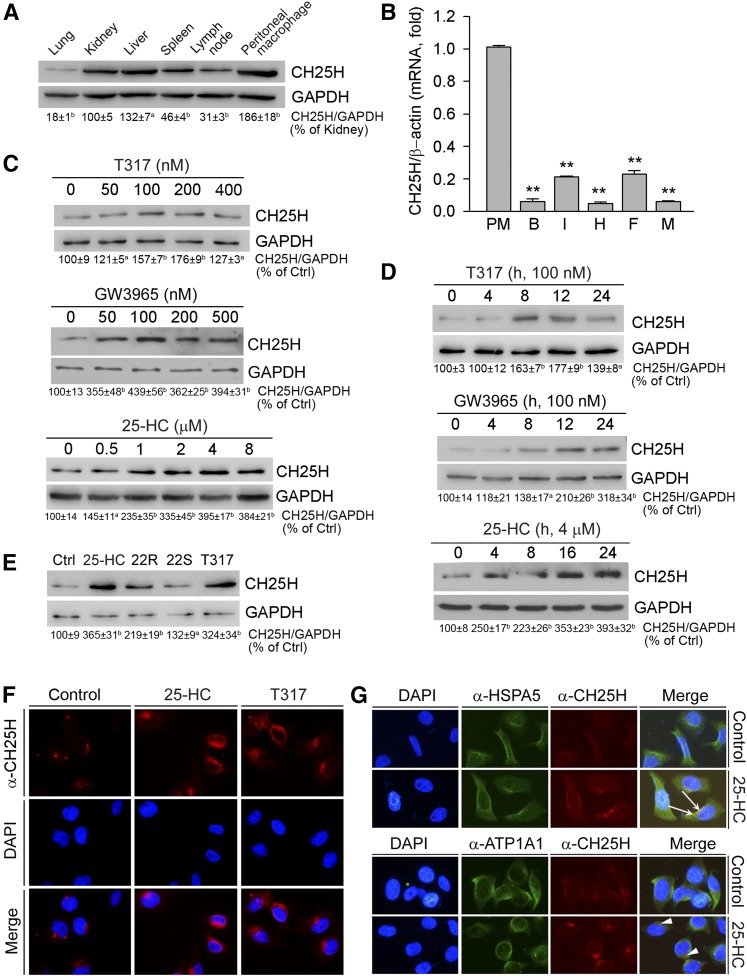
Cellular 25-HC is mainly produced from cholesterol in the reaction catalyzed by CH25H. Therefore, CH25H protein levels in tissue/cell types may determine the rate of 25-HC production. We initially extracted total cellular proteins from mouse tissues and peritoneal macrophages and then determined CH25H protein expression by Western blot. As shown in Fig. 1A, CH25H protein is expressed in multiple tissues, particularly in the liver. Meanwhile, a high expression of CH25H protein was found in mouse peritoneal macrophages. In addition, by using a qRT-PCR method, we determined that CH25H mRNA levels in mouse brain, intestine, heart, adipose tissue, and muscle were much lower than those found in peritoneal macrophages (Fig. 1B). Therefore, in mice, liver or macrophages could be one of the main sources for CH25H expression as well as 25-HC production in vivo.
Fig. 1.
Activation of LXR induces hepatic CH25H protein expression. A: Total cellular proteins were extracted from C57BL/6 mouse lung, kidney, liver, spleen, lymph node, and peritoneal macrophages. B: Total RNA was extracted from mouse peritoneal macrophages (PM), brain (B), intestine (I), heart (H), adipose tissue (F) and muscle (M). C: HepG2 cells in serum-free medium were treated with T317, GW3965, or 25-HC for 16 h. D: HepG2 cells were treated with T317 (100 nM), GW3965 (100 nM), or 25-HC (4 μM) for the indicated times. E: HepG2 cells were treated with 25-HC (2 μM), 22(R)-hydroxycholesterol (22R, 2 μM), 22(S)-hydroxycholesterol (22S, 2 μM), or T317 (200 nM) for 16 h. Expression of CH25H protein (A, C–E) was determined by Western blot. The bands from three repeated experiments were scanned and the density was quantified with statistical analysis; aP < 0.05; bP < 0.01 vs. control (n = 3). Expression of CH25H mRNA (B) was determined by quantitative real-time-PCR; **P < 0.01 vs. peritoneal macrophages (n = 4). F, G: HepG2 cells were treated with 25-HC (2 μM) or T317 (200 nM) for 16 h. Expression of CH25H protein (F, G), or HSPA5 and ATP1A1 protein (G) was determined by immunofluorescent staining. The white arrows indicate the colocalization of CH25H and HSPA5 proteins; the arrowheads indicate the different localization of CH25H protein from the plasma membrane.
By completing a sequence alignment analysis, we identified several LXREs in the promoter of the human CH25H gene. Thus, we speculated that CH25H expression can be induced by LXR activation. If this is the case, 25-HC may also activate CH25H expression in an LXR-dependent manner, based on the fact that 25-HC is an endogenous LXR ligand. To test this hypothesis, we treated HepG2 cells, a human hepatic cell line, with synthetic LXR ligands (T317 and GW3965) and 25-HC at different concentrations for 16 h. The results in Fig. 1C demonstrate that hepatic CH25H protein expression was induced by T317, GW3965, and 25-HC in a concentration-dependent manner. The time course study (Fig. 1D) shows that LXR ligands induced hepatic CH25H expression after a few hours of treatment, with the quickest being by 25-HC. In addition, the induction of CH25H expression by these LXR ligands can last for 24 h after treatment (Fig. 1D).
In addition to 25-HC, other oxysterols can also function as endogenous LXR ligands to activate expression of LXR target genes. For instance, 22(R)-hydroxycholesterol is a potent LXR ligand inducing macrophage ABCA1 expression, whereas its stereo isoform, 22(S)-hydroxycholesterol, can also induce ABCA1 expression but to a lesser extent (7). In this study, we determined that both 22(R)-hydroxycholesterol and 22(S)-hydroxycholesterol induced CH25H expression, with a greater effect by 22(R)-hydroxycholesterol (Fig. 1E). Thus, induction of CH25H expression by oxysterols may depend on their ability to activate LXR.
