Abstract
Understanding the activation and internalization of G protein-coupled receptors (GPCRs) using conditional approaches is paramount to developing new therapeutic strategies. Here, we describe the design, synthesis, and testing of ExONatide, a benzylguanine-linked peptide agonist of the glucagon-like peptide-1 receptor (GLP-1R), a class B GPCR required for maintenance of glucose levels in humans. ExONatide covalently binds to SNAP-tagged GLP-1R-expressing cells, leading to prolonged cAMP generation, Ca2+ rises, and intracellular retention of the receptor. These effects were readily switched OFF following cleavage of the introduced disulfide bridge using the cell-permeable reducing agent beta-mercaptoethanol (BME). A similar approach could be extended to a class A GPCR using GhrelON, a benzylguanine-linked peptide agonist of the growth hormone secretagogue receptor 1a (GHS-R1a), which is involved in food intake and growth. Thus, ExONatide and GhrelON allow SNAP-tag-directed activation of class A and B GPCRs involved in gut hormone signaling in a reversible manner. This tactic, termed reductively cleavable agONist (RECON), may be useful for understanding GLP-1R and GHS-R1a function both in vitro and in vivo, with applicability across GPCRs.
Short abstract
SNAP-tag-directed activation of class A and B G protein-coupled receptors can be achieved in a conditional and reversible manner using peptidic reductively cleavable agONists (RECONs).
Introduction
G protein-coupled receptors (GPCRs) transduce information encoded by external stimuli into an appropriate cell output and as such play a key role in organismal homeostasis.1 For this reason, GPCRs are important drug targets, yet many facets of their function remain enigmatic, including how they dynamically signal in space and time. Selectively targeting and interrogating GPCRs is therefore important for understanding their function.
Tethered pharmacology describes the linkage of pharmacophores in close proximity to their targets often but not necessarily in combination with recombinant engineering. By means of covalent or noncovalent high affinity binding, this approach allows the precise control of biological function as usually achieved by genetics, but with the speed of pharmacology.2 Self-labeling proteins (e.g., SNAP-, CLIP-, and Halo-tag)3 have facilitated tethered pharmacology, since they can be conditionally expressed and covalently bind molecules possessing the relevant bioconjugation handle with high selectivity and enzymatic kinetics.4,5 Moreover, these enzymes can be fused onto a variety of proteins.6,7 The SNAP-tag is ideal for targeting GPCRs, as it retains its activity to covalently bind molecules possessing an O6-benzylguanine (BG) when expressed both in vitro and in vivo,4,5 and many well-characterized SNAP_GPCR fusions exist compared to other enzyme self-labels. For instance, the SNAP-tag is the basis for photoswitchable orthogonal remotely tethered ligands (PORTLs) for class C GPCRs,2,8,9 while the Halo-tag was anchored to the membrane for the drugs acutely restricted by tethering (DART) concept.10 However, PORTLs incorporate an isomerization step for light-controlled ON/OFF responses, as shown for the metabotropic glutamate receptor 2 in vivo,11 leaving ligand attached and thus limiting investigation of receptor trafficking and surface versus internalized populations. On the other hand, DART is irreversible and does not allow a 1:1 ligand/receptor ratio, albeit the studies were performed both in vitro and in vivo and encompassed an ionotropic as well as a metabotropic receptor. Other approaches, such as bio-orthogonal ligand tethering (BOLT)12 and photoswitchable tethered ligands (PTLs),13 respectively rely on incorporating unnatural amino acids or cysteines into proteins. However, their use is limited by the requirement for site-directed mutagenesis to identify active mutants or by their inherent reactivity. We therefore reasoned that a self-labeling protein-tag bearing a cleavable linker would set the stage for studying conditional, prolonged, and reversible activation of GPCRs.
The glucagon-like peptide-1 receptor (GLP-1R) is an excellent candidate for the further development of tethered pharmacology, since it is a blockbuster drug target for type 2 diabetes treatment.14 Following ligand binding, this class B GPCR primarily activates adenylyl cyclase through Gs, leading to 3′-5′-cyclic adenosine monophosphate (cAMP) accumulation15−17 and intracellular Ca2+ fluxes.18−20 These signaling processes are terminated by postendocytotic receptor trafficking, where the GLP-1R is internalized into endosomes, followed by either lysosomal degradation or endosomal recycling to the plasma membrane.21 However, recent reports suggest that GPCR signaling continues following receptor internalization into endosomes via cytosolic cAMP generation.22−24 How internalization and subsequent trafficking influence GLP-1R function is poorly understood.25 Lastly, the GLP-1R is expressed throughout the body and displays pleiotropic activity including effects on glucose levels, locomotion, food intake, blood pressure, and inflammation.14,26−28 Despite this, the contribution of GLP-1R activation within discrete body compartments and tissues has so far relied upon Glp1r–/– animals.29−31 Key to better understanding GLP-1R, and more broadly GPCR function, is the development of tools that allow reversible receptor activation in a highly conditional manner.
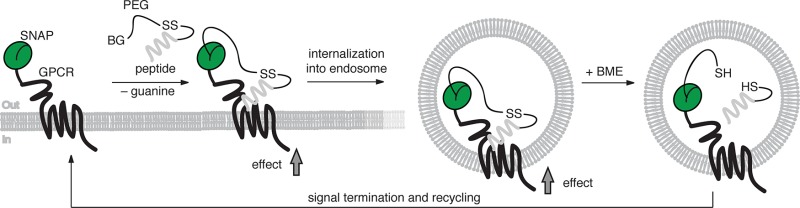
Herein, we describe the development and testing of ExONatide (Figure 1), a benzylguanine-linked and disulfide bridge-containing incretin-mimetic based upon exenatide (Byetta). ExONatide specifically labels and activates SNAP_GLP-1R, a binary response that can be switched OFF by the simple addition of reducing agent to cleave the ligand (Figure 1a,b). Using GhrelON, we also extend the concept to the growth hormone secretagogue-receptor 1a (GHS-R1a), a class A GPCR. Following fasting, ghrelin released from the stomach binds and activates GHS-R1a in neurons located in the arcuate nucleus of the hypothalamus, as well as pituitary somatotropes, leading to orexigenic (feeding) responses and growth hormone secretion.32−34 As such, ExONatide and GhrelON provide the blueprint for reductively cleavable agONist (RECON) peptides and set the scene for conditionally targeting GPCRs both in vitro and in vivo.
Figure 1.
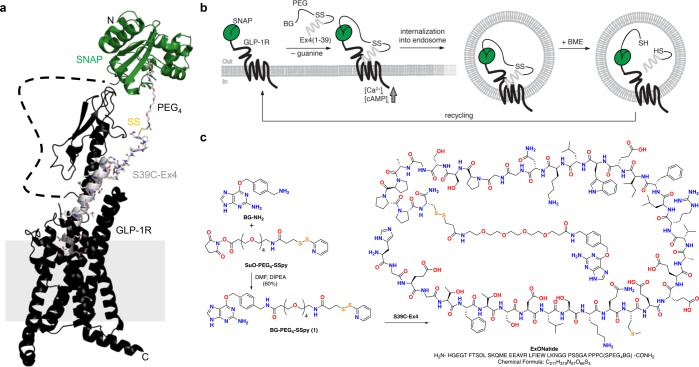
Logic and synthesis of ExONatide. (a) Crystal structure of the activated GLP-1R in complex with GLP-1 (pdb: 5VAI)74 and SNAP-tag (pdb: 3L00) resemble the molecular dimensions and design of the present study. (b) Schematic showing the reductively cleavable agONist (RECON) concept: after covalent labeling of a SNAP-tag with ExONatide, the GLP-1R can be activated, leading to Ca2+ and cAMP rises together with internalization and trafficking, which can readily be terminated by reductive cleavage using beta-mercaptoethanol (BME). (c) The cleavable disulfide bridge of ExONatide is formed through reaction of BG-PEG4-SSpy with S39C-Ex4.
Results
Design and Synthesis of ExONatide
As shown by X-ray crystal structures, incretin-mimetic peptides with agonistic activity such as exenatide (Byetta; also known as exendin-4 or Ex4(1–39)) bind to the GLP-1R with their C-terminus solvent exposed.35,36 We therefore set out to derivatize Ex4(1–39) by mutating and synthesizing the S39C-Ex4 variant by means of solid-phase peptide synthesis (SPPS) to install a free cysteine bioconjugation handle at the C-terminus. Linking BG-NH2 to a PEG4 spacer containing a pyridyl-activated disulfide from commercially available substrates, and by reacting this with S39C-Ex4, ExONatide was obtained in high purity on the milligram scale (see Supporting Information for details on synthesis and characterization) (Figure 1c). ExONatide therefore comprises a SNAP-tag reactive BG linked to a GLP-1R agonist via a reductively cleavable disulfide-containing PEG4 chain.
ExONatide Activates and Labels SNAP_GLP-1R
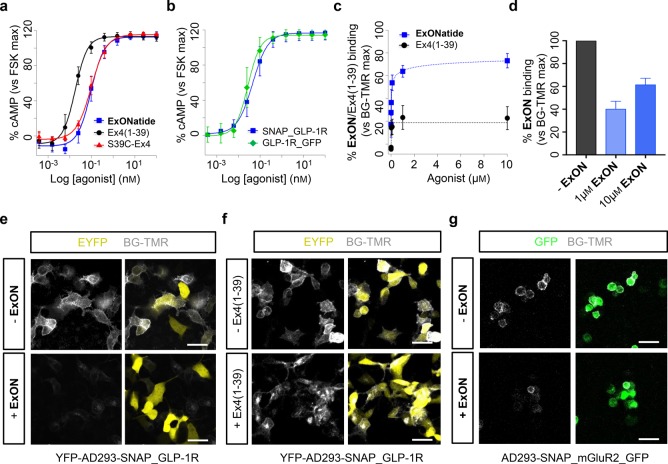
Ex4(1–39) was able to increase intracellular cAMP concentrations with an EC50 (30 min) = 17.9 ± 1.2 pM in YFP-AD293-SNAP_GLP-1R cells, as assessed using LANCE TR-FRET-based assays (Figure 2a). By contrast, cAMP concentration–responses to ExONatide were right-shifted (EC50 (30 min) = 95.2 ± 8.2 pM), with similar results seen in AD293-GLP-1R_GFP cells lacking the SNAP-tag (Figure 2a,b). Suggesting that single amino acid substitutions at the solvent exposed C-terminus of the peptide may have an effect on potency, the EC50 values for the S39C-Ex4 precursor (EC50 (30 min) = 98.8 ± 5.5 pM) and ExONatide were almost identical (Figure 2a). Nonetheless, maximal cAMP responses were near 100% (vs 5 μM forskolin) for all compounds tested, implying full agonism (Figure 2a).
Figure 2.
