Abstract
There are no known genetic variants with large effects on susceptibility to major depressive disorder (MDD). Although one proposed study approach is to increase sensitivity by increasing sample sizes, another is to focus on families with multiple affected individuals to identify genes with rare or novel variants with strong effects. Choosing the family-based approach, we performed whole-exome analysis on affected individuals (n = 12) across five MDD families, each with at least five affected individuals, early onset, and prepubertal diagnoses. We identified 67 genes where novel deleterious variants were shared among affected relatives. Gene ontology analysis shows that of these 67 genes, 18 encode transcriptional regulators, eight of which are expressed in the human brain, including four KRAB-A box-containing Zn2+ finger repressors. One of these, ZNF34, has been reported as being associated with bipolar disorder and as differentially expressed in bipolar disorder patients compared to healthy controls. We found a novel variant—encoding a non-conservative P17R substitution in the conserved repressor domain of ZNF34 protein—segregating completely with MDD in all available individuals in the family in which it was discovered. Further analysis showed a common ZNF34 coding indel segregating with MDD in a separate family, possibly indicating the presence of an unobserved, linked, rare variant in that particular family. Our results indicate that genes encoding transcription factors expressed in the brain might be an important group of MDD candidate genes and that rare variants in ZNF34 might contribute to susceptibility to MDD and perhaps other affective disorders.
Keywords: major depressive disorder, whole-exome analysis, psychiatric disorders, transcription factors, gene ontology
INTRODUCTION
Major depressive disorder (MDD) is among the most common psychiatric disorders and the leading cause of disability in the United States [Stewart et al., 2003]. Globally, depression is the third leading contributor to disease burden and is associated with serious comorbidity including increased rates of substance abuse, mortality, and numerous medical complications [Ferrari et al., 2013; Whiteford et al., 2013]. The heritability of MDD has been estimated to be as high as 70% [Kendler et al., 1993] with relative risk to first-degree relatives of probands with MDD at least two-to threefold higher than relatives of controls. This risk is further increased in relatives of probands with earlier onset of MDD, especially for relatives of children diagnosed with prepubertal MDD [Puig-Antich et al., 1989]. Therefore, samples enriched with early-onset cases yield higher power to detect genetic effects [Holmans et al., 2007]. Where exactly to draw the age cutoff, and/or whether or not age should be modeled as a continuous variable, is unclear. It is, however, quite clear from epidemiologic and family studies that prepubertal onset of MDD is uncommon suggesting that, similar to many Mendelian disorders, rare and private genetic variants play a significant role. But, although studies of some psychiatric disorders, such as schizophrenia and autism, have led to the discovery of important candidate disease variants [Rodriguez-Murillo et al., 2012; Jeste and Geschwind, 2014], genetic studies of MDD have yet to reproducibly find any variants with strong genetic effects [Flint and Kendler, 2014; Levinson et al., 2014].
The lack of reproducible genetic findings for MDD can be interpreted a number of ways. The lack of strongly associated variants (e.g., via genome-wide association studies) could mean either that the majority of MDD genetic susceptibility is not due to common variants, or that the number of common variants required to reach the MDD threshold is too large to be detected by the sample sizes reported [Flint and Kendler, 2014]. Furthermore, the presence of multiple MDD subtypes might cause substantial genetic heterogeneity between studies that affect reproducibility. Therefore, it might be helpful to apply stricter phenotypic constraints, such as requiring early onset or multiple affected relatives. We hypothesize that such an approach, when combined with a variant filtering scheme designed to enrich for putative disease-causing variants, can allow us to identify genes with strong effects on MDD susceptibility.
Here, we perform whole-exome analysis of MDD families, with early onset and multiple affected individuals, to identify novel deleterious mutations shared among affected relatives. We sampled at least two individuals from five MDD families. Each family was required to segregate MDD with high (observed) penetrance and contain at least one prepubertal MDD case, as families of such individuals show increased MDD heritability [Puig-Antich et al., 1989; Holmans et al., 2007]. All individuals were phenotypically confirmed by multiple clinical assessments, a measure that has been shown to maximize observed heritability [Kendler et al., 1993]. In addition to choosing a sample in which effect sizes are predicted to be strong, we also designed our variant filtering scheme to enrich for variants that might affect disease. It has been proposed and observed that novel non-synonymous coding variants can strongly impact disease susceptibility [Gilissen et al., 2012; Ku et al., 2013] making them high-priority candidates for being causal alleles. In this study, we focus on novel coding mutations predicted to be deleterious and consider only variants shared among affected individuals in MDD families allowing us to, additionally, use the powerful tool of cosegregation/variant-sharing analysis. We hypothesize that, given our focus on families where genetic effects are predicted to be strong and on mutations predicted to confer strong phenotypic effects, we have reasonable power to detect strong genetic effects, even with a small sample.