To further confirm the induction of hepatic CH25H protein expression by LXR activation, we conducted an immunofluorescent staining study with intact HepG2 cells following treatment with 25-HC and T317. Similar to the results found from our Western blots, Fig. 1F shows that 25-HC or T317 substantially increased CH25H protein expression in HepG2 cells.
In addition, by completing a coimmunofluorescent staining study with anti-CH25H and anti-heat-shock protein 5 [(HSPA5), a marker of endoplasmic reticulum (ER)] or anti-ATP1A1 (Na+/K+-transporting ATPase subunit α-1, a marker of plasma membrane) antibodies, we determined that CH25H colocalized with HSPA5 protein (upper panel, Fig. 1G), but not the ATPA1 protein (bottom panel, Fig. 1G), indicating that CH25H likely is an ER protein.
Activation of LXR induces CH25H expression at the transcriptional level
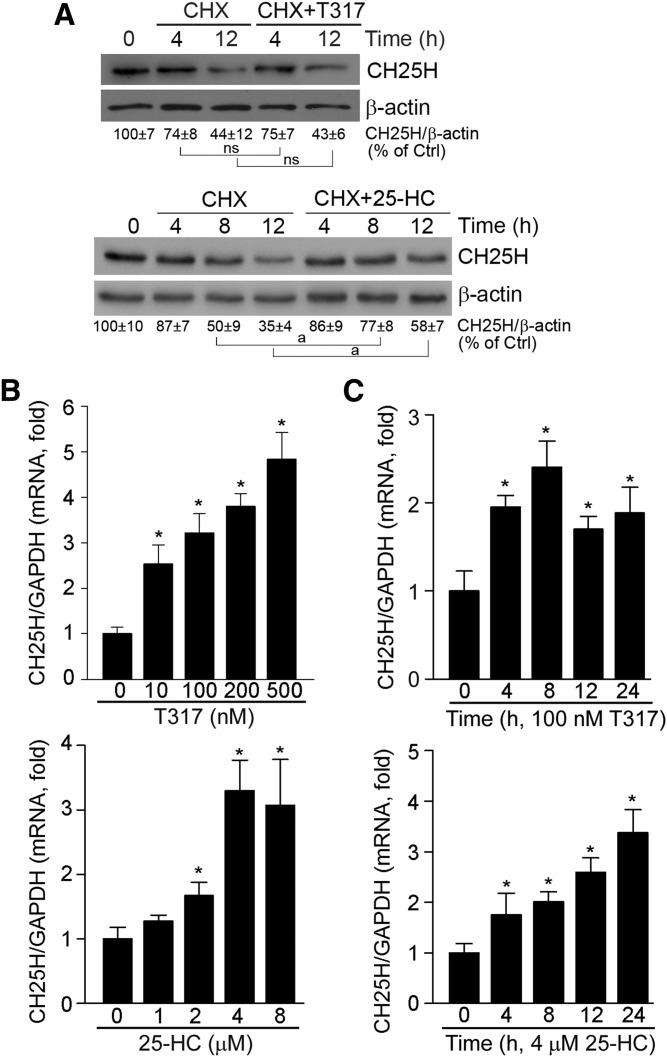
The results above demonstrate that activation of LXR by T317 and 25-HC can induce CH25H protein expression. To determine whether the induction of CH25H protein occurs at a transcriptional or/and posttranscriptional level, we treated HepG2 cells with cycloheximide to arrest protein synthesis in the absence or presence of T317 or 25-HC. We then determined the rate of CH25H protein degradation by Western blot analysis. As shown in Fig. 2A, cycloheximide alone caused cellular CH25H protein to decline with time of treatment. Meanwhile, addition of T317 to cycloheximide did not change the rate of CH25H protein degradation (Fig. 2A, upper panel). In contrast, the cotreatment of cells with cycloheximide and 25-HC caused a decrease in the rate of CH25H protein degradation (Fig. 2A, lower panel). Therefore, both transcriptional and posttranscriptional regulation of CH25H expression by LXR activation occur.
Fig. 2.
Activation of LXR regulates CH25H protein stability and induces CH25H mRNA expression. A: HepG2 cells were treated with cycloheximide (CHX, 5 μM), CHX (5 μM) plus T317 (100 nM), or 25-HC (2 μM) for the indicated times. Total cellular proteins were extracted and CH25H protein expression was determined by Western blot. aP < 0.05 vs. control; ns: not significantly different (n = 3). B, C: HepG2 cells were treated with T317 or 25-HC at the indicated concentrations for 16 h (B) or with T317 (100 nM) or 25-HC (4 μM) for the indicated times (C). Total cellular RNA was extracted and CH25H mRNA expression was determined by quantitative real-time-PCR. *P < 0.05 vs. control in the corresponding group (n = 3).
To further define how LXR activates CH25H transcription, we initially investigated the effect of LXR activation on CH25H mRNA expression. As shown in Fig. 2B and C, both 25-HC and T317 increased CH25H mRNA levels in both concentration- and time-dependent manners. To directly determine the effect of LXR activation on CH25H transcription, we constructed a CH25H promoter, and found that activity of the normal CH25H promoter was increased by T317 or 25-HC treatment (Fig. 3A); and that high-expressing LXRα or LXRβ also induced CH25H promoter activity (Fig. 3B, C).
