Abstract
The level of 5-hydroxymethylcytosine (5-hmC) converted by ten-eleven translocation (TET) family is decreased in cancers. However, whether 5-hmC level is perturbed in early stages of carcinogenesis caused by genotoxic carcinogens is not defined. 5-hmC levels and TET2 expression were measured in liver of rats treated with genotoxic carcinogens, riddelliine, or aristolochic acid. Levels of 5-hmC and TET2 expression decreased in the liver of the carcinogens-treated rats. Loss of 5-hmC correlates well with documented induction of genetic mutations by the carcinogens, suggesting that TET2-mediated 5-hydroxymethylation plays an epigenetic role in early state of carcinogenesis.
Keywords: 5-hydroxymethylcytosine, mutagenesis, carcinogenesis, riddelliine, aristolochic acid, ten-eleven translocation
INTRODUCTION
DNA methylation at the 5-carbon position of cytosine, a critical epigenetic mechanism in the regulation of the genome, is a process that impacts on a broad range of biological functions and pathological processes [1]. 5-Methylcytosine (5-mC) can be further converted to 5-hydroxymethylcytosine (5-hmC) by the product of ten-eleven translocation (TET) family genes [2]. Recent extensive data have shown that loss of 5-hmC is found in different types of cancers [3]. We reported that loss of 5-hmC is associated with increased melanoma virulence and poor survival [4]. In addition, decrease of TET2 gene expression or TET2 mutation is associated with carcinogenesis [5]. Furthermore, decrease of 5-hmC levels in rat liver exposed to a non-genotoxic carcinogen, phenobarbital, or in genotoxic carcinogen-induced hepatocellular carcinoma was recently reported [6,7]. However, the 5-hmC signature in early stages of carcinogenesis, such as initiation where mutations occur in critical tumor suppressor genes or oncogenes, has not been defined.
Genotoxic carcinogens bind to DNA and cause mutations, potentially leading to tumor formation, and accordingly, they are frequently used for modeling multiple steps of carcinogenesis. In the present study, two botanical genotoxic carcinogens, riddelliine and aristolochic acid (AA), were employed. Riddelliine is a pyrrolizidine alkaloid present in many plants worldwide and associated with human disease through contamination of staple foods, honey, milk, herbal teas, and herbal medicines [8]. Riddelliine induced a high incidence of hemangiosarcomas in rodent liver [9]. It was proved to be genotoxic and caused mutations in the transgenic cII gene, K-ras protooncongene, and p53 tumor suppressor gene in rat liver [10,11]. Thus, riddelliine has been classified as a potentially human carcinogen [12]. AA is a natural component of Aristolochia plants that have been used for medicinal purposes since antiquity. However, AA has been shown to be toxic to kidney [13] and induces genotoxicity in many assays, with demonstrated mutations in rat kidney and liver [14,15]. Consumption of AA is associated with a high incidence of urothelial cancer and other types of tumors in humans and experimental animals [16,17].
Cancer research conducted in humans often focuses on the late stages of carcinogenesis, limiting the scope of such investigation. Animal models provide a unique opportunity to understand the early stages of carcinogenesis where critical and potentially preventable genomic and epigenomic alterations may occur. In our previous studies, we have used the Big Blue transgenic mutation rat model to evaluate the mutagenesis of riddelliine and AA during early stages of carcinogenesis [11,18]. In the present study, the 5-hmC signature was studied in these animals, with the purposes gaining insight into the 5-hmC signature.
MATERIALS AND METHODS
Riddelliine (>97% pure by reversed phase high performance liquid chromatography (HPLC) analysis) was obtained from the National Toxicology Program (NTP); and AA was purchased from Sigma (St. Louis, MO). Big Blue Fisher 344 transgenic rats were purchased from Taconic Laboratories (Germantown, NY). Animal care was performed in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals and was authorized by the National Center for Toxicological Research (NCTR) Institutional Animal Care and Use Committee. The animal treatment has been described in our previous reports [11,18]. For the treatments, groups of six Big Blue rats were gavaged with 0.1, 0.3, and 1.0 mg/kg riddelliine or with 0.1, 1.0, and 10.0 mg/kg AA, as well as their vehicle controls, 5 days a week for 12 wk and sacrificed 1 d after the last treatment. By the end of the study, rats treated with 0.1, 1.0, and 10.0 mg/kg AA weighed 5%, 7%, and 15% less than the controls while the 1.0 mg/kg riddelliine dose groups were a little less (5%) than those of the vehicle control [9,19,20].