RESULTS
Sample Selection
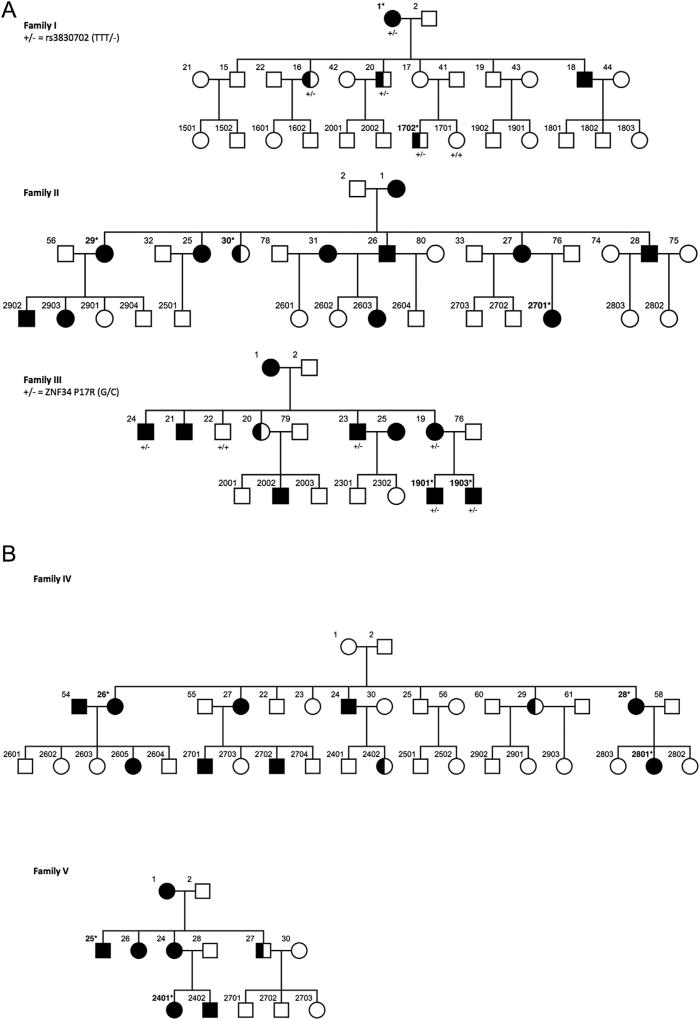
Drawing from a longitudinal study of multigenerational MDD families, in which individuals have undergone repeated and independent clinical assessments up to six times over 30 years, with final diagnosis made blind to any genotype data or clinical status of other family members, by a psychiatrist or psychologist [Warner et al., 1999; Weissman et al., 2005, 2006], we selected five families (Fig. 1) from which to sample affected relatives for variant discovery analysis. To focus on families in which the genetic effects might be particularly large, each of these families had a minimum of five affected individuals. In addition, each family had relatively low median ages of onset—that is, younger than 20 years old and ranging from 12 to 19.8 and an average median of 16.9—and at least one individual with a prepubertal MDD diagnosis (onset before age 12). We chose two individuals for whom DNA was available from each of these five families for whole-exome sequencing, with the exceptions of the large Families II and IV, where we chose to sample three affected individuals (Fig. 1).
FIG. 1.
The complete pedigrees of each of the five families from our study: half shaded shapes represent individuals with an MDD diagnosis at age 12 or below. The individuals chosen for initial exome sequencing are denoted with an asterisk and bold numbers.
Shared Novel Deleterious Variants
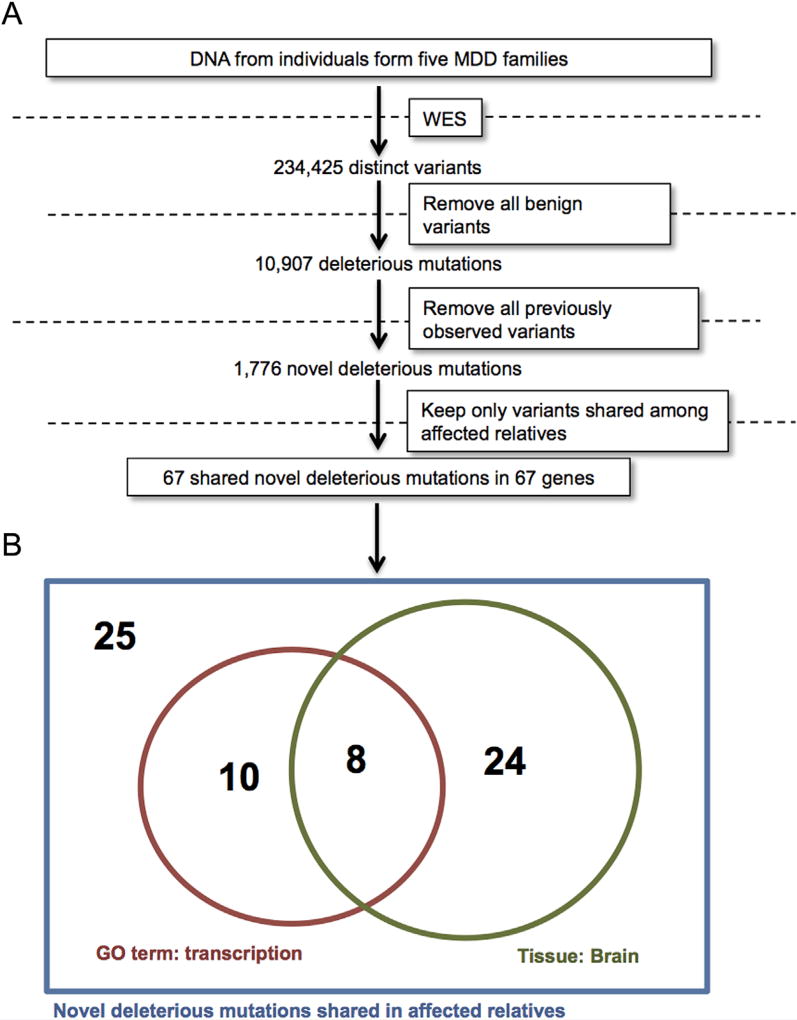
Whole exome sequencing revealed a total of 622,029 exome-wide variants across our sample, 234,425 of which were distinct. Figure 2A summarizes our subsequent filtering scheme. Filtering out all variants that were either intronic, synonymous, or predicted to be benign (see Methods section), left a total of 10,907 “deleterious,” which include variants encoding amino acid substitutions predicted to adversely affect protein function or change total protein length and/or composition (i.e., indels, frameshifts, etc.) (Table I). Novel mutations have been hypothesized and observed to have an increased chance of affecting disease phenotypes [Gilissen et al., 2012; Ku et al., 2013]. Therefore, of the 10,907 deleterious variants, we focused only on the 1,776 variants in this set not found in any examined databases (see Methods section). Of these novel deleterious mutations, only 67 were shared among affected relatives within all families where they were found (while meeting variant quality control requirements [see Methods section]). This observed sharing indicates that these novel variants are not sequencing artifacts (as the chances the same error occurred twice in a family at the same position are exceeding low) and implicates them as potentially being involved in MDD. We, therefore, refer to the