By completing a sequence alignment analysis, we found five putative LXREs in the proximal region (∼1,000 bp) of the human CH25H promoter (Fig. 3D). LXRE is also named as a direct repeat 4 because the 6 nucleotides on each side, which are separated by any four nucleotides, are repeated. We found that five of six nucleotides on each side are repeated in the LXRE1, which is more than any other LXREs, indicating that LXRE1 could be the most active. To determine the role of these LXREs in CH25H transcription, we constructed several CH25H promoters with a mutation of each LXRE, and a promoter with mutations of all the LXREs (the mutated sequences are shown in the inserted box in Fig. 3E). We determined that the normal CH25H promoter was activated by 25-HC and the high-expressing LXRα/β. Meanwhile, we found that the mutation in each LXRE reduced CH25H promoter activity, with most occurring in the LXRE1 mutation, at the basal level. In addition, we determined that the promoter with the mutation in a single LXRE was still activated by LXR activation. Consequently, the mutations of all five LXREs totally abolished the promoter basal activity, and no induction occurred to this mutated CH25H promoter by LXR activation (Fig. 3E). By comparing other LXREs, we determined that the mutation of LXRE1 reduced promoter basal activity most, and the LXRE1-mutated CH25H promoter is barely activated by LXR activation (Fig. 3E). These results further confirm that LXRE1 is important for CH25H transcription induced by LXR activation.
Next, we conducted a ChIP-qPCR to analyze the binding of LXRE1 with LXRα/β protein. In addition to anti-LXRα or anti-LXRβ antibody, we used normal IgG as a negative control antibody and anti-STAT1 as a positive control antibody because of an IFN-γ-activated site (GAS) element in the CH25H promoter. At the basal level, compared with the substantial binding of the STAT1 protein with GAS, the binding of LXRα or LXRβ protein with LXRE1 was relatively moderate in the control sample, (Fig. 3F). However, treatment of cells with 25-HC or T317 clearly increased the binding of LXRα or LXRβ protein to LXRE1 (left and middle panels, Fig. 3F).
LXR expression is critical for induction of CH25H expression by 25-HC and T317
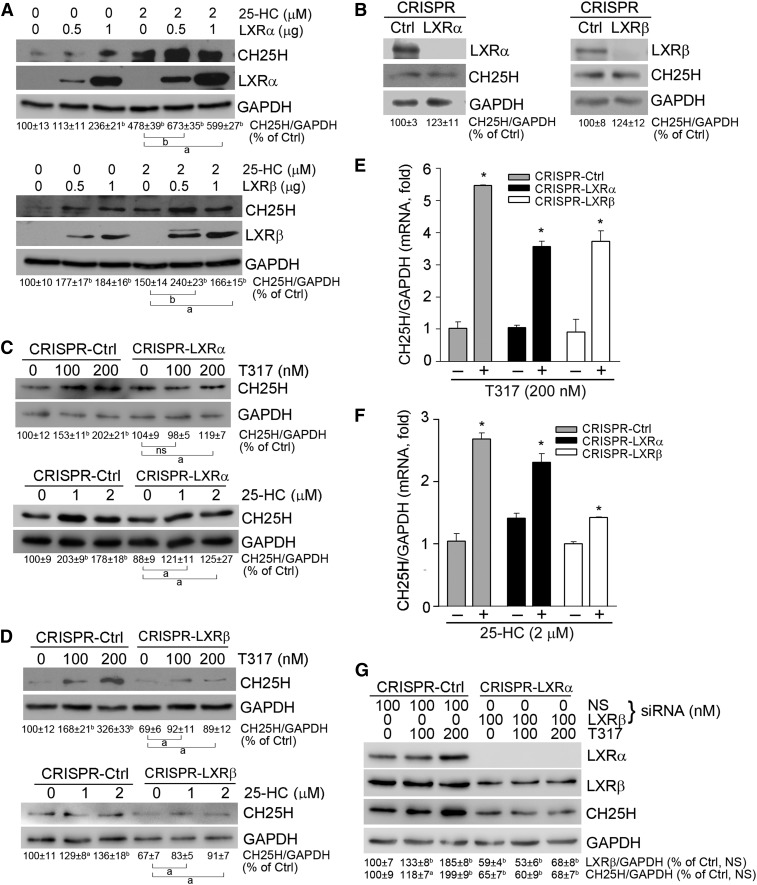
To determine whether the induction of CH25H expression by T317 or 25-HC is directly mediated by LXR, we initially transfected HepG2 cells with an LXRα or LXRβ expression vector, and then treated the transfected cells with 25-HC. Figure 4A shows that high-expressing LXRα or LXRβ induced CH25H expression, and the induction was further enhanced by 25-HC treatment.
Fig. 4.
Activation of CH25H expression by 25-hydroxycholesterol and T317 depends on LXR expression. A: HepG2 cells in 6-well plates were transfected with LXRα or LXRβ expression vector for 4 h followed by 25-HC treatment for 16 h. B: Deficiency of LXRα or LXRβ expression in CRISPR-LXRα or CRISPR-LXRβ cells was confirmed. C–F: CRISPR-Ctrl, CRISPR-LXRα, and CRISPR-LXRβ cells received the indicated treatment for 16 h. G: CRISPR-LXRα cells were transfected with LXRβ siRNA followed by the indicated treatment for 16 h. Expression of LXRα, LXRβ, and CH25H protein (A–D, G) was determined by Western blot. aP < 0.05, bP < 0.01 vs. control or as indicated; ns, not significantly different (n = 3). Expression of CH25H mRNA (E, F) was determined by quantitative real-time-PCR. *P < 0.05 vs. control in the corresponding group (n = 6).
Next, we established an LXRα or LXRβ knockout HepG2 cell line by the CRISPR/Cas9 method (23), and defined them as CRISPR-LXRα or CRISPR-LXRβ cells. The deletion of LXRα or LXRβ protein expression in these cells was confirmed by Western blot (Fig. 4B). As shown in Fig. 4C–F, both CH25H protein and mRNA expression were activated by T317 and 25-HC in the control (CRISPR-Ctrl) cells. However, the activation of CH25H protein expression was partially abolished in cells lacking either LXRα or LXRβ expression (Fig. 4C, D). Similarly, activation of CH25H mRNA expression by T317 or 25-HC was reduced in CRISPR-LXRα or CRISPR-LXRβ cells (Fig. 4F, G). These results suggest that either LXRα or LXRβ can induce CH25H expression.