Genomic DNA from liver tissue was extracted using Puregene Core Kit A according to the manufacturer’s instruction (Qiagen, Valencia, CA). The DNA concentration was determined with NanoDrop 1000 (Thermo Scientific, Waltham, MA). The immunodot blot was performed using a Bio-Dot Apparatus as previously reported [4]. Briefly, the DNA was loaded in 96-well plate in 30 μl Tris-EDTA(TE) buffer with 2-fold serial dilution (100 ng ~1 μg). The denatured DNA sample was applied to rehydrated nitrocellulose membrane and then was baked for 2 h at 80°C and then blocked with TBST containing 5% non-fat milk for 1 h at room temperature. The membrane was incubated with primary anti-5-hmC antibody (1:1000) overnight at 4°C. It was then incubated with horseradish peroxidase (HRP) conjugated secondary antibody (1:10,000) at room temperature for 1 h. The signal was developed with enhanced chemiluminescence (ECL) after wash with 20 mM Tris-buffered saline with 0.05% Tween 20 (TBS-T)
Immunohistochemical studies were performed with Tissue-Tek™ OCT Compound (OCT stands for Optimal Cutting Temperature) embedded frozen liver tissue as we reported previously [4]. Briefly, the slides were cryosectioned to 5-μm sections. After antigen retrieval, the slides were placed in 2N HCl for 30 min, rinsed in distilled water and placed in 100 mM Tris-HCl pH8.5 for 10 min. All of the slides were stained on the Dako Autostainer (Dako Corporation, Carpinteria, CA) using the EnVision (Dako) staining reagents. Sections were incubated for 60 min with rabbit-anti-5-hmC at 1:10,000 (Active motif) and then incubated with the EnVision+ Dual Link (Dako) detection reagent for 30 min. Sections were washed, treated with a solution of diaminobenzidine and hydrogen peroxide (Dako) for 10 min, and after rinsing, a toning solution (DAB Enhancer, Dako) was used for 2 min to enrich the final color.
Total RNA of liver tissue was extracted from the liver tissue by RNAeasy kit (Qiagen, Valencia, CA). The quantity of total RNA was determined by spectrophotometer using the absorbance at A260/A280 nm. The cDNAs were synthesized by SuperScript III first strand kit (Invitrogen, Carlsbad, CA) according to manufacturer’s protocol. The primer sequences for rat TET2 are 5′-AATCGCCTTCGGATTCAGACACTC-3′ (forward) and 5′- CTTGACCTCCGATACACCCATTTAGC-3′ (reverse), and the primers for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) are 5′-ATCACCATCTTCCAGGAGCGA-3′ (forward) and 5′-CCTTCTCCATGGTGGTGAA-3′ (reverse). The real-time quantitative reverse transcription PCR (qRT-PCR) reactions were performed to detect TET2 mRNA levels. The ΔΔCt method was used to analyze the TET2 mRNA expression levels.
Significant treatment differences of the study were determined by 1-way analysis of variance (ANOVA) followed by Tukey’s test. Data are presented as means ± SD. Differences were considered significant at P < 0.05.
RESULTS
5-hmC Levels were Decreased in the Livers of Riddelliine and AA-treated Big Blue Rats
The carcinogens did not induce any tumors or nodules in the livers of rats treated for three months although the treatments resulted in formation of DNA adducts and induction of mutations in rat liver at this stage [9,19–23]. In order to detect the level of 5-hmC in liver of the carcinogen-treated rats, immuno dot blot technique was used. Compared with the vehicle control group, both the riddelliine and AA treatments for 12 wk resulted in a significantly decrease in 5-hmC in liver tissue (Figure 1A and B). The 5-hmC intensities decreased from 25.8 ± 13.85 in the control group to 3.7 ± 1.51 with the highest dose of riddelliine treatment (P < 0.05), whereas the 5-hmC densities decreased from 25.0 ± 4.47 in the control group to 4.7 ± 7.51 in with the highest dose of the AA treatment (P < 0.01). The 5-hmC levels in the individual animals at the highest dose treatments were very close, suggesting a consistent effect of the carcinogen on 5-hmC level.
Figure 1.