genes where these mutations were found as putative MDD candidate genes (Table II). Gene ontology analysis shows that 18 (27%) of these genes encode transcription factors and 32 (48%) are expressed in the human brain (Table II). Joint analysis of the 18 transcription factor-encoding genes and the 32 brain-expressed genes shows that 8 of the 67 putative MDD genes encode transcription factors expressed in the brain (Fig. 2B). Compared to the number of transcription factors expressed in the brain (as designated using gene ontology [GO] analysis outlined in Methods section) this corresponds to an EASE score P-value of 0.007 suggesting significant enrichment. However, the true null distribution for transcription factors is not the list of all GO transcription factors, but rather the list of all transcription factors which would have variants that made it through our filtering scheme by chance from our exome data, which is more difficult to estimate. Therefore, this P-value can only be a, perhaps conservative, approximation. For the eight transcription factor encoding-genes expressed in the brain in our dataset, we checked whether variants of any frequency were found shared among affected relatives in our sample, as these variants might co-segregate with rare variants not captured in our exome sequencing. This was true for only two genes: ZNF34 and the X-linked TCEAL2. The ZNF34 gene (also called KOX32) is expressed in the developing brain [Lorenz et al., 2010] and is a putative bipolar disorder (BPD) candidate gene, as it (i) is differentially expressed in BPD patients relative to healthy controls [Zhao et al., 2015], (ii) contains common variants that show gene-based association with BPD [Lee et al., 2013; Zhao et al., 2015], and (iii) is found in a region of chromosome 8q24 shown by multiple studies to be linked to BPD [Cichon et al., 2001; McInnis et al., 2003].
FIG. 2.
A: Variants identified by whole-exome sequencing from five families were filtered to yield a list of 67 novel putative MDD genes. B: Among the putative novel MDD genes, many are transcriptions factors, and transcription factors expressed in the human brain [Color figure can be seen in the online version of this article, available at http://wileyonlinelibrary.com/journal/ajmgb].
TABLE I.
Filtering Cascade of Variants in Each of the Five MDD Families
| Filter | Family I | Family II | Family III | Family IV | Family V |
|---|---|---|---|---|---|
| Total number of unique variants | 119,967 | 147,031 | 120,158 | 124,920 | 121,048 |
| Exonic nonsynonymous variants | 11,485 | 15,042 | 11,492 | 13,121 | 11,476 |
| Deleterious variants | 3,915 | 5,142 | 3,860 | 4,448 | 3,940 |
| Novel variants | 382 | 438 | 319 | 464 | 333 |
| Shared heterozygous variants | 18 | 2 | 18 | 14 | 15 |
TABLE II.