To strengthen our conclusions, we conducted another experiment to determine whether LXR ligands can regulate CH25H expression in cells with inhibition of both LXRα and LXRβ expression. CRISPR-LXRα cells were transfected with LXRβ siRNA followed by T317 treatment. Compared with the T317-induced CH25H expression, which was associated with increased LXRβ expression in CRISPR-Ctrl cells (Fig. 4G, left panel), T317 barely activated CH25H expression in CRISPR-LXRα cells transfected with LXRβ siRNA (Fig. 4G, right panel). Taken together, the data shown in Figs. 3 and 4 strongly suggests that CH25H expression can be activated by 25-HC or T317 in an LXR-dependent manner.
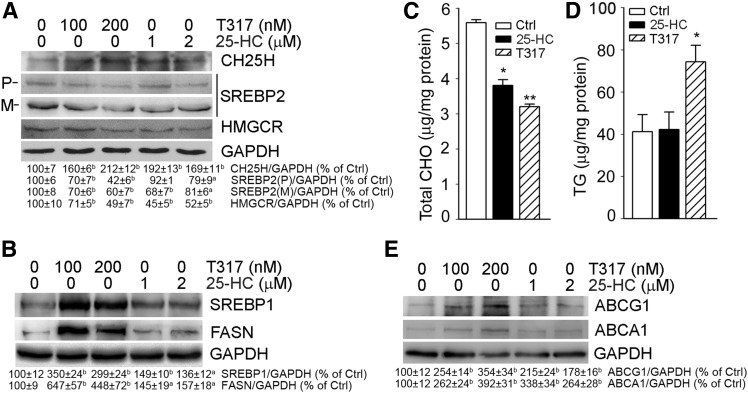
Activation of LXR has been demonstrated to play an important role in lipid metabolism. For instance, LXRα induces lipogenesis by activating SREBP1, which consequently enhances FASN expression (9). In contrast to activation of LXR and SREBP1 expression, 25-HC inactivates SREBP2 to inhibit HMGCR expression and cholesterol biosynthesis (2). In this study, we determined the effect of 25-HC or T317 on cholesterol and TG biosynthesis in HepG2 cells. After treatment with T317 or 25-HC overnight, we found that both T317 and 25-HC inactivated SREBP2 by reducing the precursor of SREBP2 and mature SREBP2. Consequently, HMGCR expression was reduced by both T317 and 25-HC (Fig. 5A). Associated with inhibition of HMGCR expression, we found that cellular total cholesterol levels were reduced (Fig. 5C). Meanwhile, we observed that T317 and 25-HC influenced expression of SREBP-1 and FASN differently. T317 substantially induced SREBP-1 and FASN expression, whereas 25-HC had a slight effect on both (Fig. 5B). As a result, cellular TG levels were increased by T317, but not by 25-HC (Fig. 5D). Activation of LXR can induce ABCA1 and ABCG1 expression to enhance cellular cholesterol efflux. We also determined that both T317 and 25-HC increased ABCA1 and ABCG1 levels in HepG2 cells (Fig. 5E), which may be another mechanism contributing to the reduction of cellular cholesterol levels in response to T317 or 25-HC treatment (Fig. 5C).
Fig. 5.
T317 and 25-HC regulate expression of the genes for biosynthesis of cholesterol or triglyceride and cholesterol efflux in HepG2 cells. A, B, E: HepG2 cells were treated with T317 and 25-HC for 16 h. Expression of cholesterol biosynthesis related genes [precursor of SREBP2 (P) and mature SREBP2 (M), HMGCR] (A), triglyceride biosynthesis related genes (SREBP1 and FASN) (B), and cholesterol efflux related genes (ABCA1 and ABCG1) (E) were determined by Western blot. aP < 0.05, bP < 0.01 vs. control (n = 3). C, D: HepG2 cells were treated with T317 (200 nM) or 25-HC (2 μM) for 16 h followed by determination of cellular total cholesterol (CHO) levels (C) and triglyceride (TG) levels (D) using assay kits. *P < 0.05; **P < 0.01 vs. control in the corresponding groups (n = 3).
Deficiency of IFN-γ expression reduces but does not block LXR-induced hepatic CH25H expression
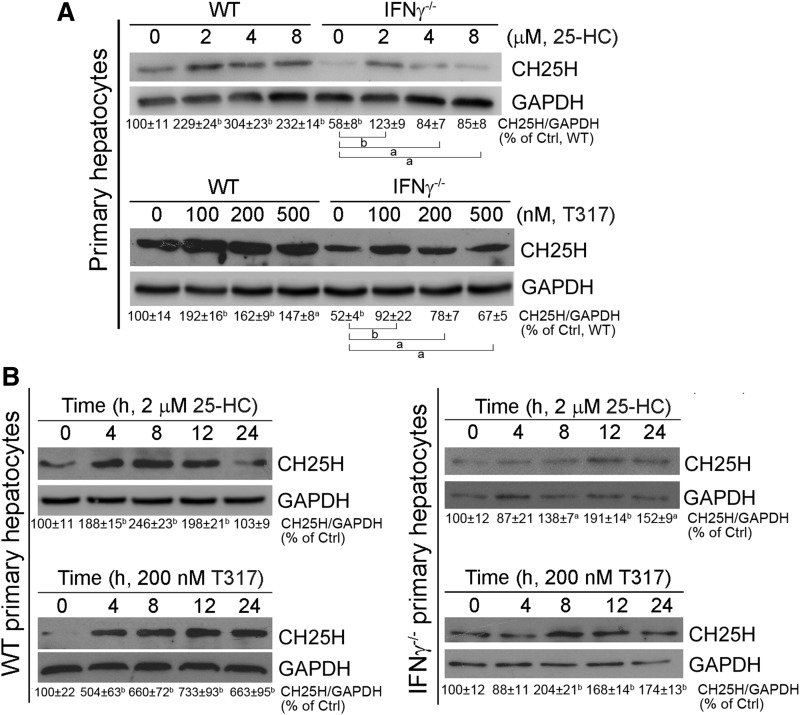
Expression of CH25H has been reported to be induced in LPS- or IFN-activated macrophages. We previously reported that IFN-γ is an LXR target gene (22). Thus, we speculated that IFN-γ might be involved in LXR-induced CH25H expression. To address this hypothesis, we isolated primary hepatocytes from wild-type and IFN-γ−/− mice, and treated these cells with 25-HC or T317. We found that deficiency of IFN-γ expression reduced CH25H expression at basal levels in control cells, indicating the importance of IFN-γ in controlling CH25H expression. However, expression of CH25H was still activated by 25-HC or T317 in IFN-γ−/− cells (Fig. 6A). In addition, we determined that 25-HC or T317 induced CH25H expression in a time-dependent manner in hepatocytes isolated from wild-type or IFN-γ−/− mice (Fig. 6B). Taken together, the results shown in Fig. 6 suggest that IFN-γ is important for CH25H expression, but the induction of CH25H expression by T317 or 25-HC occurs in an IFN-γ-independent manner.
