Abstract
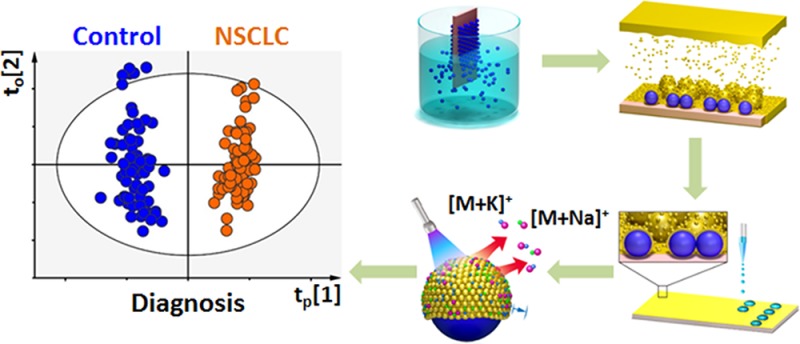
Current metabolic analysis is far from ideal to engage clinics and needs rationally designed materials and device. Here we developed a novel plasmonic chip for clinical metabolic fingerprinting. We first constructed a series of chips with gold nanoshells on the surface through controlled particle synthesis, dip-coating, and gold sputtering for mass production. We integrated the optimized chip with microarrays for laboratory automation and micro-/nanoscaled experiments, which afforded direct high-performance metabolic fingerprinting by laser desorption/ionization mass spectrometry using 500 nL of various biofluids and exosomes. Further we for the first time demonstrated on-chip in vitro metabolic diagnosis of early stage lung cancer patients using serum and exosomes. This work initiates a new bionanotechnology based platform for advanced metabolic analysis toward large-scale diagnostic use.
Short abstract
A plasmonic chip was constructed for mass spectrometry based metabolic fingerprinting of biofluids and exosomes, toward large-scale diagnostic use.
Introduction
In vitro diagnostics (IVD) serves as the “eye” of medical doctors and contributes to ∼66% of clinical diagnosis.1−3 For IVD, metabolic analysis is more distal over proteomic and genomic approaches toward precision diagnostics, and thus has been applied in clinical practice and biomedical research universally.2−6 Notably, metabolic analysis relies on rationally designed materials and device for sample treatment and metabolite detection,5,7,8 dealing with diverse biological specimens for practical diagnostic applications. Currently, there are two major challenges including (I) difficulties in direct metabolic analysis of biosamples with little pretreatment, due to the low molecular abundance and high sample complexity; and (II) construction of diagnostic tools by materials and device based platforms for real case application in clinics. Therefore, it is of key significance to develop advanced tools for metabolic analysis addressing these challenges, capable of disease detection and biomarker discovery in clinics.
Distinct from conventional techniques such as electrochemical and spectrometric methods, mass spectrometry (MS) has displayed high accuracy, sensitivity, resolution, and throughput for molecular analysis.2,7,9−12 Particularly, laser desorption/ionization (LDI) MS affords desirable speed for in-seconds detection, mass measurement for molecular identification, and high sensitivity with low costs toward large-scale use.13−15 However, owing to the broad dynamic ranges of various molecules in biological specimens, the performance of MS is affected by rigorous sample treatment and molecule detection procedures. The development of chips for LDI MS is promising to address these critical issues, but very challenging considering the following aspects: (I) preselected structures with precisely designed and tunable physiochemical parameters;16−18 (II) functional interface controlling the analytical variations and background noises for matrix-free analysis;19,20 (III) integration with microarray technology to handle ultrasmall sample volume and high-throughput screening;15,21,22 and (IV) robustness and convenience toward potential point-of-care testings23−26 (POCTs). Existing chips mostly based on silicon18,19 carbon,19,27,28 and noble metals19,29 can address some of the above aspects but have afforded limited analytical capability for real case diagnostic application thus far. Novel chip platforms with enhanced performance are in urgent demand for clinical diagnostics.
Plasmonic materials including Au, Ag, Pt, etc. enjoy characteristic surface plasmon resonances and produce hot carriers during laser irradiation.30−33 There has been tremendous interest from research groups globally on plasmonic materials and noble metals with various structures for LDI MS.16,19,34 For instance, gold nanoshells have been recently demonstrated as preferred materials for LDI MS over solid gold particles, due to the surface roughness and better hot carrier production.35,36 Notably, most approaches use in-solution seeding methods22,36,37 to synthesize plasmonic materials for LDI MS, whereas it is difficult to fine-tune their structural parameters. Conventional engineering methods,38−40 such as microelectromechanical systems (MEMS), are also very limited in terms of nanoscaled control and experimental costs for the development of LDI MS chips. Till now, there have been very few reports on defined plasmonic LDI MS chips, and new preparation methods are in pressing need to overcome these major obstacles.
Herein, we constructed a new plasmonic gold chip for metabolic fingerprinting of biofluids and exosomes in diagnostics (Figure 1a). We prepared a series of chips with gold nanoshells on the surface through controlled particle synthesis, dip-coating, and gold sputtering for mass production. We integrated the optimized chip with microarrays for laboratory automation and micro-/nanoscaled experiments, which afforded sensitive, selective, multiplex, and quantitative metabolic fingerprinting by LDI MS using 500 nL of serum, cerebrospinal fluid (CSF), urine, and exosomes. Further we for the first time demonstrated on-chip in vitro metabolic diagnosis of early stage lung cancer patients using serum and exosomes. This work initiates a novel bionanotechnology based platform for advanced metabolic analysis toward large-scale diagnostic use.
Figure 1.