Exome-Wide Novel Deleterious Mutation Shared Between Affected MDD Relatives in This Study
| CHR | BP | Class | Gene | TR | Family |
|---|---|---|---|---|---|
| 1 | 7,887,201 | Nonsynonymous SNV | PER3 | + | I |
| 1 | 15,420,741 | Nonsynonymous SNV | KAZN | V | |
| 1 | 27,210,705 | Nonsynonymous SNV | GPN2 | V | |
| 1 | 38,079,559 | Nonsynonymous SNV | RSPO1 | IV | |
| 1 | 85,648,638 | Nonsynonymous SNV | SYDE2 | I | |
| 1 | 229,600,529 | Nonsynonymous SNV | NUP133 | III | |
| 2 | 38,302,303 | Nonsynonymous SNV | CYP1B1 | I | |
| 3 | 124,732,447 | Nonframeshift deletion | HEG1 | III | |
| 3 | 189,526,170 | Nonsynonymous SNV | TP63 | + | III |
| 4 | 1,843,163 | Nonsynonymous SNV | LETM1 | IV | |
| 4 | 120,190,897 | Nonsynonymous SNV | USP53 | III | |
| 4 | 166,414,384 | Nonsynonymous SNV | CPE | I | |
| 4 | 185,941,483 | Nonsynonymous SNV | HELT | + | V |
| 6 | 39,545,925 | Nonsynonymous SNV | KIF6 | V | |
| 6 | 101,100,762 | Nonsynonymous SNV | ASCC3 | + | V |
| 7 | 6,639,520 | Nonsynonymous SNV | C7orf26 | I | |
| 8 | 19,816,875 | Nonsynonymous SNV | LPL | IV | |
| 8 | 29,195,960 | Nonsynonymous SNV | DUSP4 | I | |
| 8 | 65,493,637 | Nonframeshift insertion | BHLHE22 | + | II |
| 8 | 134,146,974 | Nonsynonymous SNV | TG | V | |
| 8 | 146,003,870 | Nonsynonymous SNV | ZNF34 | + | III |
| 9 | 113,137,717 | Nonsynonymous SNV | SVEP1 | V | |
| 9 | 139,273,619 | Nonsynonymous SNV | SNAPC4 | + | III |
| 10 | 19,676,539 | Frameshift deletion | MALRD1 | III | |
| 10 | 69,961,657 | Nonsynonymous SNV | MYPN | I | |
| 10 | 75,552,350 | Nonsynonymous SNV | ZSWIM8 | III | |
| 10 | 75,896,452 | Nonsynonymous SNV | AP3M1 | III | |
| 10 | 129,903,139 | Nonsynonymous SNV | MKI67 | V | |
| 11 | 67,926,194 | Nonsynonymous SNV | SUV420H1 | + | I |
| 11 | 73,688,018 | Nonsynonymous SNV | UCP2 | V | |
| 12 | 11,339,224 | Nonsynonymous SNV | TAS2R42 | IV | |
| 12 | 112,323,791 | Nonsynonymous SNV | MAPKAPK5 | I | |
| 12 | 113,865,812 | Nonsynonymous SNV | SDSL | III | |
| 12 | 120,984,379 | Nonframeshift deletion | RNF10 | IV | |
| 12 | 123,694,677 | Nonsynonymous SNV | MPHOSPH9 | IV | |
| 13 | 21,375,026 | Nonsynonymous SNV | XPO4 | I | |
| 13 | 24,200,859 | Nonsynonymous SNV | TNFRSF19 | V | |
| 13 | 25,840,377 | Nonsynonymous SNV | MTMR6 | III | |
| 13 | 76,179,899 | Nonsynonymous SNV | UCHL3 | I | |
| 15 | 41,339,681 | Nonsynonymous SNV | INO80 | IV | |
| 15 | 60,298,061 | Nonsynonymous SNV | FOXB1 | + | V |
| 15 | 62,170,843 | Nonsynonymous SNV | VPS13C | V | |
| 16 | 57,188,314 | Nonsynonymous SNV | FAM192A | I | |
| 16 | 67,236,149 | Nonsynonymous SNV | ELMO3 | III | |
| 16 | 67,397,493 | Nonsynonymous SNV | LRRC36 | I | |
| 16 | 68,381,548 | Frameshift deletion | PRMT7 | + | I |
| 16 | 77,246,093 | Frameshift insertion | SYCE1L | I | |
| 17 | 5,118,261 | Nonsynonymous SNV | SCIMP | V | |
| 17 | 17,697,123 | Nonframeshift deletion | RAI1 | II | |
| 17 | 38,519,353 | Nonframeshift insertion | GJD3 | I | |
| 17 | 76,109,644 | Nonsynonymous SNV | TMC6 | III | |
| 17 | 79,495,727 | Nonsynonymous SNV | FSCN2 | III | |
| 17 | 79,571,702 | Nonsynonymous SNV | NPLOC4 | III | |
| 18 | 44,398,391 | Frameshift deletion | PIAS2 | + | III |
| 19 | 4,207,909 | Nonsynonymous SNV | ANKRD24 | I | |
| 19 | 12,244,192 | Nonsynonymous SNV | ZNF20 | + | V |
| 19 | 36,230,741 | Nonframeshift deletion | IGFLR1 | IV | |
| 19 | 44,223,770 | Nonsynonymous SNV | IRGC | IV | |
| 19 | 49,416,392 | Nonsynonymous SNV | NUCB1 | IV | |
| 19 | 53,740,407 | Frameshift insertion | ZNF677 | + | III |
| 19 | 58,438,626 | Nonsynonymous SNV | ZNF418 | + | III |
| 20 | 52,199,337 | Nonsynonymous SNV | ZNF217 | + | V |
| 21 | 45,212,597 | Nonsynonymous SNV | RRP1 | IV | |
| 22 | 20,920,847 | Nonframeshift deletion | MED15 | + | IV |
| X | 53,595,767 | Nonsynonymous SNV | HUWE1 | IV | |
| X | 101,381,945 | Nonsynonymous SNV | TCEAL2 | + | I |
| X | 129,150,059 | Nonsynonymous SNV | BCORL1 | + | IV |
Transcriptional regulators (TR) expressed in the brain are highlighted and in bold.
