Abstract
Background and objective
Current surveillance strategies for colorectal cancer following polypectomy are determined by endoscopic and histopathological factors. Such a distinction has been challenged. The present study was designed to identify molecular parameters in colonic polyps potentially defining new sub-groups at risk.
Methods
One hundred patients were enrolled in this multicentre study. Polyps biopsies underwent formalin-free processing (PAXgene, PreAnalytiX) and targeted next generation sequencing (38 genes (QIAGEN), NextSeq 500 platform (Illumina)). Genetic and histopathological analyses were done blinded to other data.
Results
In 100 patients, 224 polyps were removed. Significant associations of genetic alterations with endoscopic or histological polyp characteristics were observed for BRAF, KRAS, TCF7L2, FBXW7 and CTNNB1 mutations. Multivariate analysis revealed that polyps ≥ 10 mm have a significant higher relative risk for harbouring oncogene mutations (relative risk 3.467 (1.742–6.933)). Adenomas and right-sided polyps are independent risk factors for CTNNB1 mutations (relative risk 18.559 (2.371–145.245) and 12.987 (1.637–100.00)).
Conclusions
Assessment of the mutational landscape of polyps can be integrated in the workflow of current colonoscopy practice. There are distinct genetic patterns related to polyp size and location. These results suffice to optimise individual risk calculation and may help to better define surveillance intervals.
Keywords: Colorectal polyps, polypectomy, surveillance, genetics, colorectal cancer
Key summary
What is already known about this subject?
At the moment, the focus in colorectal cancer (CRC) screening is on optical colonoscopy with conventional histopathological assessment of resected polyps. Based on current guidelines, the repeated colonoscopy interval depends on a no-risk, low-risk or high-risk situation. Recent studies brought a deeper molecular understanding of colorectal polyps that already demonstrates subtypes of adenoma with more or less aggressive behaviour. Nevertheless, molecular diagnostics of resected colonic polyps are not yet recommended in current guidelines. Correlating data between genetic alterations in resected polyps and the respective risk situations in the individual patients are lacking.
What are the new findings?
Significant associations of genetic alterations with endoscopic or histological polyp characteristics were observed for BRAF, KRAS, transcription-associated genes (TCF7L2 and/or FBXW7) and ß-Catenin (CTNNB1) mutations, already in the no-risk situation, where a repeated colonoscopy is recommended in 10 years. Multivariate analysis revealed distinct genetic patterns related to histopathology, size and location of polyps. Our data open the avenue for a molecular subtyping of resected colorectal polyps in CRC screening. We were able to show that assessment of the mutational landscape of resected polyps can easily be integrated in the workflow of current colonoscopy practice. These findings might serve as a valuable tool for individual risk calculation and thereby help to better define surveillance intervals after index colonoscopy.
Introduction
Colorectal cancer (CRC) ranks third in worldwide diagnosed cancers and represents the fourth leading cause of cancer-related deaths.1 At the moment, the focus in CRC screening is on optical colonoscopy and histopathological assessment of resected polyps. Such a strategy has proven to reduce the incidence and mortality.2,3 Therefore, primary population-based colonoscopy screening is recommended by several medical societies.4,5 Recommendations on surveillance after polypectomy are based on clinical and histopathological parameters and range from 6–12 months to 10 years.4,5 High-risk factors are polyp size of ≥10 mm that confers a 3% annual risk for CRC development, villous architecture that confers a risk of 17% or high-grade dysplasia that already has a risk of 37%.6–8 Such a distinction has also been questioned,9 particularly in consideration of (a) increasing knowledge of substantial differences for adenoma affiliation to the right or left colon10,11 and (b) the deepened molecular understanding that already demonstrates subtypes of adenoma with more or less aggressive behaviour.