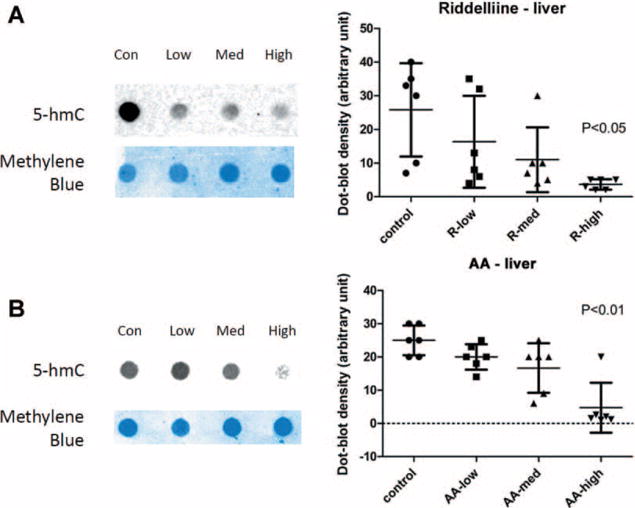
Detection of 5-hmC by immuno dot-blot in liver specimens of riddelliine or AA treated Big Blue rats. (A) A representative dot blot (left) and the bar graph of the density of the dot blots (right) in the liver sample of riddelliine treated Big Blue rats. (B) A representative dot blot (left) and the bar graph of the density of the dot blots (right) in the liver sample of AA treated Big Blue rats.
Immunofluorescence Staining Confirmed Decreased Levels of 5-hmC
Anti-5-hmC specific antibody was used to stain the liver samples and evaluate 5-hmC levels. The immunohistochemical findings confirmed the results from the immuno dot blot experiment. Positive 5-hmC immunostains highlighted the nuclei of hepatocytes in control group samples (Figure 2A and C). Very weak 5-hmC immunostains were detected in the highest dose of riddelliine or AA-treated samples compared to that of the control group samples (Figure 2). These results correlated with the immuno dot blot results (Figure 1) and provided further confirmation that these two genotoxic carcinogen treatments lowered the level of 5-hmC in rat liver.
Figure 2.
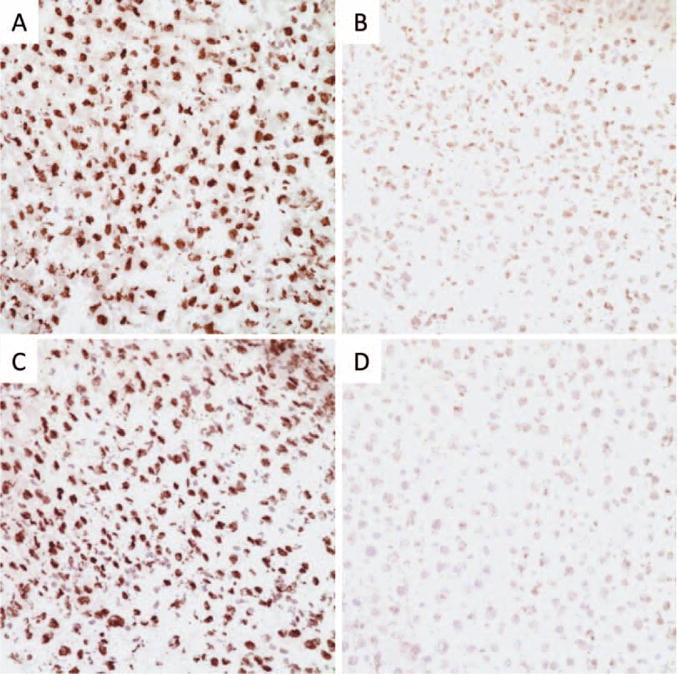
Detection of 5-hmC by immunohistochemistry in liver specimens of riddelliine or AA treated Big Blue rats. A representative IHC staining was shown for 5-hmC in liver tissue. 5-hmC in liver tissue of control group (A) and high dose riddelliine treated group (B). 5-hmC in liver tissue of control group (C) and high dose AA treated group (D).
Expression of the TET2 Gene was down-regulated by Carcinogen Treatment in Rat Liver
We further measured TET2 gene expression to explore whether the decreased level of 5-hmC in rat liver by carcinogen treatment was associated with expression change of TET2, the gene that encodes a key enzyme converting 5-mC to 5-hmC. qRT-PCR was performed to quantify the mRNA lavel of TET2 in the control and carcinogen-treated groups (highest doses). The results indicated that TET2 expression was significantly decreased in the groups receiving the highest dose of riddelliine treatment (P = 0.017, Figure 3A) or AA treatment (P = 0.003, Figure 3B) compared to the control group. TET1 and TET3 mRNA levels did not show significant difference in liver samples of rat treated with or without AA or riddelliine by qRT-PCR (data not shown).
Figure 3.