ExONatide signaling and binding. (a) ExONatide, S39C-Ex4 and Ex4(1–39) cAMP concentration–responses in YFP-AD293-SNAP_GLP-1R cells (n = 3 assays in triplicate). (b) ExONatide concentration–response curves are similar with and without the SNAP-tag (n = 3 assays in triplicate). (c) Preincubation with increasing concentrations of ExONatide exponentially decreases BG-TMR binding/fluorescence compared to Ex4(1–39) in YFP-AD293-SNAP_GLP-1R cells (n = 177–448 cells). (d) ExONatide (1–10 μM) decreases BG-TMR binding/fluorescence in AD293-SNAP_mGluR2_GFP cells (n = 137–176 cells). (e and f) Representative images showing BG-TMR fluorescence in YFP-AD293-SNAP_GLP-1R cells preincubated with and without a high concentration (1 μM) of ExONatide or Ex4(1–39) (scale bar = 33 μm). (g) Representative images showing BG-TMR fluorescence in AD293-SNAP_mGluR2_GFP cells preincubated with and without a high concentration (10 μM) of ExONatide (scale bar = 33 μm). Values are the mean ± SEM.
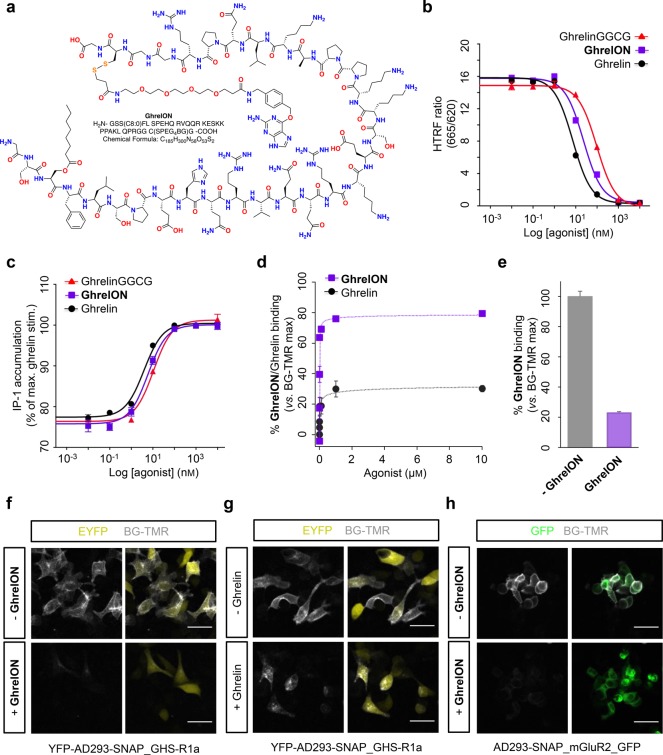
SNAP-tag labeling efficiency was determined by preincubating YFP-AD293-SNAP_GLP-1R cells with ExONatide for 30 min before washing and adding BG-TMR, a fast cell-permeable SNAP-labeling fluorophore. Increasing concentrations of ExONatide exponentially reduced BG-TMR intensity with a half-maximal binding concentration (BC50 (30 min) = 32.1 ± 22.7 nM) suggestive of near-quantitative SNAP-tag labeling at the membrane (Figures 2c,e, S1, S2a). Labeling reached 70–80%, which may reflect internalization of 20–30% GLP-1R at the time of application of ExONatide, which is non-cell permeable compared to BG-TMR, or alternatively 20–30% loss of internalized receptor due to degradation at high ExONatide concentrations.23,37 Supporting the latter, a 20–30% decrease in BG-TMR fluorescence was also seen following incubation of YFP-AD293-SNAP_GLP-1R cells with high concentrations (>1 μM) of Ex4(1–39) (Figure 2c,f). ExONatide was similarly able to label AD293-SNAP_mGluR2_GFP cells (Figure 2d,g), although labeling strength was reduced, probably due to loss of the orthosteric site that may contribute to affinity labeling (58.8 ± 2.6 vs 37.0 ± 1.5% binding, SNAP_GLP-1R vs SNAP_mGluR2_GFP cells, respectively; 1 μM ExONatide; P < 0.01, Student’s t test). No binding was detected in YFP-only transfected cells, as expected for the SNAP-tag specific BG-compound (Figure S2b). On the basis of the SNAP-tag labeling efficiency, ExONatide was used at a concentration of 800 nM for all subsequent cell biology experiments.
ExONatide Does Not Induce GLP-1R Biased Signaling
Biased signaling exists when different agonists selectively engage different signaling pathways via the same receptor. Several GLP-1R agonists engender bias between G-protein signaling (measured as cAMP production) and β-arrestin-dependent phosphorylation of ERK1/2.38 To determine whether ExONatide is a biased GLP-1R agonist, we measured signaling responses using CHO-K1-SNAP_GLP-1R cells together with the intramolecular FRET reporters TEpacvv39 and cytoplasmic EKAR,40 which respectively measure intracellular cAMP production and ERK1/2 activation via conformational changes in proximity between CFP and YFP derivatives. Ratiometric changes in fluorescence were apparent on stimulation with Ex4(1–39), S39C-Ex4, and ExONatide at nanomolar concentrations (Figure S3a,b), and concentration–response curves for each pathway were obtained (Figure S3c,d). As all compounds were full agonists in each pathway, relative potency ratios (ΔΔLogEC50) were used to calculate bias.41 Relative to Ex4(1–39), neither S39C-Ex4 nor ExONatide exhibited bias between cAMP and ERK signaling, as determined with sequential measurements over 30 min to avoid kinetic artifacts (Figure S3e).42
ExONatide Induces Conditional and Prolonged GLP-1R Signaling in Beta Cells
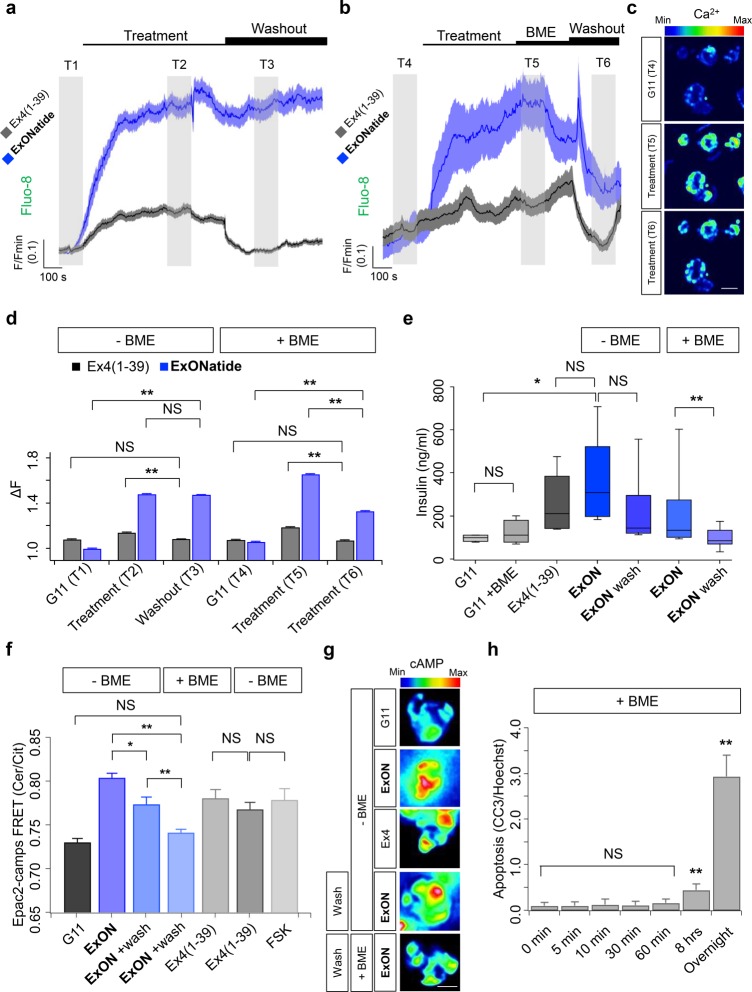
We next sought to investigate whether ExONatide would activate GLP-1R signaling in a physiologically more relevant system, i.e., in MIN6 beta cells. This was done by following GLP-1-induced Ca2+ fluxes using confocal microscopy. Both Ex4(1–39) and ExONatide induced large cytosolic Ca2+ rises in Fluo8-loaded MIN6B1-SNAP_GLP-1R cells (Figure 3a–d). While resting baseline Ca2+ levels could be restored following a washout period for Ex4(1–39), this was not the case for ExONatide where Ca2+ remained significantly elevated (Figure 3a–d). However, addition of beta-mercaptoethanol (BME) for 5 min immediately before washout allowed Ca2+ responses to ExONatide to be reduced (Figure 3b–d). In line with [Ca2+]i measurements, ExONatide stimulated insulin secretion similarly to Ex4(1–39), although this could only be halted following reductive cleavage and washout of compound (Figure 3e).
Figure 3.
ExONatide leads to prolonged Ca2+ and cAMP signaling. (a) ExONatide induces Ca2+ rises in MIN6B1-SNAP_GLP-1R cells similarly to Ex4(1–39), but this cannot be washed out with buffer (mean ± SEM traces shown) (gray shaded area T1-T3 = analysis time window) (n = 29–34 cells). (b) Application of beta-mercaptoethanol (BME) for 5 min immediately prior to washout allows Ca2+ responses to ExONatide to be subsequently reduced (mean ± SEM traces shown) (gray shaded area T4–T6 = analysis time window) (n = 34 cells). (c) Representative images showing Ca2+ responses to ExONatide in MIN6B1-SNAP_GLP-1R cells before (T4), during (T5) and after (T6) application of BME. (d) Bar graph showing amplitude of Ca2+ responses to Ex4(1–39) and ExONatide before and after washout ± BME (n = 29–34 cells) (T1-T6 relate to time windows shown in a and b). (e) Box and whiskers plot showing that ExONatide-stimulated insulin secretion can only be washed out following application of BME (n = 8 wells) (Ex4(1–39)-alone was used as a positive control) (G11; 11 mM glucose). (f) As for (a), but FRET assays for intracellular cAMP 15 min following application of ligand or washout (n = 40–71 cells). (g) Representative images showing FRET responses to ExONatide in MIN6B1-SNAP_GLP-1R cells before and after application of BME. (h) BME was unable to significantly induce apoptosis over 60 min, as determined using immunostaining for cleaved caspase 3 (CC3) (overnight incubation serves as the positive control) (n = 3 experiments). **P < 0.01 and NS, nonsignificant, as indicated or vs control; Student’s t test, one-way ANOVA (with Bonferroni’s or Tukey’s posthoc test) or Kruskal–Wallis test (with Dunn’s multiple comparison test). ExONatide and Ex4(1–39) were applied at 800 nM and 10 nM, respectively. BME was applied at 10 mM. Values are the mean ± SEM unless otherwise stated.
