Abstract

To date, the program for the development of methods and models for atomistic and continuum simulation directed toward chemicals and materials has reached an incredible degree of sophistication and maturity. Currently, one can witness an increasingly rapid emergence of advances in computing, artificial intelligence, and robotics. This drives us to consider the future of computer simulation of matter from the molecular to the human length and time scales in a radical way that deliberately dares to go beyond the foreseeable next steps in any given discipline. This perspective article presents a view on this future development that we believe is likely to become a reality during our lifetime.
Short abstract
With the rapid advances in computing, artificial intelligence, and robotics, we consider the future of simulation of matter in the 21st century and provide six challenges for the field.
1. Matter Simulation Methods
You probably are reading this article in your mobile phone.1 The materials that compose the electronic circuits that made it possible were related to the outcome of digital computer simulations carried out by several thousands of scientists of the span of decades. Peter Galison in Image and Logic: A material culture of microphysics (1997)2 discusses the inseparability of the physical reality around us and the virtual matter that is simulated on modern computers:
Without the computer-based simulation, the material culture of late-twentieth-century microphysics is not merely inconvenienced – It does not exist. [...] Machines [...] are inseparable from their virtual counterparts–all are bound to simulations.
Several subfields of digital simulation have contributed to the design of our physical reality. These include quantum physics, quantum chemistry, condensed matter physics, computational statistical mechanics, computational materials science, continuum modeling, circuit layout, etc. In this paper, we reflect on the status of these fields, which we call as a collective matter simulation methods, since the early days of computing to the current era. Our goal is that of answering the question, What are the emerging challenges and opportunities for these fields in the twenty-first century? By answering this question, we arrive to the conclusion that redefining the scope of their main mission may be necessary due to the rapid advances in the drivers of our society.
Writing about all these fields would go beyond our domain of expertise and would rely on too many examples. Therefore, for the remainder of the paper, we will use the field of quantum chemistry as an example, but the reader can have in mind that very similar arguments could be made for any of the other theoretical/computational fields mentioned above, as they are all connected to the developments in fundamental, physical theory in the first half of the twentieth century and the dramatic hardware and algorithm developments in its second half.
2. Quantum Chemistry in the 20th Century
In 1928, Paul Dirac proposed his fundamental covariant equation of motion that governs the relativistic dynamics of a single electron in a classical electromagnetic field. This equation became the basis not only of more fundamental theories such as quantum electrodynamics but also of endeavors to solve many-electron problems in chemistry, molecular physics, and materials science3—even if they originally set out from Schrödinger’s (nonrelativistic) formulation of quantum mechanics.
In a famous quote from his 1929 account on many-electron systems,4 Dirac emphasized the importance of his discovery and then continued to state that it “becomes desirable that approximate practical methods of applying quantum mechanics should be developed, which can lead to an explanation of the main features of complex atomic systems without too much computation.” This desire has been the motivation for generations of computational chemists and physicists to devise algorithms that allow us to solve the differential equations governing the dynamics of many-electron systems.
The aspiration to quantitatively assess molecular properties and to qualitatively understand their implications has been a driving force of computer simulations of molecular matter since that time. In quantum chemistry, the mission in the previous century was to calculate an energy and the molecular properties of a given, isolated molecular structure. As Per-Olov Löwdin has put it in a visionary perspective on the field,5
There seems to be a rather long way to go before we reach the mathematical goal of quantum chemistry, which is to be able to predict accurately the properties of a hypothetic polyatomic molecule....
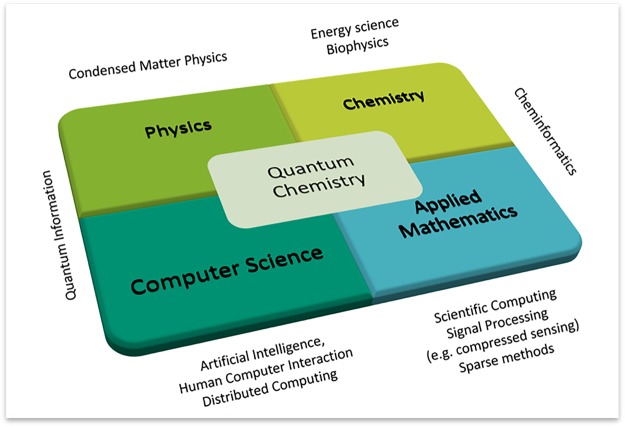
Löwdin states in his “1967-Program”6 written for the newly created International Journal of Quantum Chemistry that quantum chemistry “uses physical and chemical experience, deep going mathematical analysis and high speed electronic computers to achieve its results”. Gavroglu and Simões7 identify these as the four pillars on which the field has rested since its conception (see Figure 1).
Figure 1.
Quantum chemistry8 is an interdisciplinary field that lies at the intersection of chemistry, physics, applied math, and computer science. It borrows from several other subfields, some of which are mentioned at the borders of the diagram.
In the 20th century, the interdisciplinary field of quantum chemistry was therefore drawn from thematic aspects of the different founding disciples. It has taken from physics the laws of quantum mechanics and light-matter interaction, from chemistry the conceptual laws of molecular structure and inspiration for problems and applications, from applied math mostly computational linear algebra, and finally, from computer science a focus on high-performance computing, both parallel and using accelerators such as general-purpose graphical processing units.
Sixty years later, this enormous effort has been made and the mission has basically been accomplished. As a collective, theoretical chemists and physicists have developed methods that simulate the electronic structure of molecules up to hundreds of atoms, both in vacuum and in solvents, and that can calculate practically all observables of interest to experimentalists. Furthermore, although the so-called “chemical accuracy” cannot be obtained for all the calculations, quantum chemistry calculations have become a useful everyday tool in the arsenal of chemists.
In 1957, Löwdin5 presented an overview of various methods used in molecular and solid-state theory for the solution of the Schrödinger equation in his account. Amazingly, most of the principles and hierarchy set out in that overview prevailed until the present time. Propelled by developments in other fields (most notably the design and construction of efficient and affordable computer hardware), a huge part of Dirac’s and Löwdin’s programs set for future generations has become a reality.
In fact, we do have now computational tools at our disposal that allow us to do such calculations routinely and with relatively well-known accuracy.7 All remaining open issues of this original mission are well identified and understood. In other words, we are now able to solve the complex high-dimensional partial differential equations that govern quantum many-electron systems for arbitrary nuclear frameworks (even with rigorous error assessment9−14). Clearly, the dimension that scales with the number of elementary particles in the molecular system cannot be arbitrarily large for feasibility reasons, but the field has made the remarkable achievement of providing routine solution methods for partial differential equations whose dimension is set on input of a computation by providing the number of electrons of a reasonably sized molecular structure or unit cell. It therefore appears evident that what remains from the original program is rather straightforward to accomplish in light of the past achievements. And, in fact, it is most likely to take only a fraction of the effort invested so far to bring the original mission to an end for practical purposes.