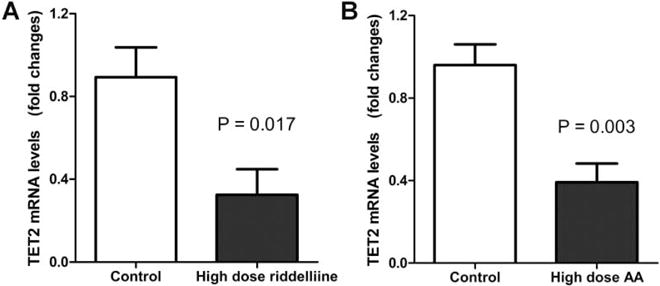
Detection of TET2 mRNA levels in liver specimens of riddelliine or AA treated Big Blue rats. (A) TET2 mRNA levels detected by qRT-PCR in liver specimens of control and riddelliine treated Big Blue rats. (B) TET2 mRNA levels in liver specimens of control and AA treated Big Blue rats.
DISCUSSION
Recent studies show that the non-genotoxic carcinogen phenobarbital induces defined changes in 5-hmC and the change of 5-hmC signature can be used as an indicator of cell states during organ maturation and drug-induced response [6]. However, the role of 5-hmC in early stages of carcinogenesis due to genotoxic carcinogen exposure is unknown. In this study, we explored the relationship between levels of 5-hmC and mutations induction by the genotoxic carcinogens, riddelliine, and AA at early stages prior to overt tumor formation. The rats were treated with doses known to induce mutations within 12 wk and hepatocellular carcinoma within 2 yr. As previously reported, the treatment regimen in this study results in dose-dependent induction of DNA adducts and mutations in rat liver [11,18,24]. Specifically, riddelliine increased mutant frequencies from 30 ×10−6 in control animals to 103 × 10−6 in the high dose groups, while AA results in 18-fold mutation induction in rat liver. In the present study, the level of 5-hmC was significantly decreased approximately 6-fold in the riddelliine-treated liver and about 10-fold in the AA-treated liver. These findings were also confirmed by 5-hmC immunohistochemistry staining in liver tissues. The results indicate that the levels of 5-hmC decrease not only in cancer cells, but also in the genotoxic carcinogen-exposed cells at pre-neoplastic stage of carcinogenesis.
We previously reported that down-regulation of TET2 is most likely the mechanism for loss of 5-hmC in melanoma, and that reintroduction of TET2 restores 5-hmC levels and decreases metastatic potential of melanoma cells [4]. In this study, we found that TET2 gene expression was significantly lower in riddelliine and AA-treated liver tissue compared with control groups, indicating that a decrease of TET2, an enzyme required for the conversion of 5-mC into 5-hmC, is at least partially a contributing factor for the reduction of 5-hmC in our model. According to our previous studies, the carcinogens induced mutant frequencies at about 10−4 to 10−3 in the cII gene of the rat livers. Therefore, the induced mutations are rare events. If one mutation occurs out of 1000 TET2 functional genes, the mutated gene will not be able to change the TET2 expression pattern. Thus, the mutations in the TET2 should not be a mechanistic event responded to the decrease of 5-hmC. The mechanisms of 5-hmC decrease by genotoxic carcinogens warrant further investigation.
In conclusion, loss of 5-hmC was detected in liver tissue of rats treated with mutagenic doses of genotoxic carcinogens, riddelliine, and AA. In addition, these genotoxic carcinogens reduced TET2 gene expression, suggesting a novel mechanism in the early stage of hepatocarcinogenesis.
Acknowledgments
We thank Dr. Qian Zhan for technical support. This work is supported in part by Karin Grunebaum cancer research foundation (CGL).
Grant sponsor: (GFM); Grant number: R01 HL84815; Grant sponsor: (GFM); Grant number: R01 CA158467
Footnotes
The views presented in this article do not necessarily reflect those of the Food and Drug Administration.
Conflict of Interest: The authors declare no conflict of interest.