MDD ZNF34 Alleles
The novel ZNF34 mutation we discovered was found shared in both (affected) exome-sequenced individuals in Family III. This mutation encodes a non-conservative P17R substitution in theZNF34 protein Krüppel-associated box-A (KRAB-A) domain, an evolutionarily conserved motif required for the activity of Zn2+ finger transcriptional repressors. This substitution has a CADD (combined annotation dependent depletion) C-score of 11.07 indicating that this variant is predicted to be more deleterious than over 90% of all possible substitutions in the human genome [Kircher et al., 2014]. In the 1,617 bp of coding sequence for the ZNF34 gene, the exome variant server (http://evs.gs.washington.edu/EVS/) lists only two missense mutations reported more than once in the European American population, and even these variants are rare (rs149062206 = 17/8211 alleles, rs371047009 = 2/8596 alleles). We confirmed the presence of the P17R mutation in the two affected Family III relatives by Sanger sequencing. We, then, Sanger sequenced the locus in all Family III individuals for whom DNA was available (Supplemental Fig. S1) and found that all five affected members were carriers of the novel ZNF34 P17R mutation and that the single available unaffected relative was not (Fig. 2). We calculated the exact probability of observing the pattern of rare allele sharing among affected individuals (n = 5) in Family III for ZNF34 P17R using the RVsharing algorithm [Bureau et al., 2014], and found the presence of this rare allele among affected relatives to be statistically significant (P-value = 0.026). Furthermore, the ZNF34 P17R mutation showed complete segregation with MDD in Family III, as it was not observed in the unaffected individual examined. These data provide evidence that this rare variant, in a brain-expressed, putative BPD gene, might be involved in determining MDD susceptibility in, at least, this single family.
When removing the requirement that coding mutations be novel, the only other ZNF34 variant that remained in our dataset was the common indel rs3830702 (TTT/−) found in both exome-sequenced (affected) individuals of Family I. We Sanger sequenced this variant in all members of Family I for whom DNA was available and observed complete segregation of the deletion (minus) allele with MDD (Fig. 1). It is improbable that this common variant (or some common variant in strong linkage disequilibrium) plays a role in MDD per se, as it would have likely already been detected by GWAS. However, it is possible that rs3830702 might be linked to some unobserved (perhaps non-coding) MDD variant in this particular family, a hypothesis that bears further investigation. Additionally, it can also not be ruled out that both the novel ZNF34 P17R-encoding variant in Family III and the common ZNF34 rs3830702 variant in Family I show complete segregation with MDD because of some unobserved (perhaps structural and/or non-exonic) variant nearby. Nonetheless, in summary, our results from the P17R-encoding variant in Family III and rs3830702 in Family I show two ZNF34 variants in our study segregating completely with MDD and with full, observed, penetrance in all individuals for whom DNA was available.
DISCUSSION
The identification of new candidate genes using longitudinally followed multigenerational families affected with MDD may be useful because the heritability is higher in these families than in population or twin samples. The inclusion of families studied with repeated measures may also increase the likelihood of identifying causative variants as evidence for heritability is increased with multiple phenotypic assessments [Kendler et al., 1993] (this may be due to increased accuracy of the diagnosis with repeated assessments and also having assessments obtained as the subject lives through the full age of risk.). Furthermore, families with prepubertal MDD cases have an increased chance of having other MDD relatives [Puig-Antich et al., 1989; Holmans et al., 2007]. We hypothesized that taking these considerations into account in our study design could afford us increased power to detect medium to strong genetic effects. Although large families have been studied in several psychiatric disorders, the current ability to rapidly sequence entire exomes allows us to identify rare and novel variants that might have been missed in previous studies. Through whole-exome sequencing of a relatively small family-based sample and selecting for families in which genetic effect sizes are hypothesized to be large, we were able to generate a list of novel putative MDD candidate genes/variants. Although the variants we report are novel and are predicted to introduce non-conservative changes in amino-acid sequence, their ability to affect disease phenotypes is particularly strong compared to other classes of variants [Gilissen et al., 2012; Ku et al., 2013]. Furthermore, since we required variants to be shared among affected MDD relatives, they are candidates for influencing the MDD phenotype. We found that many of the novel MDD candidate genes encode brain-expressed transcription factors and we validated extended segregation of a variant in one of these genes, the putative bipolar disorder (BPD) gene ZNF34.