According to his former students, Nicholas Handy, John Pople, and Isaiah Shavitt, during the 1959 Conference on Molecular Quantum Mechanics held in Boulder Colorado,2 Samuel F. Boys
[...] produced a paper tape of his whole computer program and unrolled it along the length of the chemical lecture bench. There, in one roll, was something, of which one could ask a chemical question at one end and it would produce an answer at the other! .... [M]ost of the audience probably thought the demonstration bizarre. But it was prescient.
In the narrow sense of the quote above, the field of quantum chemistry has practically solved the mission of the twentieth century quantum chemistry that we may summarize as follows:
Given a molecular geometry, obtain its energy and/or other molecular properties in the gas phase or a solvent model efficiently on a modern digital computer.
Having considered the great accomplishments of the field to date, we are ready to present our vision of what is possible next due to the rapid developments in the fields of computer science and robotics. Depending on one’s own progressive mind set, this vision may be considered either linear and therefore an Evolution of the current state of the art, or rather radical and hence a potential Revolution of the field.
3. Matter Simulation in the 21st Century
3.1. Societal Drivers
The connection between science and the current drivers for society is deep and cannot be ignored. This century poses several severe challenges that range from the rapid rise of income inequality and the apparent cracks of the neoliberal structure to the stresses on the environment due to industrialization. The work of simulation scientists therefore is linked directly or indirectly to this societal context. In particular, the solutions to many of the challenges related to this century, ranging from the discovery of novel materials for renewable energy to that of environmentally friendly pesticides or next-generation antibiotics, require tools to be developed by our field.
A characteristic of these challenges is that their solution is time-critical, which turns them into societal threats. For instance, if the search for new antibiotics cannot keep pace with the development and spread of bacterial resistance or if the development of sustainable energy cycles cannot outpace global warming, severe consequences for our society will be unavoidable. Whereas we have seen in the past decades a remarkably accurate prediction of our computational capabilities by Moore’s law, which was an original prediction of their exponential growth with time, nothing similar has been found so far for scientific discovery. However, the time-critical threats to society in the 21st century require us to find viable solutions for pressing global problems at an increasingly faster pace than what was sufficient in the past. In a sense, as suggested by Benji Maruyama15 a “Moore’s law for scientific discovery is required to increase the success for systematic as well as serendipitous discovery”.
However, tied to the current exponential growth of technology, such an exponential pace of scientific discovery could be achieved! An exponential increase in scientific throughput and lowering in cost via automation has been achieved in the field of gene sequencing. The National Institutes of Health also uses a comparison with exponential decrease in cost when comparing the cost of sequencing a human genome.16 We hope that, in certain areas of chemical discovery, an increase in throughput of calculation, synthesis, and characterization will result in an exponential increase in the rate of discovery.
3.2. Science and Technology Drivers
In this century, we are witnessing the introduction of novel technologies at a pace never seen before. Technoeconomically, the twenty-first century is deeply linked to what Klaus Schwab from the World Economic Forum (WEF) has coined as “The Fourth Industrial Revolution”.17 The WEF identified six technology drivers18 that will significantly impact society. To focus on the accelerated discovery of matter,19 we modify and expand this list to nine science and technology drivers that will deeply transform the speed and way of discovery of new chemicals and matter.
Driver 1. Human–Machine Interaction and the Internet
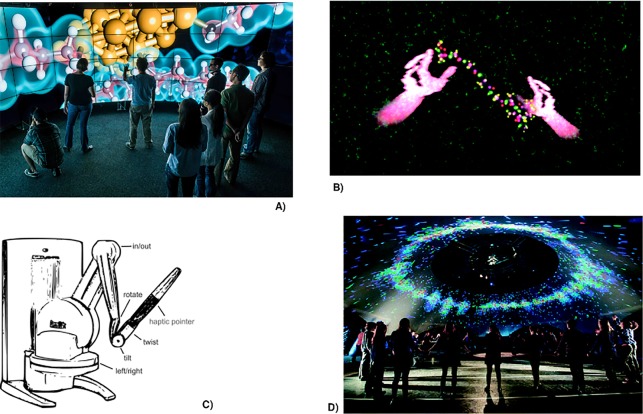
Technology will continue to enable the connection between people, enhancing their digital presence by enabling them to interact with objects and one another in new ways. Emerging technologies that enhance the human–machine interaction will allow for an unprecedented immersion into the virtual atomistic world that is otherwise inaccessible to the human senses. Already today, we see haptic and tracking devices, virtual realities, and caves adding a new level of intuition to the virtual experience of the molecular world that goes far beyond its archaic and fractured perception through computer mouse and keyboard.20−41 A perfect and seamless immersion of the scientist into the mesoscopic environment of her/his object of study is key to an accelerated understanding and manipulation of (molecular) matter in a virtual laboratory. For instance, immersion is about literally feeling the softness of a functional group in a molecule and about experiencing how it feels to push a hydrogen atom into a metal surface. By enhanced immersion and real-time data flow, one can cope with the immense data provided by computations finishing in real-time (see Figure 2)
Figure 2.
Examples of already existing tools that enhance the immersion of professional and amateur scientists into a molecular world. (A) A cave for data exploration (Electron density of a molecular data set image provided by the Electronic Visualization Laboratory (EVL) at the University of Illinois at Chicago and Argonne National Laboratory. Photo: L. Long, EVL). (B) An operator’s pair of hands manipulate a peptide during a molecular dynamics simulation (taken from ref (27); Creative Common License). (C) A simple haptic force-feedback device by which the tactile human sense can be addressed (Reproduced with permission from ref (42). Copyright 2011 Wiley-VCH Verlag GmbH & Co. KgaA). (D) Interactive atmospheric molecular dynamics simulation in an immersive projection dome (taken from ref (27); Creative Common License).
Driver 2. Computing, Communication, and Storage Everywhere
Everybody will have access to a supercomputer in their pocket (currently already a reality as a link of the already powerful existing smartphones to cloud services), with nearly unlimited storage capacity. This driver will grant the research community access to cheap computational power and ability to store and share chemical information. It will also allow amateur scientists to access important research problems much more easily, which will strengthen the general acceptance of the field in society and increase the pace of discovery (cf., recent web-based online platforms, serious gaming, and gamification are existing examples).43,44 Considering that the chemical space is unfathomably large and much information for a given problem may not be available, such restricted, unavailable, or nonexistent information will be computed on the spot as needed.