Similar results were observed using the FRET-based biosensor Epac2-camps.43 While ExONatide induced an increase in cAMP, which could be partially washed out in the absence of reducing agent, baseline cAMP levels were only achieved following prior application of BME for 10 min. We note that MIN6B1-SNAP_GLP-1R cells endogenously express GLP-1R that may give rise to background adenylyl cyclase activity, although this did not appear to be a major issue, since cAMP responses to ExONatide were reduced to control levels by BME (Figure 3f,g). An effect of reducing agent per se is unlikely, as Ca2+ responses to Ex4(1–39) were unaffected by BME (Figure 3b–d). Moreover, BME did not influence insulin secretion at 11 mM d-glucose (Figure 3e), or cAMP responses to Ex4(1–39) (Figure 3f). Together, these data provide evidence for reversible signaling through the two main GLP-1R activation pathways in pancreatic beta cells.
BME was used for reductive cleavage, since it is cell permeable and the GLP-1R undergoes internalization following activation.37 While BME may conceivably induce toxicity and loss of function through modification of cell proteins including the receptor itself, this did not seem to be an issue in the present studies. Even following 60 min incubation with 10 mM BME, cells did not display significant signs of necrosis (Figure S4a) or apoptosis (Figure 3h), although cleaved caspase 3 was significantly upregulated after overnight exposure (Figure 3h). Furthermore, MIN6 cells remained proliferative and viable in culture 2 days after 0–60 min exposure to BME, suggesting that reductive alterations to proteins are unlikely to influence cell phenotype over the long-term (Figure S4b,c). ATP responses to d-glucose were similar in control and BME-treated MIN6 cells, demonstrating normal metabolism (Figure S4d). Lastly, cAMP concentration–responses to Ex4(1–39), ExONatide, and S39C-Ex4 were all unaffected by 10 min preincubation with BME (Figure S5).
ExONatide Allows Long-Lasting and Reversible GLP-1R Internalization
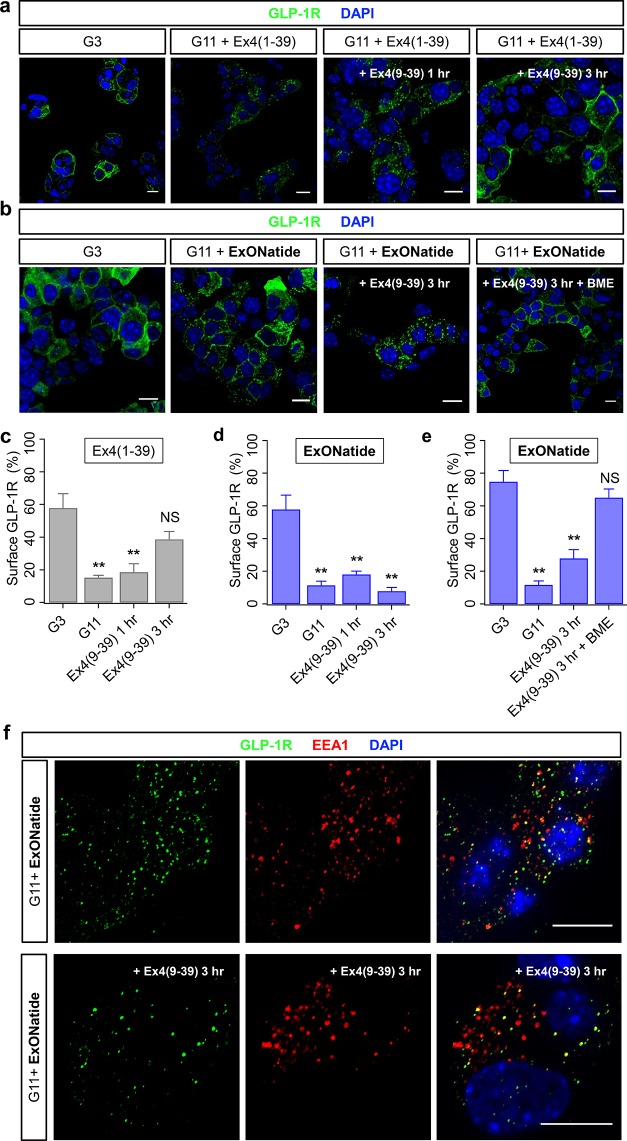
Following agonist binding, the GLP-1R undergoes internalization before ligand removal and endosomal sorting, either for recycling back to the plasma membrane or lysosomal degradation.23,25 Receptor recycling is a highly regulated process, and receptors that cannot disengage ligands are expected to remain sequestered within the endosome. To examine the effects of constitutive activation on receptor internalization, MIN6B1-SNAP_GLP-1R cells were treated with either Ex4(1–39) or ExONatide before immunohistochemistry for GLP-1R localization using a monoclonal antibody. Both Ex4(1–39) and ExONatide induced receptor internalization, as shown by a decrease in cell surface GLP-1R localization and an increase in punctate intracellular staining (Figures 4a–d, S6). However, this was only reversed by the specific antagonist for Ex4(9–39), with no significant plasma membrane recycling detected for ExONatide (Figures 4a–d, S6). Supporting a role for SNAP-tag-binding in irreversible internalization, recycling of the GLP-1R to the plasma membrane was seen following addition of both BME and Ex4(9–39) (Figure 4b,e). As for Ca2+ and cAMP signaling, BME alone did not alter receptor internalization (Figure S7).
Figure 4.
Reversible GLP-1R internalization by ExONatide. (a) Representative images showing that application of the agonist Ex4(1–39) to MIN6B1-SNAP_GLP-1R cells at high glucose concentration (11 mM; G11) induces GLP-1R internalization, which can be partially reversed with the antagonist Ex4(9–39) (G3; 3 mM glucose) (scale bar = 10 μm). (b) As for (a), but following application of ExONatide. Note that GLP-1R internalization can only be reversed by Ex4(9–39) following application of beta-mercaptoethanol (BME). (c) Surface GLP-1R expression is significantly reduced following application of Ex4(1–39), and this is reversed by application of Ex4(9–39). (d) Surface GLP-1R expression in MIN6B1-SNAP_GLP-1R cells is significantly reduced following application of ExONatide, but this is not reversed by application of Ex4(9–39). (e) As for (d), but showing plasma membrane recycling of GLP-1R following treatment with BME. (f) ExONatide-internalized GLP-1R in MIN6B1-SNAP_GLP-1R cells partially colocalizes with early endosome antigen 1 (EEA1) (top panels), and this is maintained in the presence of Ex4(9–39) (scale bar = 10 μm). **P < 0.01 and NS, nonsignificant vs G3 (one-way ANOVA with Tukey’s posthoc test). In (c) and (d), samples were run in parallel, hence the same control value (n = 3–7 experiments). ExONatide, Ex4(1–39), and Ex4(9–39) were applied at 800 nM, 10 nM, and 10 μM, respectively. BME was applied at 10 mM. Values are the mean ± SEM.
Following 1 h application of ExONatide, internalized GLP-1R showed partial (∼50%) colocalization with early endosome antigen 1 (EEA1), which is broadly in line with that previously reported at the same time point.37 Pertinently, this association was maintained even in the presence of Ex4(9–39) after 3 h (Figure 4f). Not all early endosomes were GLP-1R immunopositive (Figure 4f), however, indicating that either the receptor passes via EEA1+ endosomes before accessing other populations, or some EEA1+ endosomes no longer contain receptor at this stage.
Design and Synthesis of GhrelON
To demonstrate the broad applicability of our approach across class A and B GPCRs, we decided to install a bioconjugation handle on ghrelin, an orexigenic hormone involved in food intake and growth through GHS-R1a binding.32 By contrast to the convenient serine–cysteine substitution in S39C-Ex4 as the precursor for ExONatide, ghrelin comprises a C-terminal arginine that is highly conserved throughout mammalian ghrelin peptides.44 As the N-terminus is crucial for ghrelin potency and specificity, with the N-terminal pentapeptide GSS(C8:0)FL being the minimal fragment equipotent to ghrelin,45 we decided to retain the full-length sequence extended by a GGCG fragment. The benzyl guanine moiety was then merged with the ghrelinGGCG peptide by formation of a disulfide bond with BG-PEG4-SSpy. By using the same strategy and chemical building blocks as for ExONatide, we obtained GrehlON on the milligram scale (see Supporting Information for details on synthesis and characterization) (Figure 5a).
Figure 5.
GhrelON structure, signaling, and binding. (a) Structure of GhrelON showing the cleavable disulfide bridge and PEG-linker. (b) GhrelinGGCG and GhrelON display minimal loss of binding affinity for the GHS-R1a, as measured using specific Tag-lite competition assay (n = 2 assays in triplicate). (c) Ghrelin, ghrelinGGCG, and GhrelON demonstrate similar potencies for IP-1 generation (n = 2 assays in triplicate). (d) Preincubation of YFP-AD293-SNAP_GHS-R1a cells with GhrelON decreases BG-TMR binding/fluorescence intensity compared to native ghrelin (n = 230–385 cells). (e) GhrelON decreases BG-TMR binding/fluorescence in AD293-SNAP_mGluR2_GFP cells (n = 338–435 cells). (f and g) Representative images showing BG-TMR fluorescence in YFP-AD293-SNAP_GHS-R1a cells preincubated with and without a high concentration (1 μM) of GhrelON or ghrelin (scale bar = 33 μm). (h) As for (f) but AD293-SNAP_mGluR2_GFP cells treated with or without 10 μM GhrelON. Values are the mean ± SEM.
GhrelON Activates and Labels SNAP_GHS-R1a
Ghrelin binds the GHS-R1a with high affinity (Ki = 1.8 ± 0.3 nM), measured using HTRF-based competition assays (Figure 5b). GhrelON showed a small loss in binding affinity at the GHS-R1a (Ki = 6.1 ± 0.6 nM), which may be related to modification of the C-terminus to accept the bioconjugation handle (ghrelinGGCG also showed decreased binding affinity) (Ki = 19.6 ± 8.1 nM) (Figure 5b). Similar results were seen for IP-1 accumulation, with a small loss in potency detected for GhrelON (EC50 = 5.5 ± 0.5 nM) and ghrelinGGCG (EC50 = 11.15 ± 0.4 nM) versus ghrelin (EC50 = 3.2 ± 0.8 nM) (Figure 5c). GhrelON labeled the SNAP_GHS-R1a with almost equal efficiency (Figure 5d,f,g) to that detected with ExONatide and the SNAP_GLP-1R. While 100% labeling was not reached, again this probably reflected a reduction in the number of cell-surface receptors available for peptide labeling due to GHS-R1a internalization in the absence of ligand (Figure 5d).46 Indicating the presence of affinity labeling, SNAP-tag binding was also reduced in AD293-SNAP_mGluR2_GFP cells lacking the orthosteric site for ghrelin (Figure 5e,h).