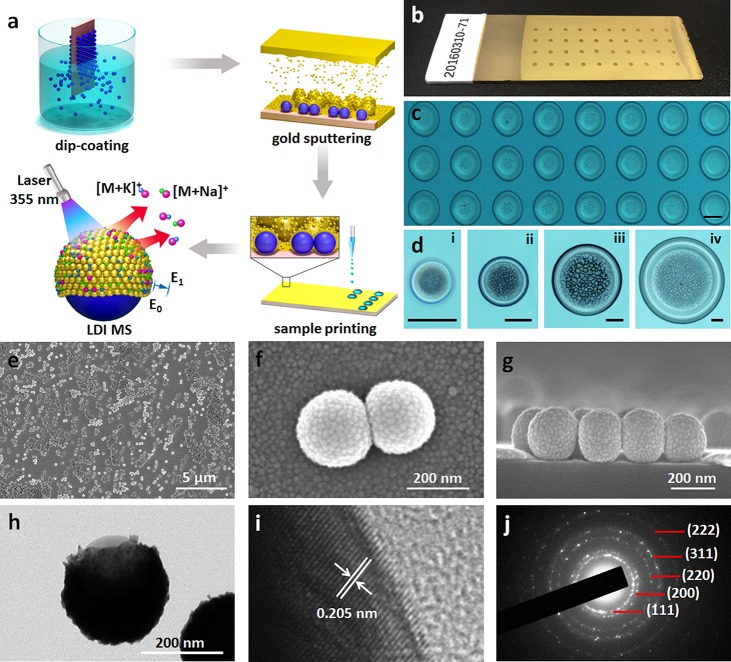
Construction of plasmonic chips. (a) Scheme of preparation and application of the plasmonic chip for LDI MS based metabolic analysis. (b) Digital and (c) microscope images of the chip after microarray printing with 400 nL and 4 nL of samples, respectively. (d) Microscope images of (i) 400 pL, (ii) 4 nL, (iii) 40 nL, and (iv) 400 nL of serum plotted on the chip. Top view (e) SEM of the chip surface and (f) high-resolution SEM of the particle surface. (g) Cross section SEM of the chip. (h) TEM of particles and (i) high-resolution TEM showing the gold crystal lattice. (j) The crystalline structure by selected area electron diffraction (SAED) pattern. The scale bar of insets in panels c and d is 200 μm.
Results and Discussion
Construction and Characterization of the Plasmonic Chip
We first constructed the plasmonic chip through a three-step process including particle synthesis, dip-coating, and gold sputtering (see Methods in the Supporting Information for details), facile for mass production as displayed in Figure 1a. The as-made chip is uniform for practical use (Figure 1b) and compatible with microarray printing (Figure 1c), toward reproducible and high-throughput metabolic analysis using tiny amounts of biosamples (down to 400 pL, Figure 1d). The chip format for application not only achieves laboratory automation for easy and labor-efficient operation, but also enables precise manipulation and handling of droplets for micro-/nanoscaled experiments.18,21,22,38
Owing to the unique preparation technique, the plasmonic chip was decorated by gold nanoshells on the surface (Figure 1a). We observed particles (∼150 nm in diameter) with nanoshells packed by small nanoparticles (∼10–20 nm, Figure 1e,f) uniformly dispersed on chip by top-view scanning electron microscopy (SEM). We demonstrated the structure of nanoshells by cross-section SEM (Figure 1g), agreeing with transmission electron microscopy (TEM, Figure 1h) and elemental mapping results in Figure S1. The nanoshell particles were negatively charged with zeta potentials of −27.4 ± 0.2 mV, according to zeta potential distributions (Figure S2), beneficial for the formation of ion layers on the surface toward efficient Na+/K+ adduction during the LDI process. We validated the gold nanoshells with the typical interplanar spacing of 2.05 Å for gold along the [200] direction by high-resolution (HR) TEM (Figure 1i) and the crystalline structure by selected area electron diffraction (SAED) pattern36,37,41 (Figure 1j). Notably, the chip afforded distinct hierarchical surface properties, including microscaled and nanoscaled surface roughness from the on-chip core-shell particles and on-shell gold nanoparticles, respectively.
Control and Optimization of Chip Structural Parameters
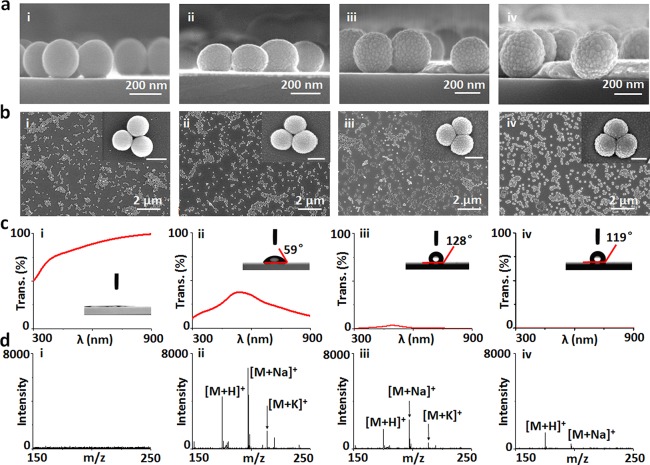
We prepared a series of chips with tunable structural parameters by controlling the gold sputtering, dip-coating conditions, and particle synthesis (see Methods in the Supporting Information for details). The surface of on-chip particles was smooth without sputtering (denoted chipt0) and became rough with increased sputtering time of 2/4/8 min (denoted chipt2/t4/t8) as displayed in Figure 2a,b. The reduction of transmittance was caused by the increased thickness of gold nanoshells with longer sputtering time (Figure 2c). The contact angle initially increased to 128° and then decreased to 119° (inset of Figure 2c), due to introduction of gold and change of surface roughness. We conducted LDI MS detection of arginine (typical spectra in Figure 2d, triplicate results in Figure S3) and other metabolites (Figures S4 and S5), and chipt2 afforded the highest peak intensity and signal-to-noise ratio (p < 0.05). The area density of particles on-chip decided the average interparticle distance for plasmon coupling,42,43 and the size of silica spheres also affected the structures of nanoshells for plasmon resonance,42,44 both of which were involved in the process of hot carrier production, local heating, and photodesorption for plasmonics enhanced LDI MS. Therefore, we further performed the selection of area density (∼12, 20, 37 particles per μm2) and size (∼100–200 nm) of particles on-chip by controlling the dip-coating conditions and particle synthesis in Figures S6 and S7 and Figures S8 and S9, respectively. We demonstrated the optimized chip with area density of ∼20 particles per μm2 and particle size of ∼150 nm for the next stage applications (p < 0.05).