Driver 3. The Internet of Things
The introduction of sensors and processors that are connected to the Internet for most of the objects around us will offer society and the science community a trivial route to build, for example, networks of off-the-shelf sensors and processors to monitor and control artificial intelligence (AI) environments and robotics systems. For example, it is not too hard to imagine a 3D printer that prints parts of a synthesis robot that itself may then be put online to receive control commands from a virtual reality devoted to chemical or materials synthesis to produce a specific chemical or material. This is closely connected to the next driver.
Driver 4. Artificial Intelligence, Big Data, and Robotics
The exponential creation of more data from the sensors and processors around us requires its organization and processing using artificial intelligence. Already now, we witness the rapid rise of machine learning tools for such purposes (see refs (45−57) for some examples). Robotics, empowered by such tools, is already making an impact in the automation of jobs, decision making, and research. It is only a matter of time before synthesis robots, which are already employed in many chemistry laboratories, will be generalized (see the work by Burke on his synthesis machine58−60) and coupled to adequate software. Much work has been devoted to devising and implementing algorithms for the automated exploration of chemical reaction networks (see, e.g., refs (61−67)). In the future, the evolution of these achievements may team up with the latest expert systems for the planning of chemical synthesis (see driver 6) to generate a reliable platform that can map out complex chemical reaction networks (based on the big data provided by reaction libraries and vast quantum chemical explorations) under predefined conditions that are then eventually realized by a relatively general synthesis robot (see Figure 3).
Figure 3.
Left: A MakerBot 3D printer (picture from https://en.wikipedia.org/wiki/3D_printing). Middle: HERMAN the High-throughput Experimentation Robot for the Multiplexed Automation of Nanochemistry (taken from https://www.youtube.com/watch?v=J0VlCItpI5s). Right: Martin Burke’s synthesis machine (taken from ref (68); permission to print this picture granted by L. Brian Stauffer, University of Illinois at Urbana–Champaign).
Driver 5. The Sharing Economy and Distributed Trust
This driver enables new social and business models. Tools like the blockchain promise to change the way we think about money and transactions in the real world. This will have an impact on how data is acquired or generated, stored, and managed in a complete reproducible and controlled way: smart evidence. It will ensure that data is immutable and protected; the user will no longer have to, for example, trust service providers as Google, Dropbox, etc. to not manipulate the data. Blockchaining will also affect how research results are published. For instance, controversial data or results can be published anonymously while the integrity of the presented data acquisition is completely verifiable. Blockchaining will create new ways of funding: smart contracts that will potentially disrupt the way science is funded.
Driver 6. The Digitization of Matter from the Macroscopic to the Atomic Scale
The continued development of printers of physical objects due to 3D printing and additive manufacturing technologies allows for new creative opportunities at the scale of meters to micrometers. The increased control over the synthesis and characterization of precise nanoscale assemblies of matter such as nanoparticles, chemical and atomic layer deposition, and lithography provides a bottom-up approach to create matter that can further be probed efficiently. Purely synthetic approaches have reached a level of sophistication in chemistry such that, for any molecule that is calculated to be stable and viable, a synthetic path can be designed. Expert systems for such purposes have a long history69−72 and experienced a fresh boost by modern technology.73−77
Driver 7. Precise Control of Molecular Biology and Nanochemistry
Our increasing ability to manipulate and control the molecular biology of cells is an example of successful molecular engineering exploiting an existing molecular machinery to produce new molecules, often with predefined function. The recently developed CRISPR/Cas gene editing78−80 technique is an easy to use approach with incredibly far reaching consequences. It is not only an amazing example for our understanding and capabilities to modify molecular processes in functional and living matter, it is also a remarkable advance in bioengineering technology that allows us to modify the genome of an organism at single-nucleotide precision. In this respect it straightforwardly extends the more traditional biochemical toolbox bioengineering that can be used to produce specific molecules. Combined with experimental automation and control through a virtual reality setup, this is a way to actually produce molecular structures designed in silico. Naturally, such a level of sophistication in understanding and manipulating the biochemical machinery of cells is an example for what would like to achieve in artificial soft and hard nanochemistry settings. And surely, matter simulation will be a decisive tool to point the way.
Driver 8. Disruptive Computing Technologies
Quantum computers employ the rules of quantum physics such as entanglement and superposition to carry out computations. The earliest suggested application for them is the simulation of quantum matter. Quantum computers can provide an exponential advantage over classical computers for the simulation of chemistry.81−83 This has been shown theoretically and experimentally realized in several occasions in demonstration experiments.84−86 Although no quantum computer with error-corrected qubits has been built so far, the severe efforts in major companies, start-ups, and academia make the emergence of moderately sized quantum computers for actual applications in chemistry in the near future rather likely.
Driver 9. Matter Imaging Technology at the Atomistic and Larger Length Scales
Coupled to driver 6, the emergence of advanced spectroscopy techniques for the study of matter has led to an increase in the available experimental data in terms of resolution, area studied, and variety of signals that are recorded. This makes characterization a prime source of data that can directly interact with the output of simulation. In this way, it can help to constrain models “on the fly” and refine digital chemical hypotheses.
The nine science and technology drivers mentioned above could help us generate a contemporary close reading of the quote of the students of Samuel Boys. He mentions a computer program, where “one could ask a chemical question at one end and it would produce an answer at the other!”2 In the context of the new frontiers of human–computer interaction and artificial intelligence, the quote of Boys can be expanded into the revised mission for the field of computer simulation:
These questions could imply complex tasks and a dialogue with a computer program (see also ref (87)) such as the following one:
Jane, the chemist: Dear Organa, good morning. Could you please suggest to me an organic molecule with an estimated synthetic cost of less than a hundred dollars per gram and three synthetic steps from available synthons that has an emissive color of 450 nm and stability against oxidation? I am thinking of organic light emitting diode emitters.
Organa: Jane, I will get back to you in 2 h.
Organa: Hello Jane, I am back! I have a set of 50 potential candidates based on your constraints and inspired by the recent literature on the subject. You can purchase 20 of them and use the matter computer to synthesize the other 30 that you have with the synthon library available in the matter computer.
Jane: Dear Organa, can you display them for me in AR (augmented reality) ?
Organa: My pleasure, Jane.
Jane: (As she waves her hand and flips through compounds and their associated properties shown to her in augmented reality) Could you synthesize all the 30 compounds and test them for emission properties? Can you order the other 20? When the compounds arrive, I will install cartridges for the 20 compounds so you can use them for automated characterization. Thank you, Organa!
To enable a computer program and associated “matter computer”, or synthesis and characterization machine, the simulation community should overcome six grand challenges that we identify in the following.
4. Grand Challenges for the Simulation of Matter in the 21st Century
Contemplating the nine drivers, we may condense the goals of future research efforts in the field of matter simulation to six grand challenges.