GhrelON Allows Prolonged but Reversible GHS-R1a Activation and Internalization
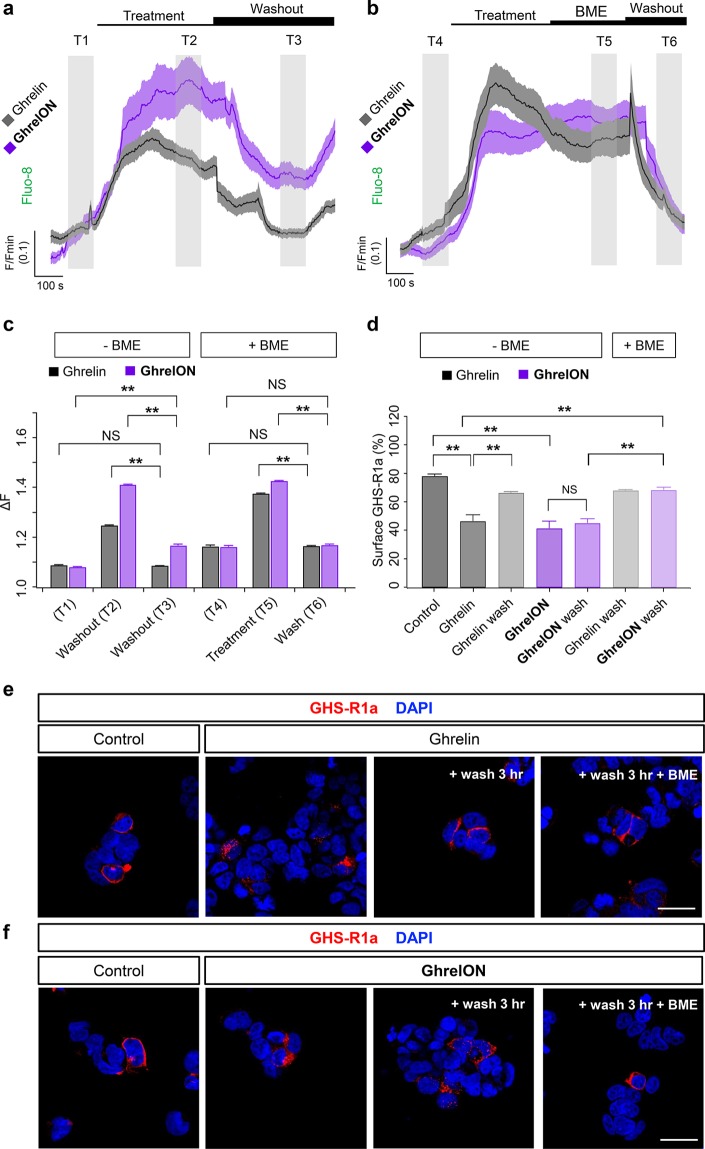
Both ghrelin and GhrelON induced large and sustained increases in cytoplasmic Ca2+ levels in YFP-AD293-SNAP_GHS-R1a cells, most likely through IP3-dependent liberation of Ca2+ from intracellular stores (Figure 6a).32,47 Whereas Ca2+ responses to ghrelin could be completely reversed following washout (Figure 6a,c), those of GhrelON were more persistent, showing an approximately 50% decrease (Figure 6a,c). Prior incubation with BME for 5 min before washout allowed Ca2+ levels to be subsequently restored to baseline levels in GhrelON-treated YFP-AD293-SNAP_GHS-R1a cells (Figure 6b,c). To examine receptor trafficking, YFP-AD293-SNAP_GHS-R1a cells were treated with ghrelin or GhrelON for 1 h before FLAG-immunostaining for the SNAP_GHS-R1a. Ghrelin induced GHS-R1a internalization (Figure 6d,e), with recycling back to the cell surface evident 3 h after agonist washout (Figure 6d,e). While GhrelON also induced GHS-R1a internalization (Figure 6d,f), this was only reversed following application of BME for 10 min before the wash step (Figure 6d,f). Treatment with BME for 10 min before application of ghrelin did not influence GHS-R1a internalization (Figure S8).
Figure 6.
GhrelON reversibly activates and internalizes the GHS-R1a. (a) Both ghrelin and GhrelON induce large increases in intracellular Ca2+ concentrations in YFP-AD293-SNAP_GHS-R1a, although washout of ligand only restores baseline Ca2+ levels for ghrelin (mean ± SEM traces shown) (gray shaded area T1–T3 = analysis time window) (n = 27–54 cells). (b) Application of beta-mercaptoethanol (BME) for 5 min immediately prior to washout reduces Ca2+ responses to GhrelON to baseline levels (mean ± SEM traces shown) (gray shaded area T4-T6 = analysis time window) (n = 27–54 cells). (c) Bar graph showing amplitude of Ca2+ responses to ghrelin and GhrelON before and after washout ± BME (n = 27–54 cells) (T1–T6 relate to time windows shown in a and b). (d) Treatment of YFP-AD293-SNAP_GHS-R1a cells with ghrelin for 1 h leads to GHS-R1a internalization, which can be partially reversed following washout and incubation for a further 3 h. GhrelON exerts similar effects, but these can only be washed out following prior application of BME for 10 min (n = 10–12 images from two experiments). (e) Representative images showing that ghrelin reduces surface GHS-R1a expression (detected via the FLAG-tag), which is reversed by a wash step (scale bar = 33 μm). f) As for (e), but showing that plasma membrane GHS-R1a recycling can only be achieved by application of BME to GhrelON-treated YFP-AD293-SNAP_GHS-R1a cells. **P < 0.01 and NS, nonsignificant, as indicated; Student’s t test or one-way ANOVA (with Bonferroni’s posthoc test). Ghrelin and GhrelON were applied at 100 nM and 800 nM, respectively. BME was applied at 10 mM. Values are the mean ± SEM.
Discussion
In the present study, we describe an incretin-mimetic termed ExONatide that allows tethered activation and internalization of the GLP-1R, a class B GPCR, when N-terminally fused to a SNAP-tag. We also show that this technology is applicable to a class A GPCR using GhrelON, which targets the SNAP-tagged GHS-R1a. In both cases, use of a disulfide bridge allows the reductive release of ligand and resumption of normal signaling processes, an approach called RECON. Thus, we further develop tethered pharmacology by using peptide ligands of class A and B GPCRs involved in the regulation of metabolism.
Prolonged activation of GPCRs has been described previously by means of cloning activating peptides onto the N-termini48,49 or by coexpression of membrane-anchored peptides for probing ion channel or GPCR function.50,51 However, using a SNAP-tagged receptor in conjunction with a BG-linked ligand bears several advantages: (i) the ligand/receptor ratio is defined as 1:1 in terms of binding and potency; (ii) local concentration with a PEG4 chain can be considered high10,52 and does not rely on membrane fluctuations; (iii) preactivation during expression and culture is absent; (iv) disturbances in vitro and in vivo are limited solely by SNAP-tag fusion; (v) it is bidirectional and can be switched ON and OFF in a binary fashion by virtue of the incorporated cleavage site; (vi) ligand can be freed from covalent tethering, allowing the study of normal trafficking processes mediated by orthosteric binding; and (vii) receptor subtypes can potentially be targeted.
RECON compares favorably to other tethered approaches, such as PORTL and DART,8−10 and provides two major advances: a chemically cleavable tether and a peptidic drug. However, PORTL and DART still possess distinct advantages including spatiotemporally precise photoswitching or proven efficacy in vivo, the latter allowing investigation of behavioral neuropharmacology.10 Thus, future studies will seek to use RECON as a platform to introduce these aspects, for example, via photoswitch incorporation, targeting ion channels, or intracellular proteins and in vivo testing.
Suggesting that the modifications required for tethering are well-tolerated, the potency of ExONatide and GhrelON for cAMP and IP-1 generation was only reduced 1 order of magnitude compared to native Ex4(1–39) and ghrelin, respectively. This may be due to the amino acid substitutions, since a similar loss of potency was detected for both the S39C variant of Ex4(1–39) and ghrelinGGCG. It can be assumed that further derivatization at this position to obtain the highly modified ExONatide or GhrelON would lower potency even more, but that any loss may be offset by covalent attachment at the receptor. Importantly, the ligand concentration required for orthosteric activation was much less than that required for full SNAP-tag labeling (pM versus μM), meaning that ExONatide and GhrelON bound to SNAP_GLP-1R and SNAP_GHS-R1a fusions are always active.
The AD293-SNAP_GLP-1R cells used in the present study likely represent an amplified system, since the EC50 values were in the pM range. While this makes calculation of the Emax difficult, ExONatide is still able to drive cAMP/Ca2+ rises and insulin release in MIN6 cells despite an apparently lowered EC50 versus Ex4(1–39). Moreover, the EC50 for ExONatide was similar in CHO cells stably expressing SNAP_GLP-1R and the high dynamic range FRET sensor TEpacvv. Suggesting that activation of the GLP-1R may occur in a cooperative manner, the Hill coefficient for cAMP generation by ExONatide was 1.42 ± 0.16 (1.25 ± 0.07 for S39C-Ex4).
ExONatide induced long-lasting GLP-1R redistribution to the endosomal compartment that could only be reversed following addition of a cell-permeable reducing agent. Although SNAP-labeled fluorophores with cell surface-restricted disulfide cleavage sites have been reported,53 they still rely on receptor activation by a native ligand. By contrast, ExONatide provides a physiological relevant tool for probing how class B GPCRs signal within organelles (e.g., endosomes), as well as how alterations in kinetics may influence second messenger recruitment (i.e., signal bias), all based upon intracellular manipulation. Indeed, ExONatide, Ex4(1–39) and S39C-Ex4 displayed similar EC50’s for cAMP and ERK, suggesting that (i) differences in agonist behavior are unlikely to be due to signal bias in the context of transmembrane versus intracellular activation; and (ii) GLP-1R internalization induced by ExONatide likely reflects signaling duration rather than intensity.
GLP-1R and GHS-R1a signaling have broad-ranging physiological functions. While conditional knockout mouse models exist,54 methods for selectively activating the receptors with ligands are lacking. CRISPR/Cas9 genome editing has, however, allowed the generation of epitope-tagged mice,55 and expressing endogenous SNAP-fusion proteins in rodents can be envisioned. Accordingly, ExONatide and GhrelON in combination with SNAP_GLP-1R or SNAP_GHS-R1a animals may provide a powerful platform for dissecting out the role of GLP-1R and GHS-R1a signaling in a cell-specific manner in vivo. The pharmacokinetics of ExONatide and GhrelON will need careful assessment before any in vivo workup, and noncleavable congeners lacking the disulfide bridge may be required to avoid reactivity with glutathione and other cysteine-containing proteins including serum albumin. However, recent studies have shown that disulfides are well-tolerated in vivo when incorporated into ligands as a backbone for cell-selective dual agonists.56 As an alternative to BME, which may be difficult to administer to an animal at high doses without apparent toxicity, the disulfide bridge can be cleaved by different modes of action in vivo, e.g., by reduced lipoic acid or harsh UV–C light.57 Furthermore, other approaches to “uncage” molecules, for instance, by using Pd-chemistry58 or bioorthogonal click and release reactions59−61 have been reported, and these molecular scaffolds may replace the disulfide bond in the future as options for in vivo application.