Figure 2.
Control and optimization of chips. (a) Cross section and (b) top view SEM of the chips with controlled sputtering time of (i) 0 min (chipt0), (ii) 2 min (chipt2), (iii) 4 min (chipt4), and (iv) 8 min (chipt8). (c) Transmittance and contact angle (inset) of the chips (chipt0/t2/t4/t8 for i–iv). (d) Typical LDI MS of arginine (200 ng/μL) on the chipt0/t2/t4/t8 (i–iv). The scale bar of insets in panel b is 200 nm.
Notably, there were Ag+ adducted molecular peaks using silver nanoshells, since silver ions can afford the ion–dipole interaction with polar functional groups like hydroxyl group in metabolites45,46 and also cationize molecules containing π-bonds.47,48 For comparison, Au+ adducted molecular peaks can be found for molecules (e.g., peptides) containing thiol groups,49,50 owing to the strong Au–S chemoaffinity. In this work, the small metabolites as detected did not contain thiol groups, and we observed no Au+ adducted molecular peaks, similar to previous literature reports.16,34
For on-chip LDI MS detection, the silica core particles within gold shells are critical to isolate the electricity/heat, retain the local heat, and produce more hot carriers.35 Notably, overloading of gold on the chip surface (chipt4/t8) affected their LDI performance, considering the overaggregation of gold nanoparticles on the surface of core silica particles and unwanted plasmon coupling of excess gold (Figure 2a,b-iii/iv). The optimized gold nanoshells as interface afforded desirable surface roughness, light absorption, and surface hydrophobicity for LDI MS.16,36 Previous noble metal material based LDI MS chips achieved detection of heavy metal ions (e.g., lead ions) in biofluids29 and imaging of metabolites in fingerprints,19 but afforded few successful examples dealing with small metabolites in complex biofluids with salts and proteins. Considering the broad definition of plasmonic materials including but not limited to noble metals,51−53 LDI MS chips based on carbon (graphene),20,27,28 silicon,17,18 and their hybrids may have unwanted fragmentation during laser radiation or need specific MEMS for device fabrication, which have also been rarely applied for metabolic analysis in biofluids. In our work, specific nanogaps and nanocrevices of gold shells on-chip may selectively trap small metabolite molecules and transfer the laser energy,6 toward advanced metabolic analysis of complex biosamples in real case.
On-Chip Metabolic Fingerprinting of Biofluids and Exosomes
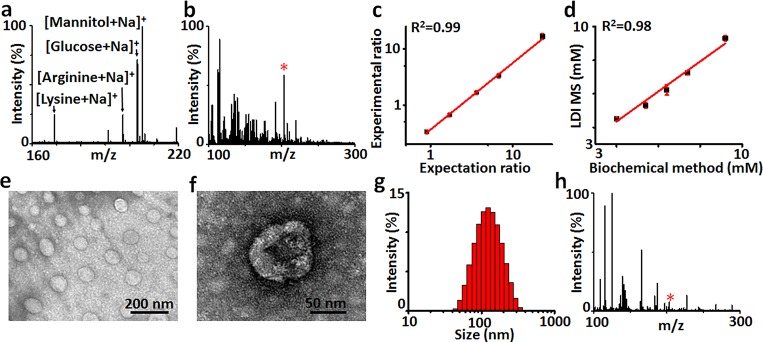
We performed on-chip metabolic fingerprinting of 500 nL of serum/CSF/urine, by direct LDI MS. We demonstrated the selectivity of chip toward small metabolites in a complex biomixture. Considering the complexity of biofluids, we first studied the salt tolerance and protein endurance of the optimized chip for small low-abundance metabolite detection (Figure 3a), using a mixture containing four small metabolites (2 nmol each), salts (0.5 M NaCl), and proteins (5 mg/mL bovine serum albumin). We observed strong Na+ adducted molecular signals at m/z of 169.18 for lysine, 197.19 for arginine, 203.15 for glucose, and 205.16 for mannitol, demonstrating the selectivity of the gold chip toward metabolites over other molecules for real-case applications in complex biofluids. Next we detected serum (Figure 3b), CSF (Figure S10a), and urine (Figure S10b) on-chip, yielding a series of molecular peaks for typical metabolites (Tables S1–3). Notably, we coupled on-chip detection with isotopic quantification to address the qualitative or semiquantitative analysis by traditional LDI MS due to the random analyte dependent ionization.6,13 As a result, we can quantitate selected metabolites (e.g., glucose, R2 = 0.99 with CV < 9.1%, Figure 3c, MS/MS in Figure S11) consistent with the biochemical method (the current gold standard to quantitate metabolites in clinics, R2 = 0.98, Figure 3d).
Figure 3.
LDI MS fingerprinting of biosamples. (a) Mass spectra of lysine, arginine, glucose, and mannitol in 5 mg/mL bovine serum albumin solution with NaCl concentration of 0.5 M and (b) 0.5 μL of human serum on the chipt2. (c) The calibration curve obtained by plotting experimental ratio of analyte/isotope (A/I) as a function of expected ratio of A/I for glucose. (d) Linear fit of LDI MS and biochemical method for quantification of glucose in serum. (e) TEM of exosomes extracted from serum and (f) the zoomed image showing the hollow structure of an exosome. (g) Size distribution and (h) mass spectrum of the exosomes. The star refers to glucose in panels b and h. Three independent experiments were performed for each sample to calculate the standard deviation (SD) as error bars in panels c and d. Data are shown as the mean ± SD (n = 3).