The most frequently detected colon lesions are hyperplastic polyps (HP), considered to be benign leading to extended surveillance intervals after resection.12 However, pathologists may misclassify sessile serrated adenoma/polyps (SSA/P) as HP and minimum criteria necessary to establish a diagnosis of an SSA/P have not been set,13 resulting in a large interobserver variation.14 Recent sequencing studies revealed new insights into basic evolutional processes in conventional adenomas and SSA/P. Two major pathways are in focus,15 associated with either chromosomal or microsatellite instability. One of the first genetic events in colorectal carcinogenesis is the inactivation of the APC/β-catenin signalling pathway, followed by molecular alterations in KRAS and TP53. This pathway is the basis of the adenoma–carcinoma sequence. Significantly less CRCs develop out of the alternatively mutated pathway termed as serrated neoplasia pathway, representing an important contributor to CRC, and a disproportionate contributor to interval or post colonoscopy CRC, particularly in the proximal colon.16 The serrated pathway includes a variety of possible driver mutations and cause in a high-frequency microsatellite instability. Moreover, CpG island hypermethylation is frequently found that can further be linked to the CpG island methylator phenotype (CIMP) of CRC and a proximal colon localisation.17,18 BRAF mutations are a hallmark for CIMPs.18 Taken all these changes together, chromosomal or microsatellite instability expressed by detection of various genetic mutations might therefore be important to calculate the risk of CRC development.
The Genetic Biopsy for Prediction of Surveillance Intervals after Endoscopic Resection of Colonic Polyps (GENESIS) study was designed to further characterise colonic polyps by adding these new molecular stratification parameters to the current state of the art recommendations and thereby potentially set up new sub-groups at risk.
Patients and methods
Institutional review board and general information
The study was approved by the ethical review board of Ulm University (Ulm University, approval date: 07.07.2015, approval number: 128/15). Participation in the study was voluntary. Written, informed consent was obtained from each patient included in this study. The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in prior approval by the institution’s Human Research Committee. All authors had access to the study data and reviewed and approved the final manuscript.
Study design and patient selection
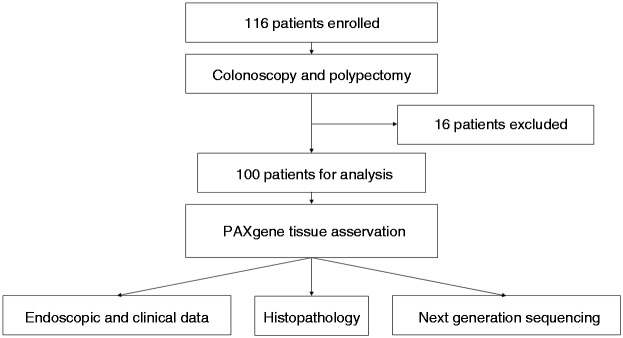
Patients within this multicentre study (www.clinicaltrials.gov number, NCT02595645) were recruited at two high-volume endoscopic centres in Germany: Department of Internal Medicine I, Ulm University and an outpatient clinic for gastroenterology in the Ulm region (Dornstadt). Data were collected prospectively and analysed retrospectively. All patients that underwent a routine colonoscopy, signed a written informed consent prior to colonoscopy and, in the case of having resectable polyps, were included in the study. Reasons for exclusion were chronic inflammatory bowel diseases and known malignancies. The recruitment period was July 2015–December 2015. One hundred and sixteen patients were enrolled, and of these 15 had to be excluded afterwards because of not having any resectable polyp and one patient was excluded because the resected polyp turned out to be a CRC (Figure 1). From each patient, we took up to six biopsies from up to six respective polyps. For final analysis, 224 polyp biopsies from 100 patients were available. Detailed patient characteristics are provided in Supplementary Material, Table S1.
Figure 1.
Flow chart of the Genetic Biopsy for Prediction of Surveillance Intervals after Endoscopic Resection of Colonic Polyps (GENESIS) study.
Sample collection
Before a detected polyp was resected, the endoscopist took a small extra biopsy of the respective polyp and stored it in a separate biopsy cassette, enabling storage of up to six biopsies from various respective polyps per patient. Each biopsy was numbered and several data, regarding localisation, diameter, morphology and removal technique were noted. Each specimen was fixed with the formalin-free PAXgene Tissue Fix (PreAnalytix GmbH, Hombrechtikon, Switzerland) for 2–4 h. Subsequently, PAXgene-fixed samples were transferred into PAXgene Tissue Stabilizer to stop the fixation process. Using a standard protocol, PAXgene-treated tissues were dehydrated and embedded in low-melting temperature paraffin in a formalin-free embedding processor. Sections from all samples were stained with haematoxylin and eosin (HE) for histopathological evaluation.