Compared to other pharmacological approaches, the RECON system affords unique opportunities in terms of understanding GPCR function in vitro: (i) endogenous GPCR signaling can be activated in a specific cell population within a complex tissue (e.g., catecholaminergic GLP-1R-expressing neurons in brain slices62); (ii) receptors with closely related ligands can be engaged with almost 100% specificity (i.e., GLP-1R, GLP-2R, and GCGR, which all bind hormones derived from post-translational processing of the proglucagon polypeptide63); (iii) the effects of intracellular ligand activity (e.g., transcriptional regulation64 and functional selectivity65) can be studied by controlling receptor recycling and degradation; and (iv) it becomes possible to study the influence of ligand dissociation constant (kon and koff) and residence time of ligands on biased behavior in diverse GPCRs.66 As such, we expect RECON and more widely tethered pharmacology to reveal novel facets of GPCR function.
Lastly, high concentrations of BME were required for reductive cleavage of both ExONatide and GhrelON. While apoptosis, necrosis, and metabolic indices were all apparently normal—even after prolonged 60 min exposure to BME—we cannot exclude an effect of reducing agent on cell viability. Moreover, BME did not influence GLP-1R or GHS-R1a signaling and trafficking, but other parameters such as exact ligand dissociation kinetics could not be easily investigated in the present studies using Tag-lite assays due to SNAP-tag occupancy, although this could feasibly be done with radioligands. Further studies are thus needed to understand whether and how BME would interfere more widely with cell proteins/receptor function, especially since the GLP-1R and GHS-R1a contain N-terminal disulfides essential for proper receptor folding. It should be noted that the cytosolic environment is highly reducing, with reported glutathione levels of ∼8 mM.67 Bioreductive cleavage is unlikely to influence activity of internalized ExONatide and GhrelON, since surface expression of the GLP-1R and GHS-R1a was not significantly altered 3 h after a wash step or application of antagonist. However, we cannot exclude that bioreduction of the disulfide bridge may alter other aspects of internalized ligand behavior not evaluated in the present study.
In summary, ExONatide and GhrelON provide templates for the design and production of agonists and constitute, to the best of our knowledge, the largest tethered drugs produced to date. Both allow prolonged yet reversible activation of cell surface receptor proteins, such as GPCRs, bearing a fused self-labeling protein tag. Ligands with other distinct properties, such as antagonists and/or modulators, or even branched versions bearing reporters, including dyes or MRI/contrast agents, can now be envisioned, and this is ongoing research in our laboratories.
Methods
Synthesis
Solid phase peptide synthesis and characterization of ExONatide and GhrelON is detailed in the Supporting Information.
Modeling
A structural model for the ExONatide bound SNAP_GLP-1R was built using pymol with the pdb structures 3L00 (SNAP-tag reacted with BG) and 5VAI (GLP-1 bound to GLP-1R). The 28 N-terminal amino acids of the GLP-1R were not resolved and are therefore depicted as a dashed line. The extended GLP-1 to resemble ExONatide, and the linker (disulfide bridge and PEG4 spacer) was built using the residue and fragment tool. We note that the structure only resembles molecular dimensions and is not energy optimized.
Cell Lines
AD293 cells were kept in Dulbecco’s Modified Eagles medium (DMEM) with 10% fetal calf serum (FCS), 1% l-glutamine and 1% penicillin/streptomycin, and incubated at 37 °C, 5% CO2. HEK293T cells stably expressing the human GHS-R1a (HEK293T-GHS-R1a) were maintained in DMEM Glutamax + high glucose supplemented with 10% heat-inactivated FCS, 50 U/mL penicillin, 50 μg/mL streptomycin, 1 mg/mL G418, 2 mM HEPES and 1% nonessential amino acids. CHO-K1 cells stably expressing the SNAP_GLP-1R (CHO-K1-SNAP_GLP-1R) were maintained in DMEM supplemented with 10% FCS, 1% penicillin/streptomycin, 500 μg/mL G418, 5 mM glucose, 10 mM HEPES and 1% nonessential amino acids. MIN6 beta cells stably expressing the SNAP_GLP-1R (MIN6B1-SNAP_GLP-1R) were grown in DMEM supplemented with 15% FCS, 25 mM d-glucose, 71 μM BME, 2 mM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin supplemented with 100 U/mL G418, and incubated at 37 °C, 5% CO2.
SNAP-Tag Binding Assays
AD293 cells were cotransfected with either SNAP_GLP-1R and YFP (YFP-AD293-SNAP_GLP-1R) or SNAP_GHS-R1a and YFP (YFP-AD293-SNAP_GHS-R1a) (both Cisbio) using PolyJet reagent (SignaGen) according to the manufacturer’s instructions. Cells were incubated for 30 min with Ex4(1–39), ghrelin, ExONatide, or GhrelON, before washing and counter-labeling with 0.5 μM BG-TMR for 30 min. Cells were imaged using Zeiss LSM780/880 meta-confocal microscopes configured with GaAsP PMT spectral detectors and 10×/0.45 W and 63×/1.20 W objectives. YFP was excited using a λ = 514 nm Argon laser, and emitted signals captured from λ = 524–567 nm. BG-TMR was excited at λ = 561 nm, and emitted signals captured from λ = 570–641 nm. Control experiments were performed using either mock cells, or AD293 transfected with SNAP_mGluR2_GFP (AD293-SNAP_mGluR2_GFP), GLP-1R_GFP (AD293-GLP-1R_GFP), or YFP (AD293-YFP), as above.
cAMP Assays
Cyclic adenosine monophosphate (cAMP) levels were measured using a PerkinElmer LANCE TR-FRET kit according to the manufacturer’s instructions, normalized to a 5 μM forskolin (FSK) maximal response and plotted as % change. Treatments were applied as indicated to suspended cells in a Greiner low-volume 384-well plate for 10 or 30 min in the presence of 100 μM 3-isobutyl-1-methylxanthine (IBMX), before lysis to extract total cAMP. Excitation was performed at λ = 340 nm, and emitted signals detected at both λ = 615 nm and λ = 665 nm using a BMG PHERAStar microplate reader. Control experiments were performed in AD293 cells transfected with GLP-1R_GFP or in the presence of 10 mM BME.
Biased Signaling Measurements
The FRET reporters TEpacvv (mTurquoise-Venus) (a kind gift from Kees Jalink) and cytoplasmic EKAR (CFP-YFP) (a kind gift from Karel Svoboda) were cotransfected into CHO-K1-SNAP_GLP-1R cells together with a puromycin resistance plasmid. Clones were then generated using puromycin selection and FACS. FRET measurements were performed in black plates, with freshly detached cells suspended in HBSS, using a Flexstation 3 plate reader (excitation λ = 440 nm, emission λ = 485 nm and λ = 535 nm for mTurquoise (CFP) and Venus (YFP), respectively). Serial measurements every 2 min were taken during a 10 min baseline and for 30 min after agonist addition at multiple doses. mTurquoise (CFP) and Venus (YFP) measurements were expressed ratiometrically and normalized to individual well baseline.
Relative potency ratios were used to calculate bias, as each compound was a full agonist in each pathway.41 In each assay, LogEC50 values were calculated at each time point for each agonist by 3-parameter fitting. Kinetic changes in LogEC50 were then fitted with a one-phase decay function, from which interpolated values were obtained at finer temporal resolution. Relative potencies (ΔLogEC50) were obtained by subtracting LogEC50 values for S39C-Ex4 and ExONatide from that of the reference agonist Ex4 in each assay at each interpolated time point. Bias (ΔΔLogEC50) was then determined by subtracting the relative potency of each test agonist in each pathway. As all agonists were tested for each pathway in parallel to reduce variability, bias was calculated on a per assay basis.
Calcium Imaging
MIN6B1-SNAP_GLP-1R or YFP-AD293-SNAP_GHS-R1a cells were loaded with the Ca2+ indicator Fluo8 (10 μM) for 30 min. Ca2+ imaging was performed using a Crest X-Light spinning disk head coupled to a Nikon Ti-E automated base and 10×/0.4 NA objective. Excitation was delivered at λ = 458–482 nm using a Lumencor Spectra X Light engine, and emitted signals were detected at λ = 500–550 nm using a Photometrics Evolve Delta 512 EMCCD. ExONatide, Ex4(1–39), GhrelON, or ghrelin were added to the imaging chamber for 15–20 min before washing the cells with buffer for 5 min ± BME. Recordings were then continued for a further 15 min. Intracellular Ca2+ concentration ([Ca2+]i) was determined as the mean at the time points indicated. For experiments with MIN6B1-SNAP_GLP-1R, d-glucose was added at 11 mM, which is permissive for incretin action. HEPES-bicarbonate buffer was used, containing in mM: 120 NaCl, 4.8 KCl, 24 NaHCO3, 0.5 Na2HPO4, 5 HEPES, 2.5 CaCl2, 1.2 MgCl2. For experiments with YFP-AD293-SNAP_GHS-R1a, samples were maintained in normal culture medium. Intensity-overtime traces were extracted using a region of interest (ROI) and for comparison normalized as F/Fmin where F = fluorescence at a given time point and Fmin = minimum fluorescence.
FRET Imaging
cAMP generation in MIN6B1-SNAP_GLP-1R cells was measured before and after washout of drug in the absence or presence of BME using the FRET probe Epac2-camps.68 Following 10 min incubation, snapshots were captured using the Crest X-Light spinning disk system, and excitation was performed at λ = 430–450 nm. Emitted signals were detected at emission λ = 460–500 nm and λ = 520–550 nm for Cerulean and Citrine, respectively. Results were expressed as the ratio of Cerulean/Citrine. In all cases, HEPES-bicarbonate buffer was used.
Insulin Secretion Assays
MIN6B1-SNAP_GLP-1R cells grown in 12-well plates were incubated with HEPES-bicarbonate buffer supplemented with 0.1% BSA and containing the indicated treatments for 30 min. For washout experiments, cells were treated with ExONatide, before washout with and without 10 mM BME for 10 min and reincubation with buffer for a further 30 min. Insulin concentration in the supernatant was assayed using a HTRF-based assay (Cisbio) according to the manufacturer’s instructions.
Apoptosis, Necrosis, and Proliferation Assay
For quantification of apoptosis, MIN6 cells were incubated with 10 mM BME for the indicated times and fixed in 4% paraformaldehyde. Immunostaining was performed using an antibody against Cleaved Caspase 3 (CC3; 1:400; #9661, Cell Signaling Technology), before secondary goat antirabbit Alexa Fluor 633 (1:1000) and Hoechst 33342 staining. Cells were imaged using a Zeiss LSM 880 confocal microscope, with excitation delivered at λ = 405 nm and λ = 633 nm. Emitted signals were collected at λ = 410–550 nm and λ = 641–695 nm for Hoechst 33342 and Alexa Fluor 633, respectively. Total area was quantified using the threshold plugin for Fiji, and results were expressed as the ratio CC3/Hoechst, as described.69
For quantification of necrosis, MIN6 cells were labeled with 5 μM calcein-AM and 5 μM propidium iodide for 30 min. Cells were imaged in HEPES-bicarbonate buffer using the Crest X-Light spinning disk system, and excitation was performed at λ = 458–482 nm and λ = 543–558 nm for calcein and propidium iodide, respectively. Emitted signals were detected at λ = 500–550 nm and λ = 602–662 nm for calcein and propidium iodide, respectively. Total area was quantified using the threshold plugin for Fiji and expressed as the ratio propidium iodide/calcein, as described.70
For quantification of proliferation, MIN6 cells were fixed and labeled with Hoechst 33342. Whole wells were imaged using the Crest X-Light spinning disk system, excitation performed at λ = 383–408 nm, and emitted signals were detected at λ = 435–485. Images were stitched together, and the total area was analyzed using the stitching and threshold plugins for Fiji, respectively.