Fig. 6.
Lack of IFN-γ expression reduces but does not abolish LXR-activated hepatic CH25H protein expression. Primary hepatocytes were isolated from wild-type and IFN-γ−/− mice. A: comparison of CH25H protein basal levels or induction of CH25H protein expression by 25-HC or T317 treatment between wild-type and IFN-γ−/− hepatocytes. B: Wild-type and IFN-γ−/− hepatocytes were treated with 25-HC (2 μM) and T317 (200 nM) for the indicated times. Expression of CH25H protein was determined by Western blot. aP < 0.05, bP < 0.01 vs. control or as indicated (n = 3).
Activation of LXR also induces macrophage CH25H expression
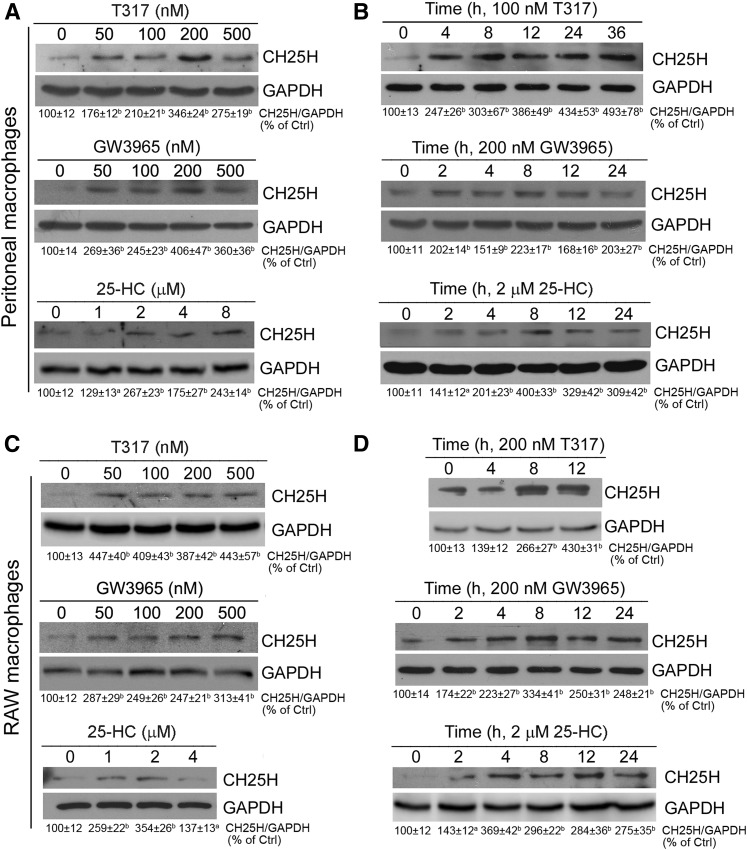
Previous reports have demonstrated that macrophage CH25H is an IFN-induced gene (15, 16). The macrophage is a major cell type producing IFNs, such as IFN-γ. Our results in this study demonstrate that the macrophage is another cell type that highly expresses CH25H (Fig. 1A). Therefore, we completed the following experiment to determine whether activation of LXR can also induce macrophage CH25H protein expression. Similar to hepatocytes, we determined that 25-HC, T317, or GW3965 induced CH25H protein expression in a dose-dependent manner (Fig. 7A), and the induction occurred quickly (Fig. 7B), in peritoneal macrophages collected from wild-type mice.
Fig. 7.
LXR activation induces macrophage CH25H protein expression. Peritoneal macrophages isolated from C57BL/6 wild-type mice (A, B) or RAW264.7 macrophages (C, D) were treated with T317, GW3965, and 25-HC at the indicated concentrations for 16 h (A, C), or with T317 (200 nM), GW3965 (200 nM) and 25-HC (2 μM) for the indicated times (B, D). Expression of CH25H protein was determined by Western blot. aP < 0.05, bP < 0.01 vs. control (n = 3).
In the RAW264.7 macrophage cell line, we also observed that CH25H protein expression was activated by T317, GW3965, or 25-HC in both concentration- and time-dependent manners (Fig. 7C, D). Combining the results we obtained from the studies with mouse primary hepatocytes (Fig. 6) and HepG2 cells (Figs. 1–4), we can make a conclusion that induction of CH25H expression by LXR activation can be independent of cell type or species.