Conventional metabolic fingerprints usually rely on nuclear magnetic resonance (NMR) spectroscopy and gas/liquid chromatography (GC/LC) MS.2,4,8 NMR spectroscopy provides quantitative information on biosamples (∼mL) on the atomic level (∼minutes) and cannot directly identify potential biomarkers on the molecular level.2,8 GC/LC MS detects the molecules but calls for rigorous pretreatment procedures (∼hours),4 due to the high complexity and low abundance of metabolites in biosamples (∼μL) that affect the efficacy of MS. Further considering the limitation of organic matrices (e.g., α-cyano-4-hydroxycinnamic acid, CHCA)6 in LDI MS that produced strong background signals in low mass range (m/z < 300, Figure S12), our on-chip analysis overcame the current major obstacles for advanced metabolic fingerprints. The LDI MS chips not only required few pretreatment procedures (e.g., enrichment and purification) of trace biosamples (down to ∼pL) for large-scale clinical use but also afforded desirable selectivity of small metabolites, speed (∼seconds), quantitation, and reproducibility.
Exosomes analysis promises precision diagnostics, but suffered from obstacles in sample harvesting and downstream detection.38,54,55 We extracted exosomes from serum by the established protocol (see Methods in the Supporting Information for details) and characterized their structures. As displayed in Figure 3e,f, TEM demonstrated hollow structures of exosomes with sizes of ∼50–150 nm, agreeing with dynamic light scattering (DLS) results (Figure 3g). We performed metabolic fingerprinting of exosomes on-chip and obtained a series of molecular peaks (Figure 3h, Table S4), distinct from that of serum (Figure 3b). There have been several bioanalytical techniques for molecular screening of cancer-derived exosomes, mainly on the proteomic and genetic levels.54−56 On the proteomic level, antibody recognition is required to be coupled with surface plasmon resonance (SPR), electrochemical test, etc. for profiling of exosome surface/lysate proteins.38,57 On the genetic level, polymerase chain reaction (PCR) is essential to amplify the signals of nucleic acids for molecular sequencing and mapping in exosomes.56,58 In contrast, the plasmonic LDI MS chips featured the profiling of small molecules on the metabolic level, free of antibody recognition and PCR amplification. Given the on-chip metabolic fingerprinting of biofluids and exosomes, we anticipated to construct novel diagnostics tools by combinational and comparative analysis.
On-Chip in Vitro Metabolic Diagnosis of Lung Cancer Patients
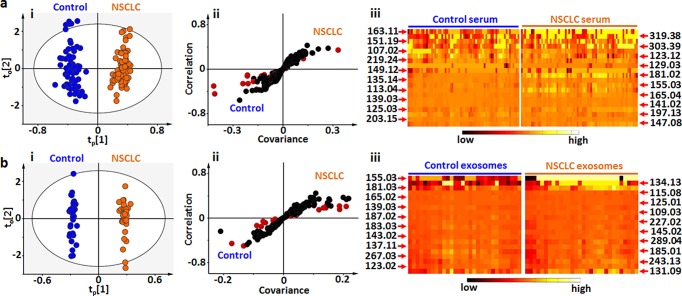
We differentiated the early stage non-small cell lung cancer (NSCLC) patients from healthy controls by on-chip metabolic analysis of serum (Table S5) and exosomes (Table S6). We performed orthogonal partial least squares discriminant analysis (OPLS-DA)9,59−61 and demonstrated the clear group separation based on the fingerprinting results of serum (Figure 4a-i, R2 = 0.955, Q2 = 0.726, p < 0.0001) and exosomes (Figure 4b-i, R2 = 0.994, Q2 = 0.716, p < 0.0001). Considering the diversity of metabolites in biospecimens, the potential identification requires the construction of a complete database for metabolites by comparing their MS/MS with standards (e.g., in Figure S11). We further performed S-plot10,59 (Figure 4a-ii and 4b-ii) based on fingerprinting of serum/exosomes and selected 20 key m/z values (according to variable importance on projection (VIP) scores, Tables S7 and S8) for the heat map (Figure 4a-iii and 4b-iii) to validate the differentiation of NSCLC patients from healthy controls. Therefore, we achieved on-chip metabolic analysis for early diagnosis of NSCLC and anticipated these key m/z values to serve as potential metabolic markers.
Figure 4.
Diagnosis of early stage NSCLC. Summary of LDI MS fingerprinting of (a) serum from 23 early stage NSCLC patients and 22 healthy controls and (b) exosomes from 10 early stage NSCLC patients and 10 healthy controls for diagnosis. (i) OPLS-DA score plots showing the global metabolic difference between NSCLC patients and healthy controls, for both serum (p < 0.0001) and exosomes (p < 0.0001). (ii) S-plots of LDI MS fingerprinting for serum and exosomes. Wings of S-plot marked by red accounting for high variance between two subpopulations. (iii) Heat maps based on selected 20 points from S-plot with high VIP scores (>1) showing the difference between the early stage NSCLC patients and healthy controls. Black represents low, and white represents high. Independent triplicate experiments were performed for each serum and exosome sample.
Lung cancer threatens human health worldwide with a mortality rate of ∼20%, and over 80% of lung cancer is NSCLC.62−64 Precise diagnosis of early stage NSCLC can raise the 5-year survival ratio and reduce the medical expenses in NSCLC management.63,65,66 Blood tests promise early diagnosis of NSCLC owing to the simple measurement using blood samples and low costs for point-of-care testing, which is noninvasive and facile for universal applications, superior to traditional biopsy and imaging methods. Traditional blood tests for NSCLC are based on the detection of selected biomarkers, which are normally employed for prognosis and very limited for diagnosis.21 Considering the metabolic variations in early stage NSCLC as demonstrated by previous reports,67 our results offer new insights for precise diagnosis of early stage NSCLC by metabolic fingerprinting of serum and exosomes. We can anticipate two major future directions for research along this line, from both materials science and biomedical science perspectives. From the materials science perspective, new types of material based chips would afford tailored interfaces toward either one molecule or a group of molecules and the further combinational use of materials can be capable of multiomics detection. From the biomedical science perspective, we may foresee the precision diagnosis of diverse physiological/pathological process using this platform and identification of novel metabolic pathways toward prevention and intervention of specific diseases.