ATP Assay
MIN6 cells were treated with 10 mM BME for 10 min at 37 °C followed by incubation with HEPES buffer containing 3 mM or 20 mM d-glucose for 30 min at 37 °C. ATP content was detected using the ATP determination kit (A22066; Thermo Fisher Scientific) and a BMG PHERAstar microplate reader, as described.71
GLP-1R Internalization Studies
MIN6B1-SNAP_GLP-1R cells were incubated in HEPES-bicarbonate buffer supplemented with 3 mM d-glucose for 2 h at 37 °C and either fixed in 4% paraformaldehyde or further stimulated with 11 mM d-glucose (G11) containing 10 nM Ex-4(1–39) or 800 nM ExONatide for 1 h at 37 °C. Cells were either fixed or washed extensively prior to incubation with 10 μM Ex-4(9–39) for a further 1 or 3 h following 0 or 10 min application of 10 mM BME. At the concentration used here, Ex4(9–39) is expected to effectively trap GLP-1R at the plasma membrane, allowing recycling to be quantified via reappearance of labeled receptor. Control experiments were performed using BME and Ex4(1–39)-alone. Immunostaining was performed using mouse antihuman GLP-1R (1:30; Mab 3F52; Developmental Studies Hybridoma Bank) and rabbit anti-EEA1 antibodies (1:50; sc-33585; Santa Cruz Biotechnology), before secondary antimouse AlexaFluor-488 staining and mounting on coverslips with Vectashield Hardset + DAPI. Images were captured using a Zeiss LSM780 microscope and a 63x/1.4 NA oil objective. Excitation was delivered at λ = 405 nm and λ = 481 nm. Emitted signals were collected at λ = 410–495 nm and λ = 493–630 nm for DAPI and AlexaFlour-488, respectively. Surface signal was quantified on binarized images using the threshold plugin for ImageJ and expressed relative to the total signal.
GHS-R1a Ligand Binding Assay
Ki values were determined from binding competition experiments performed on HEK293T-GHS-R1a cells using a homogenous time resolved fluorescence (HTRF) assay (Cisbio), as previously described.72Ki values were calculated from binding curves.
Inositol Phosphate Assay
Inositol phosphate accumulation assays were conducted 48 h after transfection using HEK293T-GHS-R1a cells (50 000 cells/well of a 96-well plate). IP-1 production was measured using the IP-One HTRF kit, as previously described.73 Values are expressed as ΔF where ΔF = (ratio 665 nm/620 nm assay – ratio 665 nm/620 nm negative control)/ratio 665 nm/620 nm negative control. The negative control, corresponding to the Lumi4-Tb blank, was used as an internal assay control. Inositol phosphate accumulation was expressed as the percentage of the maximal ghrelin response using the formula: (ΔF mock cells – ΔF receptor transfected cells)/(ΔF mock cells – ΔF maximal ghrelin stimulation for receptor transfected cells). The basal signal in the absence of any ligand stimulation corresponded to constitutive activity of GHS-R1a (representing 70–75% of maximal stimulation promoted by ghrelin).
GHS-R1a Internalization Studies
AD293-SNAP_GHS-R1a cells were incubated with 100 nM ghrelin or 800 nM GhrelON for 1 h at 37 °C. Cells were either fixed or washed before incubation for a further 3 h following 0 or 10 min application of 10 mM BME. Immunostaining was performed using mouse anti-FLAG antibody (1:200; F1804; Sigma-Aldrich), before application of secondary antimouse AlexaFluor-568 and mounting on coverslips with Vectashield Hardset + DAPI. Images were captured using a Zeiss LSM780 microscope and 63x/1.2 NA water objective. Excitation was delivered at λ = 405 nm and λ = 561. Emitted signals were collected at λ = 410–585 nm and λ = 568–691 nm, respectively. Surface/total signal was quantified as for the GLP-1R.
Statistics
Data normality was assessed using the D’Agostino-Pearson test. Nonmultifactorial comparisons were made using Student’s t test. Multifactorial comparisons were made using one-way ANOVA followed by Bonferroni’s or Tukey’s posthoc tests, or if non-Gaussian, Kruskal–Wallis test followed by Dunn’s posthoc test. Log-transformed concentration–response curves were fitted using the Hill equation to allow calculation of EC50 values. All analyses were conducted using GraphPad Prism 6.0 (GraphPad Software) and IgorPro 6.2. Results were considered significant at P < 0.05.
Acknowledgments
We are grateful to the SFB749 (T.P., M.G., and A.H.R.), the SFB TRR 152 (J.B. and D.T.), the SFB1032 (P.L. and D.T.), and the Center for Integrated Protein Science Munich (CIPSM) (T.P., J.B., P.L., M.G., D.T., and A.H.R.). T.P. was supported by an EASD Albert-Renold Young Scientist Fellowship (94571). A.T. was supported by an MRC Project Grant (MR/M012646/1) and a Diabetes UK Early-Career Small Grant (16/0005441). D.J.H. was supported by Diabetes UK. R.D. Lawrence (12/0004431), EFSD/Novo Nordisk Rising Star and Birmingham Fellowships, a Wellcome Trust Institutional Support Award, and an MRC Project Grant (MR/N00275X/1). This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (Starting Grant 715884 to D.J.H. and Advanced Grant 268795 to D.T.). MIN6B1 cells were provided by Prof. Philippe Halban (University of Geneva, Switzerland) with permission from Prof. Jun-ichi Miyazaki (University of Osaka) who produced the maternal MIN6 cell line.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.7b00237.
Chemistry and spectroscopy, synthesis, spectral data and supporting figures related to ExONatide and GhrelON (PDF)
Author Present Address
○ Max Planck Institute for Medical Research, Department of Chemical Biology, Jahnstr. 29, 69120 Heidelberg, Germany.
Author Present Address
● New York University, Department of Chemistry, Silver Center for Arts and Science, 100 Washington Square East, New York, NY 10003, USA.
Author Contributions
# T.P., J.A. and J.B. contributed equally.
Author Contributions
T.P., J.B., D.T., A.H.R., and D.J.H. conceived and designed the study. D.J.H., A.H.R., and D.T. jointly supervised the research. T.P., P.L. and M.G. performed synthesis. T.P., J.A., J.B., D.N., B.J.J., N.H.F.F, T.B., N.K., C.K., J-L.B., J.M., A.T., and D.J.H. performed the experiments. T.P., J.A., J.B., and D.J.H. wrote the paper with input from all the authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Kobilka B. K. G protein coupled receptor structure and activation. Biochim. Biophys. Acta, Biomembr. 2007, 1768 (4), 794–807. 10.1016/j.bbamem.2006.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leippe P.; Koehler Leman J.; Trauner D. Specificity and Speed: Tethered Photopharmacology. Biochemistry 2017, 56 (39), 5214–5220. 10.1021/acs.biochem.7b00687. [DOI] [PubMed] [Google Scholar]
- Xue L.; Karpenko I. A.; Hiblot J.; Johnsson K. Imaging and manipulating proteins in live cells through covalent labeling. Nat. Chem. Biol. 2015, 11 (12), 917–923. 10.1038/nchembio.1959. [DOI] [PubMed] [Google Scholar]
- Keppler A.; Gendreizig S.; Gronemeyer T.; Pick H.; Vogel H.; Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 2003, 21 (1), 86–89. 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- Yang G.; de Castro Reis F.; Sundukova M.; Pimpinella S.; Asaro A.; Castaldi L.; Batti L.; Bilbao D.; Reymond L.; Johnsson K.; Heppenstall P. A. Genetic targeting of chemical indicators in vivo. Nat. Methods 2015, 12 (2), 137–139. 10.1038/nmeth.3207. [DOI] [PubMed] [Google Scholar]
- Maurel D.; Comps-Agrar L.; Brock C.; Rives M. L.; Bourrier E.; Ayoub M. A.; Bazin H.; Tinel N.; Durroux T.; Prezeau L.; Trinquet E.; Pin J. P. Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tag technologies: application to GPCR oligomerization. Nat. Methods 2008, 5 (6), 561–567. 10.1038/nmeth.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholler P.; Moreno-Delgado D.; Lecat-Guillet N.; Doumazane E.; Monnier C.; Charrier-Savournin F.; Fabre L.; Chouvet C.; Soldevila S.; Lamarque L.; Donsimoni G.; Roux T.; Zwier J. M.; Trinquet E.; Rondard P.; Pin J. P. HTS-compatible FRET-based conformational sensors clarify membrane receptor activation. Nat. Chem. Biol. 2017, 13 (4), 372–380. 10.1038/nchembio.2286. [DOI] [PubMed] [Google Scholar]
- Levitz J.; Broichhagen J.; Leippe P.; Konrad D.; Trauner D.; Isacoff E. Y. Dual optical control and mechanistic insights into photoswitchable group II and III metabotropic glutamate receptors. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (17), E3546–E3554. 10.1073/pnas.1619652114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broichhagen J.; Damijonaitis A.; Levitz J.; Sokol K. R.; Leippe P.; Konrad D.; Isacoff E. Y.; Trauner D. Orthogonal Optical Control of a G Protein-Coupled Receptor with a SNAP-Tethered Photochromic Ligand. ACS Cent. Sci. 2015, 1 (7), 383–393. 10.1021/acscentsci.5b00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields B. C.; Kahuno E.; Kim C.; Apostolides P. F.; Brown J.; Lindo S.; Mensh B. D.; Dudman J. T.; Lavis L. D.; Tadross M. R. Deconstructing behavioral neuropharmacology with cellular specificity. Science 2017, 356 (6333), eaaj2161. 10.1126/science.aaj2161. [DOI] [PubMed] [Google Scholar]
- Berry M. H.; Holt A.; Levitz J.; Broichhagen J.; Gaub B. M.; Visel M.; Stanley C.; Aghi K.; Kim Y. J.; Trauner D.; Flannery J.; Isacoff E. Y. Restoration of patterned vision with an engineered photoactivatable G protein-coupled receptor. Nat. Commun. 2017, 8 (1), 1862. 10.1038/s41467-017-01990-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. H.; Essig S.; James J. R.; Lang K.; Chin J. W. Selective, rapid and optically switchable regulation of protein function in live mammalian cells. Nat. Chem. 2015, 7 (7), 554–561. 10.1038/nchem.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broichhagen J.; Frank J. A.; Trauner D. A Roadmap to Success in Photopharmacology. Acc. Chem. Res. 2015, 48, 1947–1960. 10.1021/acs.accounts.5b00129. [DOI] [PubMed] [Google Scholar]