Challenge 1. The Designer Challenge
While the mission of the 20th century was related to providing answers to questions pertaining to properties of specific chemical structures, the questions of the 21st century revolve around the inverse design problem:88−94 finding the best chemical structures that are associated with desired and requested properties. A potential solution for this challenge is the use of invertible models from machine learning such as generative models (GANs, autoencoders, ...)48,89 or inverting molecules from families of Hamiltonians.90−93
Challenge 2. The Chemical Turing Test
The classical Turing test95 is a gedanken experiment designed by Alan Turing to answer the question, “What is intelligence, and is it exclusive to humans?” In the test a human communicates with another human or a computer and is ultimately asked to tell the identity of the subject of the communication. The new goal of theoretical chemistry should be that of providing access to a chemical “oracle”: an AI environment which can help humans solve problems, associated with the fundamental chemical questions of the fourth industrial revolution (clean energy, efficient drugs, smart materials, green chemistry, etc.), in a way such that the human cannot distinguish between this and communicating with a human expert.
Challenge 3. The Feynman Test
In 1982 R. Feynman96 stated, “I want to talk about the possibility that there is to be an exact simulation, that the computer will do exactly the same as nature”, in his visionary article “Simulating Physics with Computers”. Computer simulations, to the best of our knowledge, applying the known rules of physics to computer-modeled particles, is an exact one-to-one mapping to reality, such that experimental and virtual data, for all practical purposes, are indistinguishable. The remaining discrepancy, between experiment and computer simulations, is an ongoing battle the computational chemists are convincingly winning inch by inch. In the not too distant future, computer simulations will be a fully adequate alternative to experiments, at which point questions like cost efficiency, environmental considerations, or other aspects will be the grounds for the choice between theory and experiment. Quantum computers97 may offer the way forward as they are known to simulate matter exactly if a suitable input state is provided.95 Enormous progress in the field has led to current experiments involving several qubits and simulating molecules as large as BeH2.81−86
Challenge 4. The Matter Computer
Let us now provide the complete 1957 quote of Per-Olov Löwdin5 mentioned in section 2:
There seems to be a rather long way to go before we reach the mathematical goal of quantum chemistry, which is to be able to predict accurately the properties of a hypothetic polyatomic molecule before it has been synthesized in the laboratories. The aim is also to obtain such knowledge of the electronic structure of matter that one can construct new substances having properties of particular value to mankind. To learn to think in terms of electrons and their quantum mechanical behaviors is probably of greater technical importance than we can now anticipate.
We can now reinterpret this quote in the context of the development of an integrated molecular discovery platform that we name a “matter computer”. These computers process chemicals instead of information. Their “registers” consist of actual chemicals, solvents, nanoparticles, etc. Their information processing subroutines are actual synthesis and characterization tools. The output of the computer is matter that can be readily analyzed and acted upon. They are the analogues of 3D printers but with molecular building blocks. All of these components should be connected to a central control system and database that can make artificial-intelligence-driven decisions about what to synthesize next. Characterization tools also should be integrated in the platform. To understand the chemical space to be explored, these matter computers employ large computer-generated compound screening libraries or generative models based on their data. This loop requires the integration of several technologies that are currently emerging in an integrated platform.15 Physical constraints are likely to make these matter computers explore a relatively small fraction of chemical space such as the number of building blocks, solvents and catalysts, capacities of the synthetic hardware, and current knowledge or prediction of reaction mechanisms. Computers themselves can help design the right robotic synthesis platforms for targeting a region of chemical space.
Challenge 5. The Immersive Chemistry Challenge
A full immersion into the virtual world of a molecular system with seamless integration of fellow researchers will boost research and education. This seamless integration is also a necessary condition to promote instantaneous computing, i.e., the fact that the computing time for some problem has been constantly shrinking over the years up to the point where starting a calculation and receiving its results can no longer be separated on the time scale of human perception, which is 60 ms for vision and 1 ms for our sense of touch. It will be necessary to accomplish an ultimate integrated hardware and software implementation of the perfect immersion of a human researcher into the molecular world. Coming back to Dirac’s quote from 1929, which ends “... without too much computation”, we need to rewrite this into “... by immediate and interactive computation” to meet the demands of the time to come.
Challenge 6. The Machine and Human Molecular Representation Learning Challenge
The natural representation of molecules from the point of view of quantum information theory is the content of a full quantum tomography98 experiment for their wave function or a quantum process tomography to explore their dynamical processes.99 The wave function contains too much information to be processed by a human brain. As we advance in our program of machine learning and automation, the question of representation learning will arise. In the field of machine learning, this pertains to finding the representation of the molecular data that the machine can learn from. From the other side of the spectrum, a human grasps to concepts that help her or him make rational decisions. We hope that, in the future, humans and machines meet in the middle. Emerging conceptual breakthroughs in chemistry may arise from humans training their computer helpers, and the computer helpers, in return, providing the raw source of inspiration to elaborate new concepts.
5. Toward a Unified Computation Science of Matter
Science as we know it today owes its classifications to how scientists viewed and understood science in the 19th century and before. As scientific understanding has progressed, the fundamental laws of chemistry and physics, as well as other areas of science, have been unified. In computational science this is more evident than anywhere else: chemistry and physics simulations use the very same fundamental numerical methods and strategies. This calls for reflection and a possible reclassification of the fields of chemistry and physics. At first sight this might seem destructive: why destroy the infrastructure that has served mankind so well in the past? Well, stone and bricks were the preferential building materials in New York after the Great Fire in 1835. However, as building space was exhausted, more living and office space could not be added by simply adding a new floor to an existing house. The stone and brick technology had come to a dead end. Buildings had to be torn down to make space for buildings using a completely different paradigm: the skyscrapers based on the use of steel frames. We suggest that the matter simulation (r)evolution will call for a healthy reclassification of the scientific fields away from classification based on structure toward functionalities.
The opposite side of the spectrum of chemical understanding lies on the scribblings by organic chemists in blackboards, also known as arrow-pushing diagrams. These conceptual tools have led to several advances in the field of organic chemistry. At the interface of quantum theory with conceptual learning, ideas such as the Woodward-Hoffman rules100 or frontier orbital theory have proven themselves invaluable tools for our understanding of chemical processes.
It goes without saying that the developments in science will have paramount implications for the educational system, too. Can we today be so complacent with respect to the expected changes in science that we continue educating students in skills which we expect to be automated in the not so distant future? Of course not! There will have to be a parallel development in the educational system. Not only will the educational system change the curriculum to adapt to the new scientific reality. At the same time the educational system will have to adapt to the new information technology (IT) reality: all information is available at your screen by the touch of a screen, a voice command, or even a thought. Pedagogic adaptation will be natural, active rather than passive learning will be an instrumental part of a progressive educational system, and the flipped classroom will be the norm rather than the exception.