Conclusions
In summary, we introduced plasmonic gold chips as new substrates for direct LDI MS detection of small metabolites in biofluids and exosomes, and further constructed a novel platform technology for metabolic fingerprinting based IVD. Notably, our nanoplasmonic platform would be able to achieve potential application for immunodiagnostics1,21,22 and translation into point-of-care devices and IVD platforms,68−70 considering the ease of surface functionalization on gold (for antibody/antigen immobilization) and low sample volumes required (down to 400 pL). Our work makes solid contributions to the design of materials and device for advanced metabolic analysis toward precision medicine, and initiates the development of various personalized diagnostic tools for diverse diseases including but not limited to lung cancer in the near future.
Acknowledgments
We gratefully acknowledge the financial support from Project 81771983, 81750110544, 81750410695, and 81650110523 by National Natural Science Foundation of China (NSFC), Project 16441909300 by Shanghai Science and Technology Commission, Project 16CR2011A by Clinical Research Plan of SHDC, and Project 2017YFC0909000 by Ministry of Science and Technology of China. This work is also sponsored by the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (TP2015015).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.7b00546.
Methods, Figures S1–S12, and Tables S1–S8 (PDF)
The authors declare the following competing financial interest(s): The authors have filed patents for both the technology and the use of the technology to detect biosamples.
Supplementary Material
References
- Zhang B.; Pinsky B. A.; Ananta J. S.; Zhao S.; Arulkumar S.; Wan H.; Sahoo M. K.; Abeynayake J.; Waggoner J. J.; Hopes C.; Tang M.; Dai H. Diagnosis of Zika virus infection on a nanotechnology platform. Nat. Med. 2017, 23, 548–550. 10.1038/nm.4302. [DOI] [PubMed] [Google Scholar]
- Wishart D. S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discovery 2016, 15, 473–484. 10.1038/nrd.2016.32. [DOI] [PubMed] [Google Scholar]
- Rohr U. P.; Binder C.; Dieterle T.; Giusti F.; Messina C. G.; Toerien E.; Moch H.; Schafer H. H. The Value of In Vitro Diagnostic Testing in Medical Practice: A Status Report. PLoS One 2016, 11, e0149856. 10.1371/journal.pone.0149856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. J.; Larson M. G.; Vasan R. S.; Cheng S.; Rhee E. P.; McCabe E.; Lewis G. D.; Fox C. S.; Jacques P. F.; Fernandez C.; O’Donnell C. J.; Carr S. A.; Mootha V. K.; Florez J. C.; Souza A.; Melander O.; Clish C. B.; Gerszten R. E. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011, 17, 448–453. 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenobi R. Single-cell metabolomics: analytical and biological perspectives. Science 2013, 342, 1243259. 10.1126/science.1243259. [DOI] [PubMed] [Google Scholar]
- Huang L.; Wan J.; Wei X.; Liu Y.; Huang J.; Sun X.; Zhang R.; Gurav D. D.; Vedarethinam V.; Li Y.; Chen R.; Qian K. Plasmonic silver nanoshells for drug and metabolite detection. Nat. Commun. 2017, 8, 220. 10.1038/s41467-017-00220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaleckis R.; Murakami I.; Takada J.; Kondoh H.; Yanagida M. Individual variability in human blood metabolites identifies age-related differences. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 4252–4259. 10.1073/pnas.1603023113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H.; An Y. J.; Xu W. J.; Kang K. W.; Park S. Real-time monitoring of cancer cell metabolism and effects of an anticancer agent using 2D in-cell NMR spectroscopy. Angew. Chem., Int. Ed. 2015, 54, 5374–5377. 10.1002/anie.201410380. [DOI] [PubMed] [Google Scholar]
- Chen W. L.; Wang J. H.; Zhao A. H.; Xu X.; Wang Y. H.; Chen T. L.; Li J. M.; Mi J. Q.; Zhu Y. M.; Liu Y. F.; Wang Y. Y.; Jin J.; Huang H.; Wu D. P.; Li Y.; Yan X. J.; Yan J. S.; Li J. Y.; Wang S.; Huang X. J.; Wang B. S.; Chen Z.; Chen S. J.; Jia W. A distinct glucose metabolism signature of acute myeloid leukemia with prognostic value. Blood 2014, 124, 1645–1654. 10.1182/blood-2014-02-554204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolee J. A.; Shrestha B.; Mengistu G.; Vertes A. Observation of subcellular metabolite gradients in single cells by laser ablation electrospray ionization mass spectrometry. Angew. Chem., Int. Ed. 2012, 51, 10386–10389. 10.1002/anie.201205436. [DOI] [PubMed] [Google Scholar]
- Macedo A. N.; Mathiaparanam S.; Brick L.; Keenan K.; Gonska T.; Pedder L.; Hill S.; Britz-McKibbin P. The Sweat Metabolome of Screen-Positive Cystic Fibrosis Infants: Revealing Mechanisms beyond Impaired Chloride Transport. ACS Cent. Sci. 2017, 3, 904–913. 10.1021/acscentsci.7b00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejeeth C.; Pang X.; Zhang R.; Xu W.; Sun X.; Liu B.; Lou J.; Wan J.; Gu H.; Yan Y.; Qian K. Extraction, Detection, and Profiling of Serum Biomarkers Using Designed Fe3O4@SiO2@HA Core-Shell Particles. Nano Res. 2018, 11, 68–79. 10.1007/s12274-017-1591-6. [DOI] [Google Scholar]