Activation of LXR induces CH25H expression in vivo
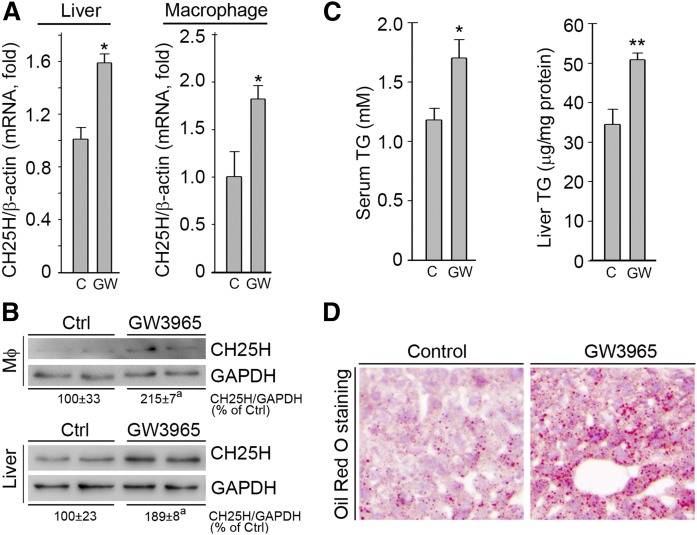
To confirm whether activation of LXR can induce CH25H expression in vivo, particularly in hepatocytes and macrophages, we fed wild-type mice normal chow or normal chow containing GW3965 at a dose of 20 mg/day/kg bodyweight for 1 week. After treatment, we collected mouse blood, liver, and peritoneal macrophage samples individually. Similar to our in vitro studies, we found that expression of CH25H mRNA in liver and macrophages was increased by GW3965 (Fig. 8A). Associated with increased CH25H mRNA expression, CH25H protein expression in liver or macrophages was also elevated by GW3965 treatment (Fig. 8B).
Fig. 8.
LXR activation induces CH25H expression in mouse liver and peritoneal macrophages in vivo. C57BL/6 mice were randomly divided into two groups. Control group was fed normal chow and the GW3965 group was fed normal chow containing GW3965 (20 mg/day/kg bodyweight) for 7 days. At the end of the experiment, mice were anesthetized and euthanized followed by collection of blood, liver, and peritoneal macrophage samples. Expression of CH25H mRNA or protein in peritoneal macrophages and the liver was determined by quantitative real-time-PCR (A) or Western blot (B). Triglyceride levels in serum and the liver were determined by quantitative TG analysis with serum samples and total lipid extracted from the liver (C). The lipid accumulation in the liver was also determined by Oil Red O staining of frozen liver sections (D). aP < 0.05 vs. control (n = 3); *P < 0.05, **P < 0.01 vs. control (n = 5).
Activation of LXR can induce hepatic lipogenesis and hypertriglyceridemia. Similar to previous reports (6), in this study, we also determined that after 1 week of GW3965 treatment, TG levels in both serum and liver were increased (Fig. 8C). Meanwhile, hepatic lipid accumulation induced by GW3965 was confirmed by Oil Red O staining of frozen liver sections (Fig. 8D).
DISCUSSION
25-HC has emerged as an important molecule in different biological processes by functioning as a negative regulator of SREBP-2 activity and cholesterol biosynthesis, a mediator of inflammation, and a potent inhibitor of viral infection. 25-HC can be generated from cholesterol by enzymatic or/and nonenzymatic reactions. Although a very small amount of 25-HC can be found in cholesterol crystals after a long period of autoxidation, it is difficult to find 25-HC in lipoproteins, such as LDL, even under strong oxidizing conditions in vitro (27). However, it is possible to produce 25-HC as a side-product in a few enzymatic reactions. For instance, 25-HC can be produced in the reactions catalyzed by cytochrome P450 27A1 (CYP27A1, sterol 27-hydroxylase), CYP46 (cholesterol 24-hydroxylase) and CYP3A4, the enzymes responsible for production of 27-hydroxycholesterol, 24(S)-hydroxylcholesterol, and 4-hydroxycholesterol, respectively (28–30). However, it still remains unclear whether these enzymes can generate 25-HC in vivo.
CH25H does not belong to the cytochrome P450 enzyme family. It mainly localizes in the ER. The following evidence supports the role of CH25H in the production and actions of 25-HC in vivo. Macrophage CH25H can be upregulated by LPS in a TLR4/TRIF-dependent manner. Compared with controls, much higher concentrations of 25-HC in serum can be determined in healthy volunteers receiving LPS injection (31). The overproduction of IL-1 family cytokines in activated macrophages isolated from CH25H deficient mice can be reduced to normal by 25-HC treatment (3). TLR activation induces immunoglobulin A production in cells lacking CH25H expression. However, the induction is blocked by extracellular 25-HC, indicating that CH25H expression is critical for the production and actions of 25-HC (13).
In this study, we demonstrated that the activated CH25H protein is localized in the ER (upper panel, Fig. 1F), which may be linked to the physiological relevance of its product, 25-HC, in the regulation of cholesterol biosynthesis. In the ER, 25-HC enhances the binding of SCAP (SREBP cleavage-activating protein) to Insigs (insulin-induced genes), thereby abrogating the movement of the SCAP-SREBP-2 complex to the Golgi apparatus, inactivating SREBP-2, and inhibiting HMGCR expression (2).
Regulation of CH25H expression in the immune system, particularly in macrophages, has been well defined. LPS can increase macrophage CH25H mRNA expression and 25-HC production in a TLR4-dependent manner (31). Activation of TLR4 by Kdo2-Lipid A and activation of other TLRs by the corresponding ligands (TLR2 by peptidoglycan from Staphylococcus aureus and TLR2/6 by lipoteichoic acid) can also activate macrophage CH25H expression (11, 13). Treatment of macrophages or dendritic cells with IFN-α-, β-, or γ-induced CH25H expression in a STAT1-dependent manner (14, 16). Taken together, these studies indicate that CH25H expression in the immune system can be regulated by multiple pathways. Interestingly, administration of Kdo2-Lipid A can activate CH25H expression in mouse tissues with the greatest effect on the liver (13), suggesting that CH25H can be expressed by different cell types, and CH25H expression can be regulated by different mechanisms. Consistently, in this study, we observed that activation of LXR by 25-HC and synthetic LXR ligands increased CH25H expression in both hepatocytes and macrophages (Figs. 1, 6, 7). And in vivo, we determined that administration of GW3965 to mice increased CH25H expression in both liver and peritoneal macrophages (Fig. 8).