ZNF34 and Transcription
By regulating the expression of many downstream effectors, transcription factors can influence a vast array of cellular events. Although candidate genes for neuropsychiatric disorders have focused on genes known to biologically affect, perhaps intuitive, disease-related processes—for example, neurotransmitter and ion channels-encoding genes—it is not surprising that variation in transcription factors can contribute strongly to complex disease phenotypes. In humans many of the C2H2 Zn2+ finger transcription factors are genomically encoded in large gene clusters and many of their biological functions remain unknown. The ZNF34 and ZNF418 (another brain-expressed transcription factor gene in found in our dataset [Table II]) gene products are closely related. They share 41% amino acid identity and each contains an N-terminal KRAB-A box and multiple C-terminal Zn2+ fingers. Both ZNF34 and ZNF418 show existing evidence for influencing affective disorder susceptibility. The Psychiatric Genetics Consortium (PGC) MDD GWAS mega-analysis found a P-value of 5 × 10−4 at the rs8102308 marker found ~25 kb downstream of ZNF418 in a chromosome 19 “ZNF “gene cluster [Lee et al., 2013]. Although this P-value fails to meet genome-wide significance after correcting for multiple tests, it likely does reach significance when correcting only for the number markers in brain-expressed transcription factors loci post hoc, an approach that our findings suggest is justifiable.
Analysis of the PCG BPD data showed ZNF34 to yield a gene-based association P-value of 0.018 in a study [Zhao et al., 2015] that also showed the ZNF34 gene to be differentially expressed between BPD patients and healthy controls. Furthermore, ZNF34 is found in a region of chr8 showing linkage to BPD in multiple studies [Cichon et al., 2001; McInnis et al., 2003]. Our validation that novel coding mutations and common variants in ZNF34 can be found co-segregating with MDD supports a hypothesis that transcription factors are plausible candidates for genetic studies of MDD. That ZNF34 is expressed in the developing brain [Lorenz et al., 2010] points to a possible developmental window through which the circuitry influencing MDD is particularly susceptible. As a transcriptional repressor, the ZNF34 gene product might be responsible for maintaining a gene-expression program in the developing brain that is protective against MDD later in life, and that the P17R variant is compromised for such a function. In addition, the potential involvement of ZNF34 in BPD may represent a kind of variable expressivity and genetic overlap between MDD and BPD, a topic of much discussion in the field of affective disorders. It is worth noting that while the complete observed segregation and penetrance of the ZNF34 P17R-encoding variant suggests a monogenic model of early-onset MDD in Family III, it is possible that other genetic and/or environmental factors also contribute to the phenotype.
Modeling MDD as a Genetic Disease
The study designs, sample sizes and resulting findings from previous MDD studies, subjects the possible genetic models for MDD susceptibility to several constraints [Flint and Kendler, 2014], including the minimum number of the genes involved, their frequencies, and effect sizes. By introducing a list of novel candidates with potentially large effect sizes, our study helps generate testable hypotheses that can further constrain these parameters to develop a more tractable model of MDD genetics than that which currently exists. Being able to estimate better the number of rare MDD variants/genes with strong effects allows us to estimate more precisely the possible number and effect sizes of common MDD variants and allows us to test and model phenotypic interactions between combinations of rare and common alleles across the genome.
One proposed explanation for the lack of reproducibility for positive findings in the MDD literature is that there are dozens or hundreds of common MDD alleles—certain combinations of which exceed some threshold for MDD expression—and thus, extremely large sample sizes are required to detect their effects. However, it is plausible to assume that both common and rare variants affect most complex disorders. Flint and Kendler [2014] discussed a model in which rare variants with large effect sizes in MDD genes might be detected by focusing on “more homogeneous heritable phenotypic groupings.” Our ability to identify ZNF34 in our study using multiple-affected, early onset families and focusing on novel variants is potential evidence for bearing this out. Another strength of our approach is our ability to assess co-segregation, which helps make up for the relatively small sample size in our study. For example, the linkage studies that discovered many causative variants for highly penetrant disorders often relied on single families to map loci of strong effect sizes.
Whether or not brain-expressed transcription factors play a more generalizable role in more common forms of depression (i.e., later onset and from families with fewer affected individuals) is yet to be determined. Nonetheless, the new candidate genes we identify here, including the putative bipolar disorder gene ZNF34, which we validate, may point to, as yet unknown, molecular substrates for MDD. Therefore, understanding the biology and function of these genes may yield useful insight that might help in the prediction and treatment of MDD and related psychiatric disorders.