Histopathology
Evaluation of the slides, histopathological grading and staging was done at the Institute of Pathology, Medical University of Graz, Austria, by a histopathologist with special expertise in colonic pathology (CL). Each evaluation was done in accordance with the current World Health Organisation (WHO) classification system of tumours of the digestive system.19
Tissue DNA extraction
For DNA isolation, we took five sections (10 µm thick) from each PAXgene-fixed human tissue sample. The samples were deparaffinised by xylene and ethanol and deoxyribonucleic acid (DNA) was isolated, using the PAXgene Tissue DNA Kit (PreAnalytiX, Hombrechtikon, Switzerland).
Next generation sequencing and molecular analyses
Isolated DNA underwent initial quality assessment using QuantiMIZE kit (QIAGEN, Hilden, Germany). For molecular analyses the GeneRead workflow (QIAGEN, Hilden, Germany) was applied (GeneRead DNAseq Targeted Panel V2 for colorectal cancer, QIAGEN, Hilden, Germany), consisting of 38 most frequently mutated genes in CRC. The GeneRead workflow comprises targeted enrichment by multiplex polymerase chain reactions (PCR), pooling and library construction with the TruSeq Nano DNA HT Sample Prep Kit (Illumina FC-121-4003). An amount of 40 ng of DNA was used for target enrichment and 16 ng of the target-enriched DNA for the library preparation. A TapeStation (Agilent 4200 TapeStation system) was used for the qualification of PCR and library fragments as well as for fragment size determination. Library concentrations were determined based on qPCR results and fragment sizes. Paired end sequencing was performed on a NextSeq 500 sequencer (Illumina) running 2 × 150 bp chemistry version 2. Variant filtering was performed in two stages. Stage 1 of the filtering marked variants that fail some of the thresholds for variant calling. Filters derived from the recommendations in the GATK best practices document were used. Stage 2 of filtering removed single nucleotide polymorphism (SNP) and indels that did not pass minimum variant frequency and coverage thresholds. Data analysis including alignment to the reference genome hg19 and variant calling was carried out using QIAGEN’s online GeneRead Variant Analysis portal (http://ngsdataanalysis.sabiosciences.com/NGS2/). Mutations found by next generation sequencing were verified against the catalogue of somatic mutations in cancer (COSMIC) database by COSMIC-ID and categorised as pathogenic according to the Functional Analysis Through Hidden Markov Models v2.3 (FATHMM prediction score).20 Only pathogenic mutations according to FATHMM prediction score were included in the final evaluation.
Statistics
All statistical analyses were performed using IBM SPSS Statistic 23. The presence of various genetic factors was tested for statistically significant association with established endoscopic or histological factors using the chi-square test. A p-value < 0.05 was considered to be statistically significant. Multivariate logistic regression analysis was performed to look for independent parameters associated with respective mutations.
Results
Clinicopathological and endoscopic data
Clinical, endoscopic, histopathological and molecular data of the entire cohort are presented in Table 1. Based on international guideline recommendations4,5 we classified the study cohort into three different risk groups: 25 (25.0%) patients were classified as no-risk, 21 (21.0%) as low- (or intermediate-) risk and 54 (54.0%) as high-risk. These recommendations are based on the histology, size and number of resected polyps and have been coined to determinate surveillance intervals following polypectomy.
Table 1.
Patient characteristics of the Genetic Biopsy for Prediction of Surveillance Intervals after Endoscopic Resection of Colonic Polyps (GENESIS) study.