As a final remark, we emphasize that we clearly see the massive effort required in future research and education that it will take to push the existing theories and technologies further to accomplish the grand challenges ahead of us. Undoubtedly, a collaborative international, interdisciplinary effort in research and education is necessary. This cannot be done alone by a few research groups. This is a call to arms for the computational scientists to continue innovating using all the new tools available to us on a daily basis.
Acknowledgments
A.A.-G. thanks Shaul Mukamel for useful discussions. A.A-G. acknowledges funding support from the Vannevar Bush Fellowship from the United States Department of Defense under Award No. N00014-16-1-2008 under the Office of Naval Research. R.L. acknowledges support by the Swedish Research Council (Grant 2016-03398).
The authors declare no competing financial interest.
References
- Sze S. M.; Ng K. K.. Physics of Semiconductor Devices; Wiley: 2006. [Google Scholar]
- Galison P.Image and Logic: A material culture of microphysics; The University of Chicago Press: 1997. [Google Scholar]
- Reiher M.; Wolf A.. Relativistic Quantum Chemistry, 2nd ed.; Wiley-VCH: 2015. [Google Scholar]
- Dirac P. A. M. Quantum Mechanics of Many-Electron Systems. Proc. R. Soc. London, Ser. A 1929, 123, 714–733. 10.1098/rspa.1929.0094. [DOI] [Google Scholar]
- Löwdin P. O. Present Situation of Quantum Chemistry. J. Phys. Chem. 1957, 61, 55–68. 10.1021/j150547a012. [DOI] [Google Scholar]
- Löwdin P.-O. Program. Int. J. Quantum Chem. 1967, 1, 1–6. 10.1002/qua.560010102. [DOI] [Google Scholar]
- Gavroglu K.; Simões A.. Neither Physics Nor Chemistry; MIT Press: 2012. [Google Scholar]
- Dykstra C.; Frenking G.; Kim K.; Scuseria G.. Theory and Applications of Computational Chemistry - The First Forty Years; Elsevier Science: 2005. [Google Scholar]
- Mortensen J. J.; Kaasbjerg K.; Frederiksen S. L.; Nørskov J. K.; Sethna J. P.; Jacobsen K. W. Bayesian error estimation in density-functional theory. Phys. Rev. Lett. 2005, 95, 216401. 10.1103/PhysRevLett.95.216401. [DOI] [PubMed] [Google Scholar]
- Wellendorff J.; Lundgaard K. T.; Jacobsen K. W.; Bligaard T. mBEEF: an accurate semi-local Bayesian error estimation density functional. J. Chem. Phys. 2014, 140, 144107. 10.1063/1.4870397. [DOI] [PubMed] [Google Scholar]
- Wellendorff J.; Lundgaard K. T.; Møgelhøj A.; Petzold V.; Landis D. D.; Nørskov J. K.; Bligaard T.; Jacobsen K. W. Density functionals for surface science: Exchange-correlation model development with Bayesian error estimation. Phys. Rev. B: Condens. Matter Mater. Phys. 2012, 85, 235149. 10.1103/PhysRevB.85.235149. [DOI] [Google Scholar]
- Simm G. N.; Reiher M. Systematic Error Estimation for Chemical Reaction Energies. J. Chem. Theory Comput. 2016, 12, 2762–2773. 10.1021/acs.jctc.6b00318. [DOI] [PubMed] [Google Scholar]
- Proppe J.; Reiher M. J. Chem. Theory Comput. 2017, 13, 3297–3317. 10.1021/acs.jctc.7b00235. [DOI] [PubMed] [Google Scholar]
- Simm G.; Proppe J.; Reiher M. Error Assessment of Computational Models in Chemistry. Chimia 2017, 71, 202–208. 10.2533/chimia.2017.202. [DOI] [PubMed] [Google Scholar]
- Tabor D. P.; Roch L. M.; Saikin S. K.; Kreisbeck C.; Sheberla D.; Montoya J.; Dwaraknath S.; Aykol M.; Ortiz C.; Tribukait H.; Amador-Bedolla C.; Brabec C. J.; Maruyama B.; Persson K. A.; Aspuru-Guzik A.. Accelerating Discovery of New Materials for Clean Energy in the Era of Smart Automation. 2017,submitted. [Google Scholar]
- DNA Sequencing Costs: Data. https://www.genome.gov/sequencingcostsdata/ (Snapshot taken January 13, 2018).
- Schwab K.The Fourth Industrial Revolution; Random House LCC: 2017. [Google Scholar]
- Deep Shift. Technological Tipping Point and Societal Impact. Survey Report. Global Agenda Council on the Future of Software & Society. World Economic Forum. http://www3.weforum.org/docs/WEF_GAC15_Technological_Tipping_Points_report_2015.pdf2015. (accessed Oct 2, 2017).