- Campbell J. E.; Drucker D. J. Pharmacology, physiology, and mechanisms of incretin hormone action. Cell Metab. 2013, 17 (6), 819–837. 10.1016/j.cmet.2013.04.008. [DOI] [PubMed] [Google Scholar]
- Gefel D.; Hendrick G. K.; Mojsov S.; Habener J.; Weir G. C. Glucagon-like peptide-I analogs: effects on insulin secretion and adenosine 3′,5′-monophosphate formation. Endocrinology 1990, 126 (4), 2164–2168. 10.1210/endo-126-4-2164. [DOI] [PubMed] [Google Scholar]
- Thorens B. Expression cloning of the pancreatic beta cell receptor for the gluco-incretin hormone glucagon-like peptide 1. Proc. Natl. Acad. Sci. U. S. A. 1992, 89 (18), 8641–8645. 10.1073/pnas.89.18.8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz G. G. t.; Leech C. A.; Habener J. F. Activation of a cAMP-regulated Ca(2+)-signaling pathway in pancreatic beta-cells by the insulinotropic hormone glucagon-like peptide-1. J. Biol. Chem. 1995, 270 (30), 17749–17757. 10.1074/jbc.270.30.17749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromada J.; Bokvist K.; Ding W. G.; Holst J. J.; Nielsen J. H.; Rorsman P. Glucagon-like peptide 1 (7–36) amide stimulates exocytosis in human pancreatic beta-cells by both proximal and distal regulatory steps in stimulus-secretion coupling. Diabetes 1998, 47 (1), 57–65. 10.2337/diab.47.1.57. [DOI] [PubMed] [Google Scholar]
- Holz G. G. Epac: A new cAMP-binding protein in support of glucagon-like peptide-1 receptor-mediated signal transduction in the pancreatic beta-cell. Diabetes 2004, 53 (1), 5–13. 10.2337/diabetes.53.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz G. G.; Leech C. A.; Heller R. S.; Castonguay M.; Habener J. F. cAMP-dependent mobilization of intracellular Ca2+ stores by activation of ryanodine receptors in pancreatic beta-cells. A Ca2+ signaling system stimulated by the insulinotropic hormone glucagon-like peptide-1-(7–37). J. Biol. Chem. 1999, 274 (20), 14147–14156. 10.1074/jbc.274.20.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roed S. N.; Nohr A. C.; Wismann P.; Iversen H.; Brauner-Osborne H.; Knudsen S. M.; Waldhoer M. Functional consequences of glucagon-like peptide-1 receptor cross-talk and trafficking. J. Biol. Chem. 2015, 290 (2), 1233–1243. 10.1074/jbc.M114.592436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calebiro D.; Nikolaev V. O.; Gagliani M. C.; de Filippis T.; Dees C.; Tacchetti C.; Persani L.; Lohse M. J. Persistent cAMP-signals triggered by internalized G-protein-coupled receptors. PLoS Biol. 2009, 7 (8), e1000172. 10.1371/journal.pbio.1000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuna R. S.; Girada S. B.; Asalla S.; Vallentyne J.; Maddika S.; Patterson J. T.; Smiley D. L.; DiMarchi R. D.; Mitra P. Glucagon-like peptide-1 receptor-mediated endosomal cAMP generation promotes glucose-stimulated insulin secretion in pancreatic beta-cells. Am. J. Physiol. Endocrinol. Metab. 2013, 305 (2), 161–170. 10.1152/ajpendo.00551.2012. [DOI] [PubMed] [Google Scholar]
- Vilardaga J. P.; Jean-Alphonse F. G.; Gardella T. J. Endosomal generation of cAMP in GPCR signaling. Nat. Chem. Biol. 2014, 10 (9), 700–706. 10.1038/nchembio.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roed S. N.; Wismann P.; Underwood C. R.; Kulahin N.; Iversen H.; Cappelen K. A.; Schaffer L.; Lehtonen J.; Hecksher-Soerensen J.; Secher A.; Mathiesen J. M.; Brauner-Osborne H.; Whistler J. L.; Knudsen S. M.; Waldhoer M. Real-time trafficking and signaling of the glucagon-like peptide-1 receptor. Mol. Cell. Endocrinol. 2014, 382 (2), 938–949. 10.1016/j.mce.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Drucker D. J. The Cardiovascular Biology of Glucagon-like Peptide-1. Cell Metab. 2016, 24 (1), 15–30. 10.1016/j.cmet.2016.06.009. [DOI] [PubMed] [Google Scholar]
- Hogan A. E.; Gaoatswe G.; Lynch L.; Corrigan M. A.; Woods C.; O’Connell J.; O’Shea D. Glucagon-like peptide 1 analogue therapy directly modulates innate immune-mediated inflammation in individuals with type 2 diabetes mellitus. Diabetologia 2014, 57 (4), 781–784. 10.1007/s00125-013-3145-0. [DOI] [PubMed] [Google Scholar]
- Dailey M. J.; Moran T. H. Glucagon-like peptide 1 and appetite. Trends Endocrinol. Metab. 2013, 24 (2), 85–91. 10.1016/j.tem.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panjwani N.; Mulvihill E. E.; Longuet C.; Yusta B.; Campbell J. E.; Brown T. J.; Streutker C.; Holland D.; Cao X.; Baggio L. L.; Drucker D. J. GLP-1 receptor activation indirectly reduces hepatic lipid accumulation but does not attenuate development of atherosclerosis in diabetic male ApoE(−/−) mice. Endocrinology 2013, 154 (1), 127–139. 10.1210/en.2012-1937. [DOI] [PubMed] [Google Scholar]
- Kim M.; Platt M. J.; Shibasaki T.; Quaggin S. E.; Backx P. H.; Seino S.; Simpson J. A.; Drucker D. J. GLP-1 receptor activation and Epac2 link atrial natriuretic peptide secretion to control of blood pressure. Nat. Med. 2013, 19 (5), 567–575. 10.1038/nm.3128. [DOI] [PubMed] [Google Scholar]
- Hansotia T.; Baggio L. L.; Delmeire D.; Hinke S. A.; Yamada Y.; Tsukiyama K.; Seino Y.; Holst J. J.; Schuit F.; Drucker D. J. Double incretin receptor knockout (DIRKO) mice reveal an essential role for the enteroinsular axis in transducing the glucoregulatory actions of DPP-IV inhibitors. Diabetes 2004, 53 (5), 1326–1335. 10.2337/diabetes.53.5.1326. [DOI] [PubMed] [Google Scholar]
- Kojima M.; Kangawa K. Ghrelin: Structure and Function. Physiol. Rev. 2005, 85 (2), 495–522. 10.1152/physrev.00012.2004. [DOI] [PubMed] [Google Scholar]
- Osterstock G.; Escobar P.; Mitutsova V.; Gouty-Colomer L. A.; Fontanaud P.; Molino F.; Fehrentz J. A.; Carmignac D.; Martinez J.; Guerineau N. C.; Robinson I. C.; Mollard P.; Mery P. F. Ghrelin stimulation of growth hormone-releasing hormone neurons is direct in the arcuate nucleus. PLoS One 2010, 5 (2), e9159. 10.1371/journal.pone.0009159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer M.; Langlet F.; Lafont C.; Molino F.; Hodson D. J.; Roux T.; Lamarque L.; Verdie P.; Bourrier E.; Dehouck B.; Baneres J. L.; Martinez J.; Mery P. F.; Marie J.; Trinquet E.; Fehrentz J. A.; Prevot V.; Mollard P. Rapid sensing of circulating ghrelin by hypothalamic appetite-modifying neurons. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (4), 1512–1517. 10.1073/pnas.1212137110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood C. R.; Garibay P.; Knudsen L. B.; Hastrup S.; Peters G. H.; Rudolph R.; Reedtz-Runge S. Crystal structure of glucagon-like peptide-1 in complex with the extracellular domain of the glucagon-like peptide-1 receptor. J. Biol. Chem. 2010, 285 (1), 723–730. 10.1074/jbc.M109.033829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge S.; Thogersen H.; Madsen K.; Lau J.; Rudolph R. Crystal structure of the ligand-bound glucagon-like peptide-1 receptor extracellular domain. J. Biol. Chem. 2008, 283 (17), 11340–11347. 10.1074/jbc.M708740200. [DOI] [PubMed] [Google Scholar]
- Roed S. N.; Wismann P.; Underwood C. R.; Kulahin N.; Iversen H.; Cappelen K. A.; Schäffer L.; Lehtonen J.; Hecksher-Soerensen J.; Secher A.; Mathiesen J. M.; Bräuner-Osborne H.; Whistler J. L.; Knudsen S. M.; Waldhoer M. Real-time trafficking and signaling of the glucagon-like peptide-1 receptor. Mol. Cell. Endocrinol. 2014, 382 (2), 938–949. 10.1016/j.mce.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Wootten D.; Reynolds C. A.; Smith K. J.; Mobarec J. C.; Koole C.; Savage E. E.; Pabreja K.; Simms J.; Sridhar R.; Furness S. G.; Liu M.; Thompson P. E.; Miller L. J.; Christopoulos A.; Sexton P. M. The Extracellular Surface of the GLP-1 Receptor Is a Molecular Trigger for Biased Agonism. Cell 2016, 165 (7), 1632–1643. 10.1016/j.cell.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarenbeek J. B.; Goedhart J.; Hink M. A.; Gadella T. W. J.; Jalink K. A mTurquoise-Based cAMP Sensor for Both FLIM and Ratiometric Read-Out Has Improved Dynamic Range. PLoS One 2011, 6 (4), e19170. 10.1371/journal.pone.0019170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey C. D.; Ehrhardt A. G.; Cellurale C.; Zhong H.; Yasuda R.; Davis R. J.; Svoboda K. A genetically encoded fluorescent sensor of ERK activity. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (49), 19264–19269. 10.1073/pnas.0804598105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenakin T.; Christopoulos A. Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat. Rev. Drug Discovery 2012, 12 (3), 205–216. 10.1038/nrd3954. [DOI] [PubMed] [Google Scholar]
- Klein Herenbrink C.; Sykes D. A.; Donthamsetti P.; Canals M.; Coudrat T.; Shonberg J.; Scammells P. J.; Capuano B.; Sexton P. M.; Charlton S. J.; Javitch J. A.; Christopoulos A.; Lane J. R. The role of kinetic context in apparent biased agonism at GPCRs. Nat. Commun. 2016, 7, 10842. 10.1038/ncomms10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett K. L.; Cooper D. M. An improved targeted cAMP sensor to study the regulation of adenylyl cyclase 8 by Ca2+ entry through voltage-gated channels. PLoS One 2013, 8 (9), e75942. 10.1371/journal.pone.0075942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delporte C. Structure and physiological actions of ghrelin. Scientifica 2013, 2013, 518909. 10.1155/2013/518909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek M. A.; Feighner S. D.; Pong S. S.; McKee K. K.; Hreniuk D. L.; Silva M. V.; Warren V. A.; Howard A. D.; Van Der Ploeg L. H.; Heck J. V. Structure-function studies on the new growth hormone-releasing peptide, ghrelin: minimal sequence of ghrelin necessary for activation of growth hormone secretagogue receptor 1a. J. Med. Chem. 2000, 43 (23), 4370–4376. 10.1021/jm0001727. [DOI] [PubMed] [Google Scholar]