- Wu J.; Wei X.; Gan J.; Huang L.; Shen T.; Lou J.; Liu B.; Zhang J. X. J.; Qian K. Multifunctional Magnetic Particles for Combined Circulating Tumor Cells Isolation and Cellular Metabolism Detection. Adv. Funct. Mater. 2016, 26, 4016–4025. 10.1002/adfm.201504184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez A. J.; Fagerer S. R.; Schmidt A. M.; Urban P. L.; Jefimovs K.; Geiger P.; Dechant R.; Heinemann M.; Zenobi R. Mass spectrometry-based metabolomics of single yeast cells. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 8790–8794. 10.1073/pnas.1209302110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban L.; Pettit N.; Li L.; Stuparu A. D.; Cai L.; Chen W.; Guan W.; Han W.; Wang P. G.; Mrksich M. Discovery of glycosyltransferases using carbohydrate arrays and mass spectrometry. Nat. Chem. Biol. 2012, 8, 769–773. 10.1038/nchembio.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C. K.; Chen W. T.; Chang H. T. Nanoparticle-based mass spectrometry for the analysis of biomolecules. Chem. Soc. Rev. 2011, 40, 1269–1281. 10.1039/C0CS00050G. [DOI] [PubMed] [Google Scholar]
- Northen T. R.; Yanes O.; Northen M. T.; Marrinucci D.; Uritboonthai W.; Apon J.; Golledge S. L.; Nordstrom A.; Siuzdak G. Clathrate nanostructures for mass spectrometry. Nature 2007, 449, 1033–1036. 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- Stopka S. A.; Rong C.; Korte A. R.; Yadavilli S.; Nazarian J.; Razunguzwa T. T.; Morris N. J.; Vertes A. Molecular Imaging of Biological Samples on Nanophotonic Laser Desorption Ionization Platforms. Angew. Chem., Int. Ed. 2016, 55, 4482–4486. 10.1002/anie.201511691. [DOI] [PubMed] [Google Scholar]
- Lim A. Y.; Ma J.; Boey Y. C. F. Development of Nanomaterials for SALDI-MS Analysis in Forensics. Adv. Mater. 2012, 24, 4211–4216. 10.1002/adma.201200027. [DOI] [PubMed] [Google Scholar]
- Kim Y.-K.; Na H.-K.; Kwack S.-J.; Ryoo S.-R.; Lee Y.; Hong S.; Hong S.; Jeong Y.; Min D.-H. Synergistic Effect of Graphene Oxide/MWCNT Films in Laser Desorption/Ionization Mass Spectrometry of Small Molecules and Tissue Imaging. ACS Nano 2011, 5, 4550–4561. 10.1021/nn200245v. [DOI] [PubMed] [Google Scholar]
- Liu B.; Li Y.; Wan H.; Wang L.; Xu W.; Zhu S.; Liang Y.; Zhang B.; Lou J.; Dai H.; Qian K. High Performance, Multiplexed Lung Cancer Biomarker Detection on a Plasmonic Gold Chip. Adv. Funct. Mater. 2016, 26, 7994–8002. 10.1002/adfm.201603547. [DOI] [Google Scholar]
- Zhang B.; Kumar R. B.; Dai H.; Feldman B. J. A plasmonic chip for biomarker discovery and diagnosis of type 1 diabetes. Nat. Med. 2014, 20, 948–953. 10.1038/nm.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.; Wan Q.; Badu-Tawiah A. K. Mass Spectrometry for Paper-Based Immunoassays: Toward On-Demand Diagnosis. J. Am. Chem. Soc. 2016, 138, 6356–6359. 10.1021/jacs.6b02232. [DOI] [PubMed] [Google Scholar]
- Zhu Z.; Guan Z.; Jia S.; Lei Z.; Lin S.; Zhang H.; Ma Y.; Tian Z. Q.; Yang C. J. Au@Pt nanoparticle encapsulated target-responsive hydrogel with volumetric bar-chart chip readout for quantitative point-of-care testing. Angew. Chem., Int. Ed. 2014, 53, 12503–12507. 10.1002/anie.201405995. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Zhou J.; Li J. Construction of Plasmonic Nano-Biosensor-Based Devices for Point-of-Care Testing. Small Methods 2017, 1, 1700197. 10.1002/smtd.201700197. [DOI] [Google Scholar]
- Furst A. L.; Hoepker A. C.; Francis M. B. Quantifying Hormone Disruptors with an Engineered Bacterial Biosensor. ACS Cent. Sci. 2017, 3, 110–116. 10.1021/acscentsci.6b00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian K.; Zhou L.; Liu J.; Yang J.; Xu H.; Yu M.; Nouwens A.; Zou J.; Monteiro M. J.; Yu C. Laser Engineered Graphene Paper for Mass Spectrometry Imaging. Sci. Rep. 2013, 3, 1415. 10.1038/srep01415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.; Kim Y. K.; Min D. H. Laser Desorption/Ionization Mass Spectrometric Assay for Phospholipase Activity Based on Graphene Oxide/Carbon Nanotube Double-Layer Films. J. Am. Chem. Soc. 2010, 132, 14714–14717. 10.1021/ja106276j. [DOI] [PubMed] [Google Scholar]
- Liu Y.-C.; Chiang C.-K.; Chang H.-T.; Lee Y.-F.; Huang C.-C. Using a Functional Nanogold Membrane Coupled with Laser Desorption/Ionization Mass Spectrometry to Detect Lead Ions in Biofluids. Adv. Funct. Mater. 2011, 21, 4448–4455. 10.1002/adfm.201101248. [DOI] [Google Scholar]
- Wu K.; Chen J.; McBride J. R.; Lian T. Efficient Hot Electron Transfer by Plasmon Induced Interfacial Charge Transfer Transition. Science 2015, 349, 632–635. 10.1126/science.aac5443. [DOI] [PubMed] [Google Scholar]