- De Luna P.; Wei J. N.; Bengio Y.; Aspuru-Guzik A.; Sargent E. H. Use Machine Learning to Find Energy Materials. Nature 2017, 552, 23–27. 10.1038/d41586-017-07820-6. [DOI] [PubMed] [Google Scholar]
- Marti K. H.; Reiher M. Haptic quantum chemistry. J. Comput. Chem. 2009, 30, 2010–2020. 10.1002/jcc.21201. [DOI] [PubMed] [Google Scholar]
- Haag M. P.; Vaucher A. C.; Bosson M.; Redon S.; Reiher M. Interactive Chemical Reactivity Exploration. ChemPhysChem 2014, 15, 3301–3319. 10.1002/cphc.201402342. [DOI] [PubMed] [Google Scholar]
- Vaucher A. C.; Haag M. P.; Reiher M. J. Comput. Chem. 2016, 37, 805–812. 10.1002/jcc.24268. [DOI] [PubMed] [Google Scholar]
- Vaucher A. C.; Reiher M. Molecular Propensity as a Driver for Explorative Reactivity Studies. J. Chem. Inf. Model. 2016, 56, 1470–1478. 10.1021/acs.jcim.6b00264. [DOI] [PubMed] [Google Scholar]
- Martínez T. J. Ab Initio Reactive Computer Aided Molecular Design. Acc. Chem. Res. 2017, 50, 652–656. 10.1021/acs.accounts.7b00010. [DOI] [PubMed] [Google Scholar]
- Luehr N.; Jin A. G. B.; Martínez T. J. Ab Initio Interactive Molecular Dynamics on Graphical Processing Units (GPUs). J. Chem. Theory Comput. 2015, 11, 4536–4544. 10.1021/acs.jctc.5b00419. [DOI] [PubMed] [Google Scholar]
- Kozlíková B.; Krone M.; Falk M.; Lindow N.; Baaden M.; Baum D.; Viola I.; Parulek J.; Hege H.-C. Visualization of Biomolecular Structures: State of the Art Revisited. Computer Graphics Forum 2017, 36, 178–204. 10.1111/cgf.13072. [DOI] [Google Scholar]
- Glowacki D. R.; O’Connor M.; Calabró G.; Price J.; Tew P.; Mitchell T.; Hyde J.; Tew D. P.; Coughtrie D. J.; McIntosh-Smith S. A GPU-accelerated immersive audio-visual framework for interaction with molecular dynamics using consumer depth sensors. Faraday Discuss. 2014, 169, 63–87. 10.1039/C4FD00008K. [DOI] [PubMed] [Google Scholar]
- Pavlov E.; Taiji M.; Scukins A.; Markesteijn A.; Karabasov S.; Nerukh D. Visualising and controlling the flow in biomolecular systems at and between multiple scales: from atoms to hydrodynamics at different locations in time and space. Faraday Discuss. 2014, 169, 285–302. 10.1039/C3FD00159H. [DOI] [PubMed] [Google Scholar]
- Anthopoulos A.; Pasqualetto G.; Grimstead I.; Brancale A. Haptic-driven, interactive drug design: implementing a GPU-based approach to evaluate the induced fit effect. Faraday Discuss. 2014, 169, 323–342. 10.1039/C3FD00139C. [DOI] [PubMed] [Google Scholar]
- Haag M. P.; Reiher M. Studying chemical reactivity in a virtual environment. Faraday Discuss. 2014, 169, 89–118. 10.1039/C4FD00021H. [DOI] [PubMed] [Google Scholar]
- Iakovou G.; Hayward S.; Laycock S. A real-time proximity querying algorithm for haptic-based molecular docking. Faraday Discuss. 2014, 169, 359–377. 10.1039/C3FD00123G. [DOI] [PubMed] [Google Scholar]
- Stone J. E.; Gullingsrud J.; Grayson P.; Schulten K., A system for interactive molecular dynamics simulation. In ACM Symposium on Interactive 3D Graphics; Hughes J. F., Séquin C. H., Eds.; 2001; pp 191–194. [Google Scholar]
- Isralewitz B.; Gao M.; Schulten K. Steered molecular dynamics and mechanical functions of proteins. Curr. Opin. Struct. Biol. 2001, 11, 224–230. 10.1016/S0959-440X(00)00194-9. [DOI] [PubMed] [Google Scholar]
- Zhu F.; Tajkhorshid E.; Schulten K. Theory and simulation of water permeation in aquaporin-1. Biophys. J. 2004, 86, 50–57. 10.1016/S0006-3495(04)74082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosson M.; Richard C.; Plet A.; Grudinin S.; Redon S. Interactive quantum chemistry: A divide-and-conquer ASED-MO method. J. Comput. Chem. 2012, 33, 779–790. 10.1002/jcc.22905. [DOI] [PubMed] [Google Scholar]
- O’Connor M.; Sage R.; Tew P.; McIntosh-Smith S.; Glowacki D. R.. The Nano Simbox: using virtual reality to interactively steer scientific simulations on high-performance computational architectures. Supercomputing 2016, accepted
- Mitchell T.; Hyde J.; Tew P.; Glowacki D. R. danceroom Spectroscopy: At the Frontiers of Physics, Performance, Interactive Art and Technology. Leonardo 2016, 49, 138–147. 10.1162/LEON_a_00924. [DOI] [Google Scholar]
- Davies E.; Tew P.; Glowacki D.; Smith J.; Mitchell T.. Evolving Atomic Aesthetics and Dynamics. In Evolutionary and Biologically Inspired Music, Sound, Art and Design: 5th International Conference, EvoMUSART 2016, Porto, Portugal, March 30–April 1, 2016, Proceedings; Johnson C., Ciesielski V., Correia J., Machado P., Eds.; Springer International Publishing: 2016; pp 17–30. [Google Scholar]
- Salvadori A.; Del Frate G.; Pagliai M.; Mancini G.; Barone V. Immersive virtual reality in computational chemistry: Applications to the analysis of QM and MM data. Int. J. Quantum Chem. 2016, 116, 1731–1746. 10.1002/qua.25207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehr N.; Jin A. G. B.; Martínez T. J. Ab Initio Interactive Molecular Dynamics on Graphical Processing Units (GPUs). J. Chem. Theory Comput. 2015, 11, 4536–4544. 10.1021/acs.jctc.5b00419. [DOI] [PubMed] [Google Scholar]
- Park S.; Schulten K. Calculating potentials of mean force from steered molecular dynamics simulations. J. Chem. Phys. 2004, 120, 5946–5961. 10.1063/1.1651473. [DOI] [PubMed] [Google Scholar]
- Haag M. P.; Marti K. H.; Reiher M. Generation of Potential Energy Surfaces in High Dimensions and Their Haptic Exploration. ChemPhysChem 2011, 12, 3204–3213. 10.1002/cphc.201100539. [DOI] [PubMed] [Google Scholar]
- http://fold.it/ (accessed Oct. 2, 2017).
- http://qmcathome.org/ (accessed Oct 2, 2017).
- Duvenaud D.; Maclaurin D.; Aguilera-Iparraguirre J.; Gómez-Bombarelli R.; Hirzel T.; Aspuru-Guzik A.; Adams R. P. Convolutional Networks on Graphs for Learning Molecular Fingerprints. NIPS’15 Proceedings of the 28th International Conference on Neural Information Processing Systems 2015, 2, 2224–2232. [Google Scholar]
- Pyzer-Knapp E. O.; Simm G. N.; Aspuru Guzik A. A Bayesian approach to calibrating high-throughput virtual screening results and application to organic photovoltaic materials. Mater. Horiz. 2016, 3, 226–233. 10.1039/C5MH00282F. [DOI] [Google Scholar]
- Wei J. N.; Duvenaud D.; Aspuru-Guzik A. Neural Networks for the Prediction of Organic Chemistry Reactions. ACS Cent. Sci. 2016, 2, 725–732. 10.1021/acscentsci.6b00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Bombarelli R.; Wei J. N.; Duvenaud D.; Hernández-Lobato J. M.; Sánchez-Lengeling B.; Sheberla D.; Aguilera-Iparraguirre J.; Hirzel T. D.; Adams R. P.; Aspuru-Guzik A. Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci. 2018, 10.1021/acscentsci.7b00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B.; von Lilienfeld O. A.. The “DNA” of chemistry: Scalable quantum machine learning with “amons”. 2017, arXiv:1707.04146.