Expression of macrophage CH25H can be induced by IFNs. Previously, we identified that IFN-γ is an LXR target gene (22). Activation of LXR by ligands induces IFN-γ expression and secretion (22, 32). Therefore, the LXR-induced CH25H expression may be involved by activation of IFN-γ. Indeed, we found that lack of IFN-γ expression reduced the basal levels of CH25H protein, indicating the importance of IFN-γ in the activation of CH25H expression. However, LXR activation still induced CH25H expression in cells lacking IFN-γ expression (Fig. 6). Thus, we conclude that IFN-γ can enhance but not determine LXR-induced CH25H expression. Our results also indicate the interaction between 25-HC and IFN-γ, which may suggest that LXR has different pathways to regulate CH25H expression in a cell type-dependent manner. For instance, in cells highly expressing IFN-γ, such as macrophages, LXR activation can induce CH25H expression in two ways: by direct induction via activation of LXRE in CH25H, and by indirect induction via activation of IFN-γ. However, in cells with high cholesterol metabolic activity that have little IFN-γ expression, the direct induction may dominate the effects of LXR on CH25H expression.
In summary, based on the fact that 25-HC is an endogenous LXR ligand, we identified LXREs in the CH25H promoter, and determined that activation of LXR by ligands including 25-HC can induce CH25H expression at the transcriptional level. In addition, we determined that induction of CH25H expression by LXR activation is independent of cell type, species, and IFN-γ expression. Induction of CH25H expression can occur in vivo in response to LXR ligand administration. We believe that the results in this study advance our understanding of the regulation of CH25H expression as well as 25-HC biological actions.
Footnotes
Abbreviations:
- ACC1
- acetyl-CoA carboxylase 1
- Cas9
- CRISPR-associated protein 9
- ChIP
- chromatin immunoprecipitation
- CH25H
- cholesterol 25-hydroxylase
- CRISPR
- clustered regulatory interspaced short palindromic repeat
- ER
- endoplasmic reticulum
- GAS
- IFN-gamma-activated site
- HMGCR
- HMG-CoA reductase
- 25-HC
- 25-hydroxycholesterol
- IL
- interleukin
- LPS
- lipopolysaccharide
- LXR
- liver X receptor
- LXRE
- LXR response element
- qPCR
- quatitative PCR
- SCAP
- SREBP cleavage-activating protein
- TG
- triglyceride
- TLR
- Toll-like receptor
- T317
- T0901317
This work was supported by the National Natural Science Foundation of China Grants 81473204 and 81773727 to J.H., 81573427 and 81722046 to Y.D., and 31770863 to Y.C.; the Program for Changjiang Scholars and Innovative Research Team in University (IRT13023) and 111 Project B08011 to J.H.; the Tianjin Municipal Science and Technology Commission of China Grant 16JCZDJC34700 to J.H. and 17JCYBJC25000 to Y.C.; the National Institutes of Health (R01DK112971) to Q.R.M; and the Rosenfeld Heart Foundation of New York to D.P.H. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Diczfalusy U. 2013. On the formation and possible biological role of 25-hydroxycholesterol. Biochimie. 95: 455–460. [DOI] [PubMed] [Google Scholar]
- 2.Adams C. M., Reitz J., De Brabander J. K., Feramisco J. D., Li L., Brown M. S., and Goldstein J. L.. 2004. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J. Biol. Chem. 279: 52772–52780. [DOI] [PubMed] [Google Scholar]
- 3.Reboldi A., Dang E. V., McDonald J. G., Liang G., Russell D. W., and Cyster J. G.. 2014. Inflammation. 25-Hydroxycholesterol suppresses interleukin-1-driven inflammation downstream of type I interferon. Science. 345: 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng D., Chang C. C., Qu X., and Chang T. Y.. 1995. Activation of acyl-coenzyme A:cholesterol acyltransferase by cholesterol or by oxysterol in a cell-free system. J. Biol. Chem. 270: 685–695. [DOI] [PubMed] [Google Scholar]
- 5.Calkin A. C., and Tontonoz P.. 2012. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat. Rev. Mol. Cell Biol. 13: 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joseph S. B., McKilligin E., Pei L., Watson M. A., Collins A. R., Laffitte B. A., Chen M., Noh G., Goodman J., Hagger G. N., et al. . 2002. Synthetic LXR ligand inhibits the development of atherosclerosis in mice. Proc. Natl. Acad. Sci. USA. 99: 7604–7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou X., He W., Huang Z., Gotto A. M. Jr., Hajjar D. P., and Han J.. 2008. Genetic deletion of low density lipoprotein receptor impairs sterol-induced mouse macrophage ABCA1 expression. A new SREBP1-dependent mechanism. J. Biol. Chem. 283: 2129–2138. [DOI] [PubMed] [Google Scholar]
- 8.Gold E. S., Ramsey S. A., Sartain M. J., Selinummi J., Podolsky I., Rodriguez D. J., Moritz R. L., and Aderem A.. 2012. ATF3 protects against atherosclerosis by suppressing 25-hydroxycholesterol-induced lipid body formation. J. Exp. Med. 209: 807–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grefhorst A., Elzinga B. M., Voshol P. J., Plösch T., Kok T., Bloks V. W., van der Sluijs F. H., Havekes L. M., Romijn J. A., Verkade H. J., et al. . 2002. Stimulation of lipogenesis by pharmacological activation of the liver X receptor leads to production of large, triglyceride-rich very low density lipoprotein particles. J. Biol. Chem. 277: 34182–34190. [DOI] [PubMed] [Google Scholar]
- 10.Englund M. C., Karlsson A. L., Wiklund O., Bondjers G., and Ohlsson B. G.. 2001. 25-hydroxycholesterol induces lipopolysaccharide-tolerance and decreases a lipopolysaccharide-induced TNF-alpha secretion in macrophages. Atherosclerosis. 158: 61–71. [DOI] [PubMed] [Google Scholar]