- Cortes E.; Xie W.; Cambiasso J.; Jermyn A. S.; Sundararaman R.; Narang P.; Schlucker S.; Maier S. A. Plasmonic hot electron transport drives nano-localized chemistry. Nat. Commun. 2017, 8, 14880. 10.1038/ncomms14880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D. L.; Cortes E.; Giannini V.; Torok P.; Roschuk T.; Maier S. A. Decoupling absorption and emission processes in super-resolution localization of emitters in a plasmonic hotspot. Nat. Commun. 2017, 8, 14513. 10.1038/ncomms14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W.; Sikdar D.; Yap L. W.; Guo P.; Premaratne M.; Li X.; Cheng W. Matryoshka-caged gold nanorods: Synthesis, plasmonic properties, and catalytic activity. Nano Res. 2016, 9, 415–423. 10.1007/s12274-015-0922-8. [DOI] [Google Scholar]
- Ocsoy I.; Gulbakan B.; Shukoor M. I.; Xiong X.; Chen T.; Powell D. H.; Tan W. Aptamer-Conjugated Multifunctional Nanoflowers as a Platform for Targeting, Capture, and Detection in Laser Desorption Ionization Mass Spectrometry. ACS Nano 2013, 7, 417–427. 10.1021/nn304458m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjavacas A.; Liu J. G.; Kulkarni V.; Nordlander P. Plasmon-induced hot carriers in metallic nanoparticles. ACS Nano 2014, 8, 7630–7638. 10.1021/nn502445f. [DOI] [PubMed] [Google Scholar]
- Gan J.; Wei X.; Li Y.; Wu J.; Qian K.; Liu B. Designer SiO2@Au nanoshells towards sensitive and selective detection of small molecules in laser desorption ionization mass spectrometry. Nanomedicine: NBM 2015, 11, 1715–1723. 10.1016/j.nano.2015.06.010. [DOI] [PubMed] [Google Scholar]
- Fan Z.; Bosman M.; Huang X.; Huang D.; Yu Y.; Ong K. P.; Akimov Y. A.; Wu L.; Li B.; Wu J.; Huang Y.; Liu Q.; Png C. E.; Gan C. L.; Yang P.; Zhang H. Stabilization of 4H hexagonal phase in gold nanoribbons. Nat. Commun. 2015, 6, 7684. 10.1038/ncomms8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im H.; Shao H.; Park Y. I.; Peterson V. M.; Castro C. M.; Weissleder R.; Lee H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495. 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui Y.; Gomez-Diaz J. S.; Qian Z.; Alu A.; Rinaldi M. Plasmonic piezoelectric nanomechanical resonator for spectrally selective infrared sensing. Nat. Commun. 2016, 7, 11249. 10.1038/ncomms11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente J.; Ou J. Y.; Plum E.; Youngs I. J.; Zheludev N. I. A magneto-electro-optical effect in a plasmonic nanowire material. Nat. Commun. 2015, 6, 7021. 10.1038/ncomms8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.; Xia Y. Triangular Nanoplates of Silver: Synthesis, Characterization, and Use as Sacrificial Templates For Generating Triangular Nanorings of Gold. Adv. Mater. 2003, 15, 695–699. 10.1002/adma.200304652. [DOI] [Google Scholar]
- Vazquez-Mena O.; Sannomiya T.; Villanueva L. G.; Voros J.; Brugger J. Metallic Nanodot Arrays by Stencil Lithography for Plasmonic Biosensing Applications. ACS Nano 2011, 5, 844–853. 10.1021/nn1019253. [DOI] [PubMed] [Google Scholar]
- Hentschel M.; Saliba M.; Vogelgesang R.; Giessen H.; Alivisatos A. P.; Liu N. Transition from Isolated to Collective Modes in Plasmonic Oligomers. Nano Lett. 2010, 10, 2721–2726. 10.1021/nl101938p. [DOI] [PubMed] [Google Scholar]
- Lin L.; Zapata M.; Xiong M.; Liu Z.; Wang S.; Xu H.; Borisov A. G.; Gu H.; Nordlander P.; Aizpurua J.; Ye J. Nanooptics of Plasmonic Nanomatryoshkas: Shrinking the Size of a Core-Shell Junction to Subnanometer. Nano Lett. 2015, 15, 6419–6428. 10.1021/acs.nanolett.5b02931. [DOI] [PubMed] [Google Scholar]
- Zakett D.; Schoen A. E.; Cooks R. G.; Hemberger P. H. Laser-desorption mass spectrometry-mass spectrometry and the mechanism of desorption ionization. J. Am. Chem. Soc. 1981, 103, 1295–1297. 10.1021/ja00395a086. [DOI] [Google Scholar]
- McLuckey S. A.; Schoen A. E.; Cooks R. G. Silver ion affinities of alcohols as ordered by mass spectrometry-mass spectrometry. J. Am. Chem. Soc. 1982, 104, 848–850. 10.1021/ja00367a035. [DOI] [Google Scholar]
- Grace L. I.; Abo-Riziq A.; deVries M. S. An in situ silver cationization method for hydrocarbon mass spectrometry. J. Am. Soc. Mass Spectrom. 2005, 16, 437–440. 10.1016/j.jasms.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Jackson A. U.; Shum T.; Sokol E.; Dill A.; Cooks R. G. Enhanced detection of olefins using ambient ionization mass spectrometry: Ag+ adducts of biologically relevant alkenes. Anal. Bioanal. Chem. 2011, 399, 367–376. 10.1007/s00216-010-4349-5. [DOI] [PubMed] [Google Scholar]
- Nicolardi S.; van der Burgt Y. E. M.; Codee J. D. C.; Wuhrer M.; Hokke C. H.; Chiodo F. Structural Characterization of Biofunctionalized Gold Nanoparticles by Ultrahigh-Resolution Mass Spectrometry. ACS Nano 2017, 11, 8257–8264. 10.1021/acsnano.7b03402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z.; de Paiva R. E. F.; Nelson K.; Farrell N. P. Diversity in Gold Finger Structure Elucidated by Traveling-Wave Ion Mobility Mass Spectrometry. Angew. Chem., Int. Ed. 2017, 56, 4464–4467. 10.1002/anie.201612494. [DOI] [PubMed] [Google Scholar]
- Nicoletti O.; de la Pena F.; Leary R. K.; Holland D. J.; Ducati C.; Midgley P. A. Three-dimensional imaging of localized surface plasmon resonances of metal nanoparticles. Nature 2013, 502, 80–84. 10.1038/nature12469. [DOI] [PubMed] [Google Scholar]