METHODS
Subjects, Sequencing, and Genotyping
All subjects were recruited from the longitudinal study of extensively phenotyped mutli-generational families, the methods are fully described in [Weissman et al., 2006]. The purpose of the study was to determine the transmission of MDD and other disorders in offspring and then grandchildren in high versus low risk families with risk defined as depression in Generation 1 of the identified proband. Probands (Generation 1) were identified with moderate to severe major depression and were attending a medication clinic for treatment. The controls (not depressed probands) were selected from a population-based study in the same community and were matched on gender and age. All subjects reported Caucasian ancestry. All offspring and then grandchildren were interviewed by clinically trained personnel blind to the clinical status of the probands or previous interviews. Extensive assessments included refined research methods developed for this sample and have been fully described [Weissman et al., 2005, 2006]. The data were then reviewed by a clinical psychiatrist or psychologist and a best estimate diagnosis was made blind to the status of the proband in Generation 1. The subjects were interviewed up to six times over 30 years. DNA was collected in the fifth and sixth wave which was the 25 and 30 year follow-ups.
We performed exome sequencing on all 12 individuals from our discovery phase (see Results section). Capture, sequencing, alignment, mapping, and variant calling were performed by Axeq Technologies (Rockville, MD). Capture was performed using Agilent SureSelect V4+UTR, which targets 71 Mb from 20,965 genes and the resulting libraries sequenced using an Illumina HiSeq 2000. We targeted to obtain 100× raw data in order to achieve 50× on target depth. BWA algorithm was then used to align the sequenced reads to the UCSC hg19 reference genome and variants detected using SAMTOOLS. The large list of variants was then subject to multiple rounds of filtering to narrow it down to the most probable candidate set of causative variants. The average Phred (Q) score across all individuals was 119.3, corresponding to an average 1.2 × 1012 probability of erroneous variant calling, and the average total read depth was 57.4. The ZNF34 variants found in our discovery phase were confirmed and further interrogated in family members by Sanger sequencing.
Variant Selection and Statistical Analysis
The variant lists, output by SAMTOOLS, were subject to a two way filtering process. The first filtering process was to produce a list of shared, novel, deleterious mutations. In order to get this list, we filtered out all nonsynonynous and splice variants from the 12 individuals in the discovery sample. We, then, chose only variants designated as predicted disease causing variants by either SIFT, LRT, or MutationTaster. Novel variants from each dataset were then selected by excluding all variants present in either 1000 Genomes, The Exome Sequencing Project, Complete Genomics, dbSNP, COS-MIC, NCI databases. All variants that met the novel and deleteriousness criteria also had to be shared among all affected members within a family. Finally, for novel mutations that were shared, we removed variants with a read depth < 10 in all individuals for whom the variant was found. Deleteriousness of the ZNF34 P17R-encoding mutation was assessed by combined annotation dependent depletion (CADD), which scores the effect of individual substitutions relative to all possible substitutions in the human genome [Kircher et al., 2014]. Functional annotation of the genes harboring the novel, deleterious variants (obtained in the above step) was retrieved using DAVID [Huang da et al., 2009a, 2009b] online gene ontology function (http://david.abcc.ncifcrf.gov), and the reported EASE score P-values were used to determine significance [Hosack et al., 2003]. Used in gene enrichment analysis, the EASE score is a version of a Fisher exact test modified to be conservative, but not as conservative as implementing Boferroni or Benjamini–Hochberg FDR multiple test correction techniques, which can compromise sensitivity in gene enrichment applications [Huang da et al., 2009a]. For calculation of the EASE score P-value for the enrichment of brain-expressed transcription factors the background value for this group in the genome was determined using two steps. All genes annotated with the gene ontology term “transcription” (GO:0006351) in Uniprot (http://www.uniprot.org) were retrieved (n = 2,500). This list was then used as input for DAVID which reported the number expressed in the brain to be n = 1009.
Supplementary Material
Acknowledgments
This work was supported by Brain and Behavior Research Foundation (formerly NARSAD) Young Investigator Award 17832, NIH grants R01 MH036197; P50 MH090966; U01 MH099225; and R01 NS 061829-04; T32-MH65213, The Sackler Institute for Developmental Psychobiology, and by Nationwide Children’s Hospital, Columbus, OH. We thank David A. Greenberg for computational and other resources, Iulinana Ionita-Laza and Esther N. Drill for insightful input and members of the Greenberg lab for editorial assistance.
Footnotes
Conflicts of interest: The authors have no conflicts of interest to report.