- Mear Y.; Enjalbert A.; Thirion S. GHS-R1a constitutive activity and its physiological relevance. Front. Neurosci. 2013, 7 (87), 1–5. 10.3389/fnins.2013.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima M.; Hosoda H.; Date Y.; Nakazato M.; Matsuo H.; Kangawa K. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature 1999, 402 (6762), 656–660. 10.1038/45230. [DOI] [PubMed] [Google Scholar]
- Nielsen S. M.; Nielsen L. Z.; Hjorth S. A.; Perrin M. H.; Vale W. W. Constitutive activation of tethered-peptide/corticotropin-releasing factor receptor chimeras. Proc. Natl. Acad. Sci. U. S. A. 2000, 97 (18), 10277–10281. 10.1073/pnas.97.18.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y.; Zhou X. E.; Hou L.; Zhao L.-H.; Liu B.; Wang G.; Jiang Y.; Melcher K.; Xu H. E. An intrinsic agonist mechanism for activation of glucagon-like peptide-1 receptor by its extracellular domain. Cell Discovery 2016, 2, 16042. 10.1038/celldisc.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C.; Nitabach M. N. Membrane-tethered ligands: tools for cell-autonomous pharmacological manipulation of biological circuits. Physiology 2013, 28 (3), 164–171. 10.1152/physiol.00056.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J. P.; Zhu Y.; Choi C.; Beinborn M.; Nitabach M. N.; Kopin A. S. Membrane-tethered ligands are effective probes for exploring class B1 G protein-coupled receptor function. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (19), 8049–8054. 10.1073/pnas.0900149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy V. M.; Semetey V.; Bracher P. J.; Shen N.; Whitesides G. M. Dependence of effective molarity on linker length for an intramolecular protein-ligand system. J. Am. Chem. Soc. 2007, 129 (5), 1312–1320. 10.1021/ja066780e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole N. B.; Donaldson J. G. Releasable SNAP-tag probes for studying endocytosis and recycling. ACS Chem. Biol. 2012, 7 (3), 464–469. 10.1021/cb2004252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. P.; An Z.; Wagner C.; Lewis A. G.; Cohen E. B.; Li B.; Mahbod P.; Sandoval D.; Perez-Tilve D.; Tamarina N.; Philipson L. H.; Stoffers D. A.; Seeley R. J.; D’Alessio D. A. The Role of β Cell Glucagon-like Peptide-1 Signaling in Glucose Regulation and Response to Diabetes Drugs. Cell Metab. 2014, 19 (6), 1050–1057. 10.1016/j.cmet.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt R. J.; Chen S.; Zhou Y.; Yim M. J.; Swiech L.; Kempton H. R.; Dahlman J. E.; Parnas O.; Eisenhaure T. M.; Jovanovic M.; Graham D. B.; Jhunjhunwala S.; Heidenreich M.; Xavier R. J.; Langer R.; Anderson D. G.; Hacohen N.; Regev A.; Feng G.; Sharp P. A.; Zhang F. CRISPR-Cas9 Knockin Mice for Genome Editing and Cancer Modeling. Cell 2014, 159 (2), 440–455. 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarta C.; Clemmensen C.; Zhu Z.; Yang B.; Joseph S. S.; Lutter D.; Yi C. X.; Graf E.; Garcia-Caceres C.; Legutko B.; Fischer K.; Brommage R.; Zizzari P.; Franklin B. S.; Krueger M.; Koch M.; Vettorazzi S.; Li P.; Hofmann S. M.; Bakhti M.; Bastidas-Ponce A.; Lickert H.; Strom T. M.; Gailus-Durner V.; Bechmann I.; Perez-Tilve D.; Tuckermann J.; Hrabe de Angelis M.; Sandoval D.; Cota D.; Latz E.; Seeley R. J.; Muller T. D.; DiMarchi R. D.; Finan B.; Tschop M. H. Molecular Integration of Incretin and Glucocorticoid Action Reverses Immunometabolic Dysfunction and Obesity. Cell Metab. 2017, 26 (4), 620–632e6. 10.1016/j.cmet.2017.08.023. [DOI] [PubMed] [Google Scholar]
- Lundblad R. L.Chemical Reagents for Protein Modification, CRC Press, 2014. [Google Scholar]
- Friedman Ohana R.; Levin S.; Wood M. G.; Zimmerman K.; Dart M. L.; Schwinn M. K.; Kirkland T. A.; Hurst R.; Uyeda H. T.; Encell L. P.; Wood K. V. Improved Deconvolution of Protein Targets for Bioactive Compounds Using a Palladium Cleavable Chloroalkane Capture Tag. ACS Chem. Biol. 2016, 11 (9), 2608–2617. 10.1021/acschembio.6b00408. [DOI] [PubMed] [Google Scholar]
- Azoulay M.; Tuffin G.; Sallem W.; Florent J. C. A new drug-release method using the Staudinger ligation. Bioorg. Med. Chem. Lett. 2006, 16 (12), 3147–3149. 10.1016/j.bmcl.2006.03.073. [DOI] [PubMed] [Google Scholar]
- Versteegen R. M.; Rossin R.; ten Hoeve W.; Janssen H. M.; Robillard M. S. Click to release: instantaneous doxorubicin elimination upon tetrazine ligation. Angew. Chem., Int. Ed. 2013, 52 (52), 14112–14116. 10.1002/anie.201305969. [DOI] [PubMed] [Google Scholar]
- Bernard S.; Audisio D.; Riomet M.; Bregant S.; Sallustrau A.; Plougastel L.; Decuypere E.; Gabillet S.; Kumar R. A.; Elyian J.; Trinh M. N.; Koniev O.; Wagner A.; Kolodych S.; Taran F. Bioorthogonal Click and Release Reaction of Iminosydnones with Cycloalkynes. Angew. Chem., Int. Ed. 2017, 56 (49), 15612–15616. 10.1002/anie.201708790. [DOI] [PubMed] [Google Scholar]
- Cork S. C.; Richards J. E.; Holt M. K.; Gribble F. M.; Reimann F.; Trapp S. Distribution and characterisation of Glucagon-like peptide-1 receptor expressing cells in the mouse brain. Mol. Metab. 2015, 4 (10), 718–731. 10.1016/j.molmet.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brubaker P. L.; Drucker D. J. Structure-function of the glucagon receptor family of G protein-coupled receptors: the glucagon, GIP, GLP-1, and GLP-2 receptors. Recept. Channels 2002, 8 (3–4), 179–188. 10.1080/10606820213687. [DOI] [PubMed] [Google Scholar]
- Tsvetanova N. G.; von Zastrow M. Spatial encoding of cyclic AMP signaling specificity by GPCR endocytosis. Nat. Chem. Biol. 2014, 10 (12), 1061–1065. 10.1038/nchembio.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Otero J.; Ahn K. H.; Delgado-Peraza F.; Mackie K.; Kendall D. A.; Yudowski G. A. Ligand-specific endocytic dwell times control functional selectivity of the cannabinoid receptor 1. Nat. Commun. 2014, 5, 4589 10.1038/ncomms5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein Herenbrink C.; Sykes D. A.; Donthamsetti P.; Canals M.; Coudrat T.; Shonberg J.; Scammells P. J.; Capuano B.; Sexton P. M.; Charlton S. J.; Javitch J. A.; Christopoulos A.; Lane J. R. The role of kinetic context in apparent biased agonism at GPCRs. Nat. Commun. 2016, 7, 10842 10.1038/ncomms10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang C.; Sinskey A. J.; Lodish H. F. Oxidized redox state of glutathione in the endoplasmic reticulum. Science 1992, 257 (5076), 1496–1502. 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- Hodson D. J.; Mitchell R. K.; Marselli L.; Pullen T. J.; Gimeno Brias S.; Semplici F.; Everett K. L.; Cooper D. M.; Bugliani M.; Marchetti P.; Lavallard V.; Bosco D.; Piemonti L.; Johnson P. R.; Hughes S. J.; Li D.; Li W. H.; Shapiro A. M.; Rutter G. A. ADCY5 couples glucose to insulin secretion in human islets. Diabetes 2014, 63 (9), 3009–3021. 10.2337/db13-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broichhagen J.; Podewin T.; Meyer-Berg H.; von Ohlen Y.; Johnston N. R.; Jones B. J.; Bloom S. R.; Rutter G. A.; Hoffmann-Röder A.; Hodson D. J.; Trauner D. Optical Control of Insulin Secretion Using an Incretin Switch. Angew. Chem., Int. Ed. 2015, 54 (51), 15565–15569. 10.1002/anie.201506384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodson D. J.; Mitchell R. K.; Bellomo E. A.; Sun G.; Vinet L.; Meda P.; Li D.; Li W. H.; Bugliani M.; Marchetti P.; Bosco D.; Piemonti L.; Johnson P.; Hughes S. J.; Rutter G. A. Lipotoxicity disrupts incretin-regulated human beta cell connectivity. J. Clin. Invest. 2013, 123 (10), 4182–4194. 10.1172/JCI68459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodson D. J.; Tarasov A. I.; Gimeno Brias S.; Mitchell R. K.; Johnston N. R.; Haghollahi S.; Cane M. C.; Bugliani M.; Marchetti P.; Bosco D.; Johnson P. R.; Hughes S. J.; Rutter G. A. Incretin-modulated beta cell energetics in intact islets of Langerhans. Mol. Endocrinol. 2014, 28 (6), 860–871. 10.1210/me.2014-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyris J. P.; Roux T.; Trinquet E.; Verdie P.; Fehrentz J. A.; Oueslati N.; Douzon S.; Bourrier E.; Lamarque L.; Gagne D.; Galleyrand J. C.; M’Kadmi C.; Martinez J.; Mary S.; Baneres J. L.; Marie J. Homogeneous time-resolved fluorescence-based assay to screen for ligands targeting the growth hormone secretagogue receptor type 1a. Anal. Biochem. 2011, 408 (2), 253–262. 10.1016/j.ab.2010.09.030. [DOI] [PubMed] [Google Scholar]
- M’Kadmi C.; Leyris J. P.; Onfroy L.; Gales C.; Sauliere A.; Gagne D.; Damian M.; Mary S.; Maingot M.; Denoyelle S.; Verdie P.; Fehrentz J. A.; Martinez J.; Baneres J. L.; Marie J. Agonism, Antagonism, and Inverse Agonism Bias at the Ghrelin Receptor Signaling. J. Biol. Chem. 2015, 290 (45), 27021–27039. 10.1074/jbc.M115.659250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Sun B.; Feng D.; Hu H.; Chu M.; Qu Q.; Tarrasch J. T.; Li S.; Sun Kobilka T.; Kobilka B. K.; Skiniotis G. Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature 2017, 546 (7657), 248–253. 10.1038/nature22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.