- Smith J. S.; Isayev O.; Roitberg A. E. ANI-1: an extensible neural network potential with DFT accuracy at force field computational cost. Chem. Sci. 2017, 8, 3192–3203. 10.1039/C6SC05720A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B.; von Lilienfeld O. A. Communication: Understanding molecular representations in machine learning: The role of uniqueness and target similarity. J. Chem. Phys. 2016, 145, 161102. 10.1063/1.4964627. [DOI] [PubMed] [Google Scholar]
- Montavon G.; Rupp M.; Gobre V.; Vazquez-Mayagoitia A.; Hansen K.; Tkatchenko A.; Müller K.-R.; von Lilienfeld O. A. Machine learning of molecular electronic properties in chemical compound space. New J. Phys. 2013, 15, 095003. 10.1088/1367-2630/15/9/095003. [DOI] [Google Scholar]
- Rupp M.; Tkatchenko A.; Müller K.-R.; von Lilienfeld O. A. Fast and Accurate Modeling of Molecular Atomization Energies with Machine Learning. Phys. Rev. Lett. 2012, 108, 058301. 10.1103/PhysRevLett.108.058301. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan R.; von Lilienfeld O. A. Many Molecular Properties from One Kernel in Chemical Space. Chimia 2015, 69, 182–186. 10.2533/chimia.2015.182. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan R.; Dral P. O.; Rupp M.; von Lilienfeld O. A. Big Data Meets Quantum Chemistry Approximations: The Δ-Machine Learning Approach. J. Chem. Theory Comput. 2015, 11, 2087–2096. 10.1021/acs.jctc.5b00099. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan R.; von Lilienfeld O. A. Machine Learning, Quantum Chemistry, and Chemical Space. Rev. Comp. Chem. 2017, 30, 225–256. 10.1002/9781119356059.ch5. [DOI] [Google Scholar]
- Chang K. Y. S.; von Lilienfeld O. A. Quantum Mechanical Treatment of Variable Molecular Composition: From ’Alchemical’ Changes of State Functions to Rational Compound Design. Chimia 2014, 68, 602–608. 10.2533/chimia.2014.602. [DOI] [PubMed] [Google Scholar]
- Li J.; Ballmer S. G.; Gillis E. P.; Fujii S.; Schmidt M. J.; Palazzolo A. M. E.; Lehmann J. W.; Morehouse G. F.; Burke M. D. Synthesis of many different types of organic small molecules using one automated process. Science 2015, 347, 1221–1226. 10.1126/science.aaa5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service R. F. The synthesis machine. Science 2015, 347, 1190–1193. 10.1126/science.347.6227.1190. [DOI] [PubMed] [Google Scholar]
- Service R. F. Billion-dollar project would synthesize hundreds of thousands of molecules in search of new medicines. Science 2017, 10.1126/science.aal1073. [DOI] [Google Scholar]
- Maeda S.; Ohno K.; Morokuma K. Systematic exploration of the mechanism of chemical reactions: the global reaction route mapping (GRRM) strategy using the ADDF and AFIR methods. Phys. Chem. Chem. Phys. 2013, 15, 3683–3701. 10.1039/c3cp44063j. [DOI] [PubMed] [Google Scholar]
- Zimmerman P. M. Automated discovery of chemically reasonable elementary reaction steps. J. Comput. Chem. 2013, 34, 1385–1392. 10.1002/jcc.23271. [DOI] [PubMed] [Google Scholar]
- Zubarev D. Y.; Rappoport D.; Aspuru-Guzik A. Uncertainty of Prebiotic Scenarios: The Case of the Non-Enzymatic Reverse Tricarboxylic Acid Cycle. Sci. Rep. 2015, 5, 8009. 10.1038/srep08009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeler M.; Simm G. N.; Proppe J.; Reiher M. Heuristics-Guided Exploration of Reaction Mechanisms. J. Chem. Theory Comput. 2015, 11, 5712–5722. 10.1021/acs.jctc.5b00866. [DOI] [PubMed] [Google Scholar]
- Habershon S. Automated Prediction of Catalytic Mechanism and Rate Law Using Graph-Based Reaction Path Sampling. J. Chem. Theory Comput. 2016, 12, 1786–1798. 10.1021/acs.jctc.6b00005. [DOI] [PubMed] [Google Scholar]
- Sameera W. M. C.; Maeda S.; Morokuma R. Computational Catalysis Using the Artificial Force Induced Reaction Method. Acc. Chem. Res. 2016, 49, 763–773. 10.1021/acs.accounts.6b00023. [DOI] [PubMed] [Google Scholar]
- Simm G. N.; Reiher M. Context-Driven Exploration of Complex Chemical Reaction Networks. J. Chem. Theory Comput. 2017, 13, 6108–6119. 10.1021/acs.jctc.7b00945. [DOI] [PubMed] [Google Scholar]
- Sanderson K. Complex molecules made to order in synthesis machine. Nature News 2015, 10.1038/nature.2015.17113. [DOI] [Google Scholar]
- Pensak D. A.; Corey E. J. LHASA—Logic and Heuristics Applied to Synthetic Analysis. ACS Symp. Ser. 1977, 61, 1–32. 10.1021/bk-1977-0061.ch001. [DOI] [Google Scholar]
- Gasteiger J.; Ihlenfeldt W. D.; Röse P.; Wanke R. Computer-assisted reaction prediction and synthesis design. Anal. Chim. Acta 1990, 235, 65–75. 10.1016/S0003-2670(00)82062-0. [DOI] [Google Scholar]
- Gasteiger J.; Ihlenfeldt W. D.; Röse P. A collection of computer methods for synthesis design and reaction prediction. Recl. Trav. Chim. Pays-Bas 1992, 111, 270–290. 10.1002/recl.19921110605. [DOI] [Google Scholar]
- Kowalik M.; Gothard C. M.; Drews A. M.; Gothard N. A.; Weckiewicz A.; Fuller P. E.; Grzybowski B. A.; Bishop K. J. M. Parallel Optimization of Synthetic Pathways within the Network of Organic Chemistry. Angew. Chem., Int. Ed. 2012, 51, 7928–7932. 10.1002/anie.201202209. [DOI] [PubMed] [Google Scholar]
- Szymkuć S.; Gajewska E. P.; Klucznik T.; Molga K.; Dittwald P.; Startek M.; Bajczyk M.; Grzybowski B. A. Computer-Assisted Synthetic Planning: The End of the Beginning. Angew. Chem., Int. Ed. 2016, 55, 5904. 10.1002/anie.201506101. [DOI] [PubMed] [Google Scholar]
- Segler M. H. S.; Preuss M.; Waller M. P.. Learning to Plan Chemical Syntheses. 2017, arXiv:1708.04202.