| Variable | All (%) |
|---|---|
| Clinical data | |
| Number of patients | 100 (100) |
| Gender | |
| Female | 50 (50) |
| Male | 50 (50) |
| Average age (years) | 62.9 |
| Risk-group | |
| No-risk | 25 (25) |
| Low-risk | 21 (21) |
| High-risk | 54 (54) |
| Endoscopic data | |
| Number of polyps | 224 (100) |
| NICE classification | |
| NICE type I | 146 (65.2) |
| NICE type II | 63 (28.1) |
| NICE type III | 5 (2.2) |
| No data | 10 (4.5) |
| Localization | |
| Localization left colon | 90 (40.2) |
| Localization right colon | 126 (56.3) |
| No data | 8 (3.6) |
| Polyp size | |
| <10 mm | 121 (54.0) |
| ≥10 mm | 71 (31.7) |
| No data | 32 (14.3) |
| Histopathological data | |
| Adenomatous yes/no | |
| Adenomatous polyps | 112 (50.0) |
| Non-adenomatous polyps | 110 (49.1) |
| No data | 2 (0.9) |
| Dysplasia no/low/high | |
| No dysplasia | 110 (49.1) |
| Hyperplastic polyp | 75 (33.5) |
| Hamartomatous polyp | 4 (1.8) |
| Leiomyoma | 1 (0.4) |
| Normal colonic mucosa | 24 (10.7) |
| Sessil serrated adenoma | 6 (2.7) |
| Low-grade dysplasia | 109 (48.7) |
| Tubular adenoma | 106 (47.3) |
| Tubulo-villous adenoma | 3 (1.3) |
| High-grade dysplasia | 3 (1.3) |
| Tubular adenoma | 3 (1.3) |
| No data | 2 (0.9) |
| Molecular data (mutations) | |
| Oncogene | |
| NRAS | 1 (0.4) |
| BRAF | 44 (19.6) |
| PIK3CA | 5 (2.2) |
| KRAS | 29 (12.9) |
| Cell-cycle | |
| MSH2 | 2 (0.9) |
| MLH1 | 128 (57.1) |
| ATM | 85 (37.9) |
| GPC6 | 69 (30.8) |
| ERBB2 | 1 (0.4) |
| CDC27 | 1 (0.4) |
| EP300 | 2 (0.9) |
| Tumor suppressor | |
| APC | 8 (3.6) |
| TP53 | 3 (1.3) |
| DCC | 2 (0.9) |
| Cytostructure | |
| MAP7 | 5 (2.2) |
| PTPN12 | 183 (81.7) |
| DMD | 8 (3.6) |
| Transcription | |
| TCF7L2 | 2 (0.9) |
| FBXW7 | 3 (1.3) |
| CTNNB1 (ß-catenin) | 19 (8.5) |
NICE: NBI-International-Colorectal-Endoscopic.
Molecular data
Frequency of respective mutations related to polyp characteristics
The most relevant findings of the respective mutations are provided in Table 1 and Table 2, detailed molecular data amongst all analysed genes are listed in Supplementary Material, Table S1. Amongst the 38 analysed genes, 20 genes showed alterations in the analysed polyps, in the remaining 18 genes no relevant changes could be seen. Findings were categorised by the respective gene according to its biological function: oncogenes, cell-cycle associated genes, tumour suppressor genes, cytostructure-associated genes and transcription-associated genes (Table 1). Moreover, CTNNB1 was analysed. Further analysis showed significant correlations between the respective molecular landscape, polyp histology, polyp localisation within the colon and polyp size. For BRAF mutations, a significant correlation between polyp histology and the occurrence of a distinct BRAF mutation (V600E) could be observed (adenomas vs. non-adenomas: 1.8% vs 39.1%, p < 0.001). Moreover, a trend to more BRAF mutations in right sided and larger polyps (≥10 mm) occurred, although not significant (polyp localisation right vs left 23.8% vs 15.6%, p=n.s.; polyp size < 10 vs ≥ 10 mm 19.0% vs 25.4%, p = n.s.). KRAS mutations were significantly associated with adenomatous polyps in the left colon and larger size (adenomas vs non-adenomas: 17.9% vs 8.2%, p < 0.05; polyp localisation right vs left 7.9% vs 20.0%, p < 0.01; polyp size < 10 vs ≥ 10 mm 7.4% vs 19.7%, p < 0.05). Detection of transcription-associated genes (FBXW7 and/or TCF7L2) was significantly associated with larger polyps ≥ 10 mm (0.8% vs 7.0%, p < 0.05). Mutations in CTNNB1 (ß-catenin) linked to activation of the Wnt-signaling pathway were significantly associated to adenomatous polyps in the right colon (adenomas vs non-adenomas: 16.1% vs 0.9%, p < 0.001; polyp localisation right vs left 13.5% vs 1.1%, p < 0.001). No significant differences were observed with respect to polyp histology, polyp localisation or size and detection of tumour suppressor genes (APC, TP53 and DCC), cite-structure associated genes (MAP7, PTPN12 and DMD) or cell-cycle associated genes (MSH2, MLH1, ATM, GPC6, ERBB2, CDC27 and EP300).