- Li X.; Zhu J.; Wei B. Hybrid nanostructures of metal/two-dimensional nanomaterials for plasmon-enhanced applications. Chem. Soc. Rev. 2016, 45, 3145–3187. 10.1039/C6CS00195E. [DOI] [PubMed] [Google Scholar]
- de la Rica R.; Stevens M. M. Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye. Nat. Nanotechnol. 2012, 7, 821–824. 10.1038/nnano.2012.186. [DOI] [PubMed] [Google Scholar]
- Melo S. A.; Luecke L. B.; Kahlert C.; Fernandez A. F.; Gammon S. T.; Kaye J.; LeBleu V. S.; Mittendorf E. A.; Weitz J.; Rahbari N.; Reissfelder C.; Pilarsky C.; Fraga M. F.; Piwnica-Worms D.; Kalluri R. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A.; Costa-Silva B.; Shen T. L.; Rodrigues G.; Hashimoto A.; Tesic Mark M.; Molina H.; Kohsaka S.; Di Giannatale A.; Ceder S.; Singh S.; Williams C.; Soplop N.; Uryu K.; Pharmer L.; King T.; Bojmar L.; Davies A. E.; Ararso Y.; Zhang T.; Zhang H.; Hernandez J.; Weiss J. M.; Dumont-Cole V. D.; Kramer K.; Wexler L. H.; Narendran A.; Schwartz G. K.; Healey J. H.; Sandstrom P.; Labori K. J.; Kure E. H.; Grandgenett P. M.; Hollingsworth M. A.; de Sousa M.; Kaur S.; Jain M.; Mallya K.; Batra S. K.; Jarnagin W. R.; Brady M. S.; Fodstad O.; Muller V.; Pantel K.; Minn A. J.; Bissell M. J.; Garcia B. A.; Kang Y.; Rajasekhar V. K.; Ghajar C. M.; Matei I.; Peinado H.; Bromberg J.; Lyden D. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au Yeung C. L.; Co N.-N.; Tsuruga T.; Yeung T.-L.; Kwan S.-Y.; Leung C. S.; Li Y.; Lu E. S.; Kwan K.; Wong K.-K.; Schmandt R.; Lu K. H.; Mok S. C. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat. Commun. 2016, 7, 11150. 10.1038/ncomms11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S.; Park J.; Pathania D.; Castro C. M.; Weissleder R.; Lee H. Integrated Magneto-Electrochemical Sensor for Exosome Analysis. ACS Nano 2016, 10, 1802–1809. 10.1021/acsnano.5b07584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y.; Ren Y.; Hu X.; Mu J.; Samykutty A.; Zhuang X.; Deng Z.; Kumar A.; Zhang L.; Merchant M. L.; Yan J.; Miller D. M.; Zhang H.-G. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat. Commun. 2017, 8, 14448. 10.1038/ncomms14448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E. C.; Pasikanti K. K.; Nicholson J. K. Global urinary metabolic profiling procedures using gas chromatography-mass spectrometry. Nat. Protoc. 2011, 6, 1483–1499. 10.1038/nprot.2011.375. [DOI] [PubMed] [Google Scholar]
- Kim N. H.; Cha Y. H.; Lee J.; Lee S. H.; Yang J. H.; Yun J. S.; Cho E. S.; Zhang X.; Nam M.; Kim N.; Yuk Y. S.; Cha S. Y.; Lee Y.; Ryu J. K.; Park S.; Cheong J. H.; Kang S. W.; Kim S. Y.; Hwang G. S.; Yook J. I.; Kim H. S. Snail reprograms glucose metabolism by repressing phosphofructokinase PFKP allowing cancer cell survival under metabolic stress. Nat. Commun. 2017, 8, 14374. 10.1038/ncomms14374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A. G.; Bouchet S.; Tolu J.; Bjorn E.; Mateos-Rivera A.; Bertilsson S. Molecular composition of organic matter controls methylmercury formation in boreal lakes. Nat. Commun. 2017, 8, 14255. 10.1038/ncomms14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preoperative chemotherapy for non-small-cell lung cancer: a systematic review and meta-analysis of individual participant data. Lancet 2014, 383, 1561–1571. 10.1016/S0140-6736(13)62159-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart B.; Wild C. P.. World cancer report; WHO: 2014. [Google Scholar]
- Gridelli C.; Rossi A.; Carbone D. P.; Guarize J.; Karachaliou N.; Mok T.; Petrella F.; Spaggiari L.; Rosell R. Non-small-cell lung cancer. Nat. Rev. Dis. Primers 2015, 1, 15048. 10.1038/nrdp.2015.48. [DOI] [PubMed] [Google Scholar]
- Goldstraw P.; Crowley J.; Chansky K.; Giroux D. J.; Groome P. A.; Rami-Porta R.; Postmus P. E.; Rusch V.; Sobin L. The IASLC Lung Cancer Staging Project: Proposals for the Revision of the TNM Stage Groupings in the Forthcoming (Seventh) Edition of the TNM Classification of Malignant Tumours. J. Thorac. Oncol. 2007, 2, 706–714. 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- Goldstraw P.; Ball D.; Jett J. R.; Le Chevalier T.; Lim E.; Nicholson A. G.; Shepherd F. A. Non-small-cell lung cancer. Lancet 2011, 378, 1727–1740. 10.1016/S0140-6736(10)62101-0. [DOI] [PubMed] [Google Scholar]
- Arya S. K.; Bhansali S. Lung cancer and its early detection using biomarker-based biosensors. Chem. Rev. 2011, 111, 6783–6809. 10.1021/cr100420s. [DOI] [PubMed] [Google Scholar]
- Vashist S. K.; Luppa P. B.; Yeo L. Y.; Ozcan A.; Luong J. H. T. Emerging Technologies for Next-Generation Point-of-Care Testing. Trends Biotechnol. 2015, 33, 692–705. 10.1016/j.tibtech.2015.09.001. [DOI] [PubMed] [Google Scholar]
- Vashist S. K.; Luong J. H. T. Trends in in vitro diagnostics and mobile healthcare. Biotechnol. Adv. 2016, 34, 137–138. 10.1016/j.biotechadv.2016.03.007. [DOI] [PubMed] [Google Scholar]
- Sun X.; Wan J.; Qian K. Designed Microdevices for In Vitro Diagnostics. Small Methods 2017, 1, 1700196. 10.1002/smtd.201700196. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.