- Segler M. H. S.; Waller M. P. Modelling Chemical Reasoning to Predict and Invent Reactions. Chem. - Eur. J. 2017, 23, 6118. 10.1002/chem.201604556. [DOI] [PubMed] [Google Scholar]
- Segler M. H. S.; Preuss M.; Waller M. P.. Towards “AlphaChem”: Chemical Synthesis Planning with Tree Search and Deep Neural Network Policies. 2017, arXiv:1702.00020.
- Segler M. H. S.; Waller M. P. Neural-Symbolic Machine Learning for Retrosynthesis and Reaction Prediction. Chem. - Eur. J. 2017, 23, 5966. 10.1002/chem.201605499. [DOI] [PubMed] [Google Scholar]
- Hsu P. D.; Lander E. S.; Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 2014, 157, 1262–1278. 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J. D.; Joung J. K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347–355. 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; La Russa M.; Qi L. S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264. 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- Aspuru-Guzik A.; Dutoi A. D.; Love P. J.; Head-Gordon M. Simulated Quantum Computation of Molecular Energies. Science 2005, 309, 1704–1707. 10.1126/science.1113479. [DOI] [PubMed] [Google Scholar]
- Reiher M.; Wiebe N.; Svore K. M.; Wecker D.; Troyer M. Elucidating reaction mechanisms on quantum computers. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 7555–7560. 10.1073/pnas.1619152114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassal I.; Whitfield J. D.; Perdomo-Ortiz A.; Yung M.-H.; Aspuru-Guzik A. Simulating Chemistry Using Quantum Computers. Annu. Rev. Phys. Chem. 2011, 62, 185–207. 10.1146/annurev-physchem-032210-103512. [DOI] [PubMed] [Google Scholar]
- Lanyon B. P.; Whitfield J. D.; Gillett G. G.; Goggin M. E.; Almeida M. P.; Kassal I.; Biamonte J. D.; Mohseni M.; Powell B. J.; Barbieri M.; Aspuru-Guzik A.; White A. G. Towards quantum chemistry on a quantum computer. Nat. Chem. 2010, 2, 106–111. 10.1038/nchem.483. [DOI] [PubMed] [Google Scholar]
- O’Malley P. J. J.; Babbush R.; Kivlichan I. D.; Romero J.; McClean J. R.; Barends R.; Kelly J.; Roushan P.; Tranter A.; Ding N.; Campbell B.; Chen Y.; Chen Z.; Chiaro B.; Dunsworth A.; Fowler A. G.; Jeffrey E.; Lucero E.; Megrant A.; Mutus J. Y.; Neeley M.; Neill C.; Quintana C.; Sank D.; Vainsencher A.; Wenner J.; White T. C.; Coveney P. V.; Love P. J.; Neven H.; Aspuru-Guzik A.; Martinis J. M. Scalable Quantum Simulation of Molecular Energies. Phys. Rev. X 2016, 6, 031007. 10.1103/PhysRevX.6.031007. [DOI] [Google Scholar]
- Kandala A.; Mezzacapo A.; Temme K.; Takita M.; Brink M.; Chow J. M.; Gambetta J. M. Hardware-efficient variational quantum solver for small molecules and quantum magnets. Nature 2017, 549, 242–246. 10.1038/nature23879. [DOI] [PubMed] [Google Scholar]
- Lu Y. The “OK, Molly” Chemistry. Acc. Chem. Res. 2017, 50, 647–651. 10.1021/acs.accounts.7b00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weymuth T.; Reiher M. Inverse quantum chemistry: Concepts and strategies for rational compound design. Int. J. Quantum Chem. 2014, 114, 823–837. 10.1002/qua.24687. [DOI] [Google Scholar]
- Sanchez-Lengeling B.; Outerial C.; Guimaraes G. L.; Aspuru-Guzik A.. Optimizing distributions over molecular space. An Objective-Reinforced Generative Adversarial Network for Inverse-design Chemistry (ORGANIC). 2017, Preprint: chemrxiv:5309668.
- Kuhn C.; Beratan D. Inverse strategies for molecular design. J. Phys. Chem. 1996, 100, 10595–10599. 10.1021/jp960518i. [DOI] [Google Scholar]
- Wang M.; Hu X.; Beratan D. N.; Yang W. Designing Molecules by Optimizing Potentials. J. Am. Chem. Soc. 2006, 128, 3228–3232. 10.1021/ja0572046. [DOI] [PubMed] [Google Scholar]
- Xiao D.; Yang W.; Beratan D. N. Inverse molecular design in a tight-binding framework. J. Chem. Phys. 2008, 129, 044106. 10.1063/1.2955756. [DOI] [PubMed] [Google Scholar]
- Xiao D.; Martini L. A.; Snoeberger R. C. III; Crabtree R. H.; Batista V. S. Inverse Design and Synthesis of acac-Coumarin Anchors for Robust TiO2 Sensitization. J. Am. Chem. Soc. 2011, 133, 9014–9022. 10.1021/ja2020313. [DOI] [PubMed] [Google Scholar]
- Weymuth T.; Reiher M. Gradient-driven molecule construction: An inverse approach applied to the design of small-molecule fixating catalysts. Int. J. Quantum Chem. 2014, 114, 838–850. 10.1002/qua.24686. [DOI] [Google Scholar]
- Turing A. M. Computing Machinery and Intelligence. Mind 1950, 59, 433–460. 10.1093/mind/LIX.236.433. [DOI] [Google Scholar]
- Feynman R. P. Simulating physics with computers. Int. J. Theor. Phys. 1982, 21, 467–488. 10.1007/BF02650179. [DOI] [Google Scholar]
- Nielsen M. A.; Chuang I. L.. Quantum Computation and Quantum Information; Cambridge University Press: 2011. [Google Scholar]
- Yuen-Zhou J.; Krich J. J.; Mohseni M.; Aspuru-Guzik A. Quantum State and Process Tomography of Energy Transfer Systems via Ultrafast Spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 17615–17620. 10.1073/pnas.1110642108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen-Zhou J.; Arias D. H.; Eisele D. M.; Steiner C. P.; Krich J. J.; Bawendi M.; Nelson K. A.; Aspuru-Guzik A. Coherent exciton dynamics in supramolecular light-harvesting nanotubes revealed by ultrafast quantum process tomography. ACS Nano 2014, 8, 5527–5534. 10.1021/nn406107q. [DOI] [PubMed] [Google Scholar]
- Woodward R. B.; Hoffmann R. The Conservation of Orbital Symmetry. Angew. Chem., Int. Ed. Engl. 1969, 8, 781–853. 10.1002/anie.196907811. [DOI] [Google Scholar]