References
- 1.Reddington JP, Pennings S, Meehan RR. Non-canonical functions of the DNA methylome in gene regulation. Biochem J. 2013;451:13–23. doi: 10.1042/BJ20121585. [DOI] [PubMed] [Google Scholar]
- 2.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin SG, Jiang Y, Qiu R, et al. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 2011;71:7360–7365. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lian CG, Xu Y, Ceol C, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alayed K, Patel KP, Konoplev S, et al. TET2 mutations, myelodysplastic features, and a distinct immunoprofile characterize blastic plasmacytoid dendritic cell neoplasm in the bone marrow. Am J Hematol. 2013;88:1055–1061. doi: 10.1002/ajh.23567. [DOI] [PubMed] [Google Scholar]
- 6.Thomson JP, Lempiainen H, Hackett JA, et al. Non-genotoxic carcinogen exposure induces defined changes in the 5-hydroxymethylome. Genome Biol. 2012;13:R 93. doi: 10.1186/gb-2012-13-10-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C, Liu L, Chen X, et al. Decrease of 5-hydroxymethylcytosine is associated with progression of hepatocellular carcinoma through downregulation of TET1. PLoS One. 2013;8:e 62828. doi: 10.1371/journal.pone.0062828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edgar JA, Colegate SM, Boppre M, Molyneux RJ. Pyrrolizidine alkaloids in food: a spectrum of potential health consequences. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2011;28:308–324. doi: 10.1080/19440049.2010.547520. [DOI] [PubMed] [Google Scholar]
- 9.Chan PC, Haseman JK, Prejean JD, Nyska A. Toxicity and carcinogenicity of riddelliine in rats and mice. Toxicol Lett. 2003;144:295–311. doi: 10.1016/s0378-4274(03)00240-6. [DOI] [PubMed] [Google Scholar]
- 10.Hong HL, Ton TV, Devereux TR, et al. Chemical-specific alterations in ras, p53, and beta-catenin genes in hemangiosarcomas from B6C3F1 mice exposed to o-nitrotoluene or riddelliine for 2 years. Toxicol Appl Pharmacol. 2003;191:227–234. doi: 10.1016/s0041-008x(03)00165-0. [DOI] [PubMed] [Google Scholar]
- 11.Mei N, Heflich RH, Chou MW, Chen T. Mutations induced by the carcinogenic pyrrolizidine alkaloid riddelliine in the liver cII gene of transgenic big blue rats. Chemical Research in Toxicology. 2004;17:814–818. doi: 10.1021/tx049955b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.NTP. Final report on carcinogens background document for riddelliine. Rep Carcinog Backgr Doc. 2008:i–104. 8-5977. [PubMed] [Google Scholar]
- 13.Debelle FD, Vanherweghem JL, Nortier JL. Aristolochic acid nephropathy: a worldwide problem. Kidney Int. 2008;74:158–169. doi: 10.1038/ki.2008.129. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Meng F, Arlt VM, Mei N, Chen T, Parsons BL. Aristolochic acid-induced carcinogenesis examined by ACB-PCR quantification of H-Ras and K-Ras mutant fraction. Mutagenesis. 2011;26:619–628. doi: 10.1093/mutage/ger023. [DOI] [PubMed] [Google Scholar]
- 15.Chen T. Genotoxicity of aristolochic acid: a review. J Food Drug Anal. 2007;15:387–399. [Google Scholar]
- 16.Chen CH, Dickman KG, Moriya M, et al. Aristolochic acid-associated urothelial cancer in Taiwan. Proc Natl Acad Sci U S A. 2012;109:8241–8246. doi: 10.1073/pnas.1119920109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mengs U, Lang W, Poch I. The crcinogenic action of aristolochic acid in rats. Arch Toxicol. 1982;51:109–119. [Google Scholar]
- 18.Mei N, Arlt VM, Phillips DH, Heflich RH, Chen T. DNA adduct formation and mutation induction by aristolochic acid in rat kidney and liver. Mutat Res-Fund Mol M. 2006;602:83–91. doi: 10.1016/j.mrfmmm.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mei N, Heflich RH, Chou MW, Chen T. Mutations induced by the carcinogenic pyrrolizidine alkaloid riddelliine in the liver cII gene of transgenic big blue rats. Chem Res Toxicol. 2004;17:814–818. doi: 10.1021/tx049955b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mei N, Arlt VM, Phillips DH, Heflich RH, Chen T. DNA adduct formation and mutation induction by aristolochic acid in rat kidney and liver. Mutat Res. 2006;602:83–91. doi: 10.1016/j.mrfmmm.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mei N, Chou MW, Fu PP, Heflich RH, Chen T. Differential mutagenicity of riddelliine in liver endothelial and parenchymal cells of transgenic big blue rats. Cancer Lett. 2004;215:151–158. doi: 10.1016/j.canlet.2004.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mengs U, Lang W, Poch J. The carcinogenic action of aristolochic acid in rats. Arch Toxicol. 1982;51:107–119. [Google Scholar]
- 23.National Toxicology Program. Toxicology and carcinogenesis studies of riddelliine (CAS No. 23246-96-0) in F344/N rats and B6C3F1 mice (gavage studies) Natl Toxicol Program Tech Rep Ser. 2003;508:1–280. [PubMed] [Google Scholar]
- 24.Chou MW, Jian Y, Williams LD, et al. Identification of DNA adducts derived from riddelliine, a carcinogenic pyrrolizidine alkaloid. Chem Res Toxicol. 2003;16:1130–1137. doi: 10.1021/tx030018y. [DOI] [PubMed] [Google Scholar]