- 11.Gold E. S., Diercks A. H., Podolsky I., Podyminogin R. L., Askovich P. S., Treuting P. M., and Aderem A.. 2014. 25-Hydroxycholesterol acts as an amplifier of inflammatory signaling. Proc. Natl. Acad. Sci. USA. 111: 10666–10671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morello F., Saglio E., Noghero A., Schiavone D., Williams T. A., Verhovez A., Bussolino F., Veglio F., and Mulatero P.. 2009. LXR-activating oxysterols induce the expression of inflammatory markers in endothelial cells through LXR-independent mechanisms. Atherosclerosis. 207: 38–44. [DOI] [PubMed] [Google Scholar]
- 13.Bauman D. R., Bitmansour A. D., McDonald J. G., Thompson B. M., Liang G., and Russell D. W.. 2009. 25-Hydroxycholesterol secreted by macrophages in response to Toll-like receptor activation suppresses immunoglobulin A production. Proc. Natl. Acad. Sci. US A. 106: 16764–16769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park K., and Scott A. L.. 2010. Cholesterol 25-hydroxylase production by dendritic cells and macrophages is regulated by type I interferons. J. Leukoc. Biol. 88: 1081–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S. Y., Aliyari R., Chikere K., Li G., Marsden M. D., Smith J. K., Pernet O., Guo H., Nusbaum R., Zack J. A., et al. . 2013. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity. 38: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanc M., Hsieh W. Y., Robertson K. A., Kropp K. A., Forster T., Shui G., Lacaze P., Watterson S., Griffiths S. J., Spann N. J., et al. . 2013. The transcription factor STAT-1 couples macrophage synthesis of 25-hydroxycholesterol to the interferon antiviral response. Immunity. 38: 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou X., Yin Z., Guo X., Hajjar D. P., and Han J.. 2010. Inhibition of ERK1/2 and activation of liver X receptor synergistically induce macrophage ABCA1 expression and cholesterol efflux. J. Biol. Chem. 285: 6316–6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao L., Wang C., Zhang X., Peng L., Liu W., Zhang X., Liu Y., He J., Jiang C., Ai D., et al. . 2016. Hyperhomocysteinemia activates the aryl hydrocarbon receptor/CD36 pathway to promote hepatic steatosis in mice. Hepatology. 64: 92–105. [DOI] [PubMed] [Google Scholar]
- 19.Ma X., Liu Y., Wang Q., Chen Y., Liu M., Li X., Xiang R., Wei Y., Duan Y., and Han J.. 2015. Tamoxifen induces the development of hernia in mice by activating MMP-2 and MMP-13 expression. Biochim. Biophys. Acta. 1852: 1038–1048. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y., Duan Y., Kang Y., Yang X., Jiang M., Zhang L., Li G., Yin Z., Hu W., Dong P., et al. . 2012. Activation of liver X receptor induces macrophage interleukin-5 expression. J. Biol. Chem. 287: 43340–43350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L., Chen Y., Yang X., Yang J., Cao X., Li X., Li L., Miao Q. R., Hajjar D. P., Duan Y., et al. . 2016. MEK1/2 inhibitors activate macrophage ABCG1 expression and reverse cholesterol transport-An anti-atherogenic function of ERK1/2 inhibition. Biochim. Biophys. Acta. 1861: 1180–1191. [DOI] [PubMed] [Google Scholar]
- 22.Wang Q., Ma X., Chen Y., Zhang L., Jiang M., Li X., Xiang R., Miao R., Hajjar D. P., Duan Y., et al. . 2014. Identification of interferon-gamma as a new molecular target of liver X receptor. Biochem. J. 459: 345–354. [DOI] [PubMed] [Google Scholar]
- 23.Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lacazette E. 2017. A laboratory practical illustrating the use of the ChIP-qPCR method in a robust model: Estrogen receptor alpha immunoprecipitation using Mcf-7 culture cells. Biochem. Mol. Biol. Educ. 45: 152–160. [DOI] [PubMed] [Google Scholar]
- 25.Hu W., Zhang W., Chen Y., Rana U., Teng R. J., Duan Y., Liu Z., Zhao B., Foeckler J., Weiler H., et al. . 2016. Nogo-B receptor deficiency increases liver X receptor alpha nuclear translocation and hepatic lipogenesis through an adenosine monophosphate-activated protein kinase alpha-dependent pathway. Hepatology. 64: 1559–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y., Duan Y., Yang X., Sun L., Liu M., Wang Q., Ma X., Zhang W., Li X., Hu W., et al. . 2015. Inhibition of ERK1/2 and activation of LXR synergistically reduce atherosclerotic lesions in ApoE-deficient mice. Arterioscler. Thromb. Vasc. Biol. 35: 948–959. [DOI] [PubMed] [Google Scholar]
- 27.Dzeletovic S., Babiker A., Lund E., and Diczfalusy U.. 1995. Time course of oxysterol formation during in vitro oxidation of low density lipoprotein. Chem. Phys. Lipids. 78: 119–128. [DOI] [PubMed] [Google Scholar]
- 28.Cali J. J., Hsieh C. L., Francke U., and Russell D. W.. 1991. Mutations in the bile acid biosynthetic enzyme sterol 27-hydroxylase underlie cerebrotendinous xanthomatosis. J. Biol. Chem. 266: 7779–7783. [PMC free article] [PubMed] [Google Scholar]
- 29.Lund E. G., Guileyardo J. M., and Russell D. W.. 1999. cDNA cloning of cholesterol 24-hydroxylase, a mediator of cholesterol homeostasis in the brain. Proc. Natl. Acad. Sci. USA. 96: 7238–7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Honda A., Miyazaki T., Ikegami T., Iwamoto J., Maeda T., Hirayama T., Saito Y., Teramoto T., and Matsuzaki Y.. 2011. Cholesterol 25-hydroxylation activity of CYP3A. J. Lipid Res. 52: 1509–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diczfalusy U., Olofsson K. E., Carlsson A. M., Gong M., Golenbock D. T., Rooyackers O., Flaring U., and Bjorkbacka H.. 2009. Marked upregulation of cholesterol 25-hydroxylase expression by lipopolysaccharide. J. Lipid Res. 50: 2258–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma X., Wang Q., Liu Y., Chen Y., Zhang L., Jiang M., Li X., Xiang R., Miao R. Q., Duan Y., and Han J.. 2015. Inhibition of tumor growth by U0126 is associated with induction of interferon-gamma production. Int. J. Cancer. 136: 771–783. [DOI] [PubMed] [Google Scholar]