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Bureau A, Younkin SG, Parker MM, Bailey-Wilson JE, Marazita ML, Murray JC, et al. Inferring rare disease risk variants based on exact probabilities of sharing by multiple affected relatives. Bioinformatics. 2014;30(15):2189–2196. doi: 10.1093/bioinformatics/btu198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichon S, Schumacher J, Muller DJ, Hurter M, Windemuth C, Strauch K, et al. A genome screen for genes predisposing to bipolar affective disorder detects a new susceptibility locus on 8q. Hum Mol Genet. 2001;10(25):2933–2944. doi: 10.1093/hmg/10.25.2933. [DOI] [PubMed] [Google Scholar]
- Ferrari AJ, Charlson FJ, Norman RE, Patten SB, Freedman G, Murray CJ, et al. Burden of depressive disorders by country, sex, age, and year: Findings from the global burden of disease study 2010. PLoS Med. 2013;10(11):e1001547. doi: 10.1371/journal.pmed.1001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Kendler KS. The genetics of major depression. Neuron. 2014;81(3):484–503. doi: 10.1016/j.neuron.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen C, Hoischen A, Brunner HG, Veltman JA. Disease gene identification strategies for exome sequencing. Eur J Hum Genet. 2012;20(5):490–497. doi: 10.1038/ejhg.2011.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmans P, Weissman MM, Zubenko GS, Scheftner WA, Crowe RR, Depaulo JR, Jr, et al. Genetics of recurrent early-onset major depression (GenRED): Final genome scan report. Am J Psychiatry. 2007;164(2):248–258. doi: 10.1176/ajp.2007.164.2.248. [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4(10):R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009a;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009b;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeste SS, Geschwind DH. Disentangling the heterogeneity of autism spectrum disorder through genetic findings. Nat Rev Neurol. 2014;10(2):74–81. doi: 10.1038/nrneurol.2013.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ. The lifetime history of major depression in women. Reliability of diagnosis and heritability. Arch Gen Psychiatry. 1993;50(11):863–870. doi: 10.1001/archpsyc.1993.01820230054003. [DOI] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku CS, Tan EK, Cooper DN. From the periphery to centre stage: De novo single nucleotide variants play a key role in human genetic disease. J Med Genet. 2013;50(4):203–211. doi: 10.1136/jmedgenet-2013-101519. [DOI] [PubMed] [Google Scholar]
- Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45(9):984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson DF, Mostafavi S, Milaneschi Y, Rivera M, Ripke S, Wray NR, et al. Genetic studies of major depressive disorder: Why are there no genome-wide association study findings and what can we do about it? Biol Psychiatry. 2014;76(7):510–512. doi: 10.1016/j.biopsych.2014.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz P, Dietmann S, Wilhelm T, Koczan D, Autran S, Gad S, et al. The ancient mammalian KRAB zinc finger gene cluster on human chromosome 8q24.3 illustrates principles of C2H2 zinc finger evolution associated with unique expression profiles in human tissues. BMC Genomics. 2010;11:206. doi: 10.1186/1471-2164-11-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnis MG, Lan TH, Willour VL, McMahon FJ, Simpson SG, Addington AM, et al. Genome-wide scan of bipolar disorder in 65 pedigrees: Supportive evidence for linkage at 8q24, 18q22, 4q32, 2p12, and 13q12. Mol Psychiatry. 2003;8(3):288–298. doi: 10.1038/sj.mp.4001277. [DOI] [PubMed] [Google Scholar]
- Puig-Antich J, Goetz D, Davies M, Kaplan T, Davies S, Ostrow L, et al. A controlled family history study of prepubertal major depressive disorder. Arch Gen Psychiatry. 1989;46(5):406–418. doi: 10.1001/archpsyc.1989.01810050020005. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Murillo L, Gogos JA, Karayiorgou M. The genetic architecture of schizophrenia: New mutations and emerging paradigms. Annu Rev Med. 2012;63:63–80. doi: 10.1146/annurev-med-072010-091100. [DOI] [PubMed] [Google Scholar]
- Stewart WF, Ricci JA, Chee E, Hahn SR, Morganstein D. Cost of lost productive work time among US workers with depression. JAMA. 2003;289(23):3135–3144. doi: 10.1001/jama.289.23.3135. [DOI] [PubMed] [Google Scholar]
- Warner V, Weissman MM, Mufson L, Wickramaratne PJ. Grandparents, parents, and grandchildren at high risk for depression: A three-generation study. J Am Acad Child Adolesc Psychiatry. 1999;38(3):289–296. doi: 10.1097/00004583-199903000-00016. [DOI] [PubMed] [Google Scholar]
- Weissman MM, Wickramaratne P, Nomura Y, Warner V, Verdeli H, Pilowsky DJ, Grillon C, Bruder G. Families at high and low risk for depression: A 3-generation study. Arch Gen Psychiatry. 2005;62(1):29–36. doi: 10.1001/archpsyc.62.1.29. [DOI] [PubMed] [Google Scholar]
- Weissman MM, Wickramaratne P, Nomura Y, Warner V, Pilowsky D, Verdeli H. Offspring of depressed parents: 20 years later. Am J Psychiatry. 2006;163(6):1001–1008. doi: 10.1176/ajp.2006.163.6.1001. [DOI] [PubMed] [Google Scholar]
- Whiteford HA, Degenhardt L, Rehm J, Baxter AJ, Ferrari AJ, Erskine HE, et al. Global burden of disease attributable to mental and substance use disorders: Findings from the Global Burden of Disease Study 2010. Lancet. 2013;382(9904):1575–1586. doi: 10.1016/S0140-6736(13)61611-6. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Xu J, Chen J, Kim S, Reimers M, Bacanu SA, et al. Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. Mol Psychiatry. 2015;20(5):563–572. doi: 10.1038/mp.2014.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.