Table 2.
Frequency of respective mutations related to polyp characteristics.
| Polyp histology adenomas/non-adenomas (n = 112 vs n = 110) | Polyp location right/left (n = 126 vs n = 90) | Polyp size <10/≥10 mm (n = 121 vs n = 71) | |
|---|---|---|---|
| Oncogene | 22.3%/49.1% | 34.1%/36.7% | 27.3%/50.7%a |
| NRAS | 0.9%/0.0% | 0.0%/0.8% | 0.0%/1.4% |
| BRAF | 1.8%/39.1%a | 23.8%/15.6% | 19.0%/25.4% |
| PIK3CA | 1.8%/3.6% | 2.2%/1.6% | 1.7%/4.2% |
| KRAS | 17.9%/8.2%b | 7.9%/20%c | 7.4%/19.7%b |
| Cell-cycle | 83.0%/78.2% | 78.6%/81.1% | 81.8%/87.3% |
| MSH2 | 1.8%/0.0% | 0.0%/1.6% | 0.0%/2.8% |
| MLH1 | 57.1%/57.3% | 60.3%/51.1% | 55.4%/63.4% |
| ATM | 37.5%/39.1% | 35.7%/43.3% | 39.7%/45.1% |
| GPC6 | 33.9%/28.2% | 28.9%/33.3% | 37.2%/28.2% |
| ERBB2 | 0.9%/0.0% | 0.8%/0.0% | 0.0%/1.4% |
| CDC27 | 0.9%/0.0% | 0.0%/1.1% | 0.0%/1.4% |
| EP300 | 0.9%/0.9% | 0.8%/1.1% | 0.0%/2.8% |
| Tumor suppressor | 12.5%/6.4% | 12.7%/5.6% | 9.1%/11.3% |
| APC | 5.4%/1.8% | 4.8%/2.2% | 3.3%/4.2% |
| TP53 | 2.7%/0.0% | 1.6%/1.1% | 0.0%/4.2% |
| DCC | 0.0%/1.8% | 1.6%/0.0% | 0.8%/0.0% |
| Cyto-structure | 78.6%/86.4% | 80.2%/86.7% | 80.2%/84.5% |
| MAP7 | 0.9%/3.6% | 2.4%/1.1% | 2.5%/2.8% |
| PTPN12 | 77.7%/85.5% | 78.6%/86.7% | 79.3%/83.1% |
| DMD | 5.4%/1.8% | 4.8%/2.2% | 0.0%/0.0% |
| Transcription | 4.5%/0.9% | 1.6%/2.4% | 0.8%/7.0%b |
| TCF7F2 | 1.8%/0.0% | 0.0%/2.2% | 0.0%/2.8% |
| FBXW7 | 2.7%/0.0% | 0.8%/2.2% | 0.8%/2.8% |
| CTNNB1 (ß-catenin) | 16.1%/0.9%a | 13.5%/1.1%a | 10.7%/7.0% |
p < 0.001; bp < 0.05; cp < 0.01; chi-squared method. Univariate analysis.
Association of risk factors (polyp characteristics) with respective mutations
Large polyps (≥10 mm) have a significant higher relative risk (RR) to carry any oncogene (KRAS and/or BRAF) mutation (RR 3.467; confidence interval (CI) 1.742–6.933), compared to smaller ones. Moreover, adenomas and also right-sided polyps are independent risk factors for CTNNB1 mutations (adenomas’ RR for CTNNB1 mutations (95% CI): 18.559 (2.371–145.245)); right-sided polyps’ RR for CTNNB1 mutations (95% CI): 12.987 (1.637–100.00)). For transcription-associated genes, no significant polyp related and independent risk factors could be identified by multivariate analysis (Table 3). Of interest, even small diminutive polyps (>5 mm) of different histology harboured oncogenic mutations (Figure 2).
Table 3.
Risk factors (polyp characteristics) and association with respective mutations. Multivariate analysis using logistic regression analysis.
| Adenomas RR (95% CI) | Right-sided polyps RR (95% CI) | Polyps ≥ 10 mm RR (95% CI) | |
|---|---|---|---|
| Any oncogene | 0.209 (0.101–0.414) | 0.695 (0.357–1.353) | 3.467 (1.742–6.933) |
| BRAF | 0.026 (0.006–0.111) | 1.575 (0.660–3.759) | 1.772 (0.741–4.239) |
| KRAS | 2.527 (0.914–6.988) | 0.301 (0.116–0.784) | 2.898 (1.123–7.478) |
| Any transcription factor | 3.660 (0.396–33.866) | 0.319 (0.054–1.898) | 8.631 (0.963–77.364) |
| CTNNB1 (ß-catenin) | 18.559 (2.371–145.245) | 12.987 (1.637–100.00) | 0.708 (0.219–2.286) |
CI: confidence interval; RR: relative risk.
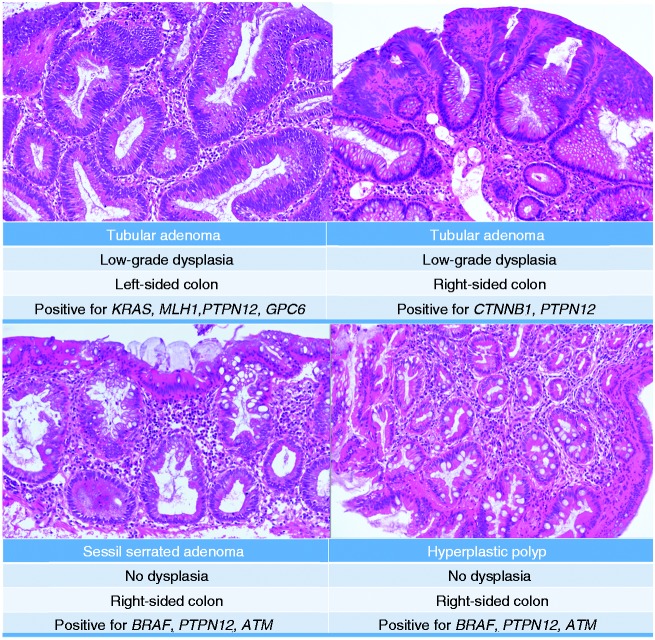
Figure 2.
Exemplary images of small diminutive polyps (<5 mm) resected by biopsy. Images are hematoxylin and eosin (HE)-stains of PAXgene-fixed specimens (see text). In addition to the histopathological results, location of polyps and mutated genes according to next generation sequencing are mentioned.
Mutational landscape of polyps in relation to cancer risk
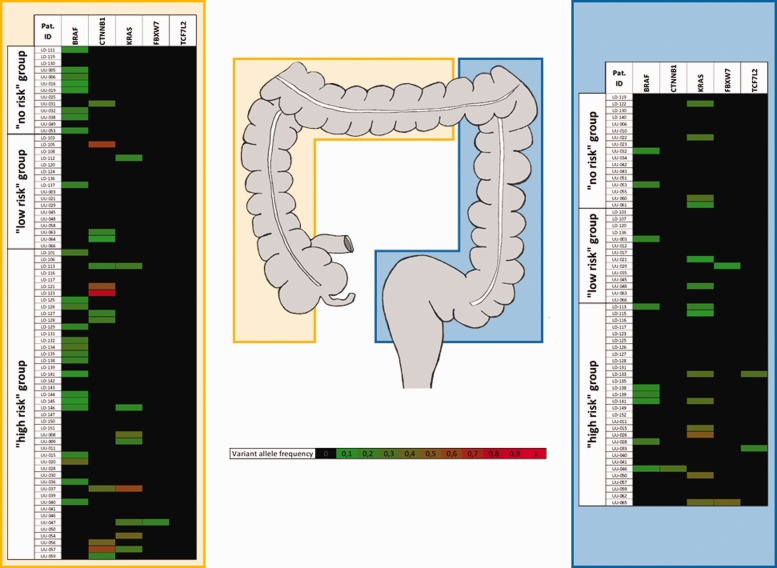
Fifty-four patients (54%) were classified as high-risk, 21 patients (21%) as low-risk and 25 patients (25%) were categorised as no-risk. Considering the respective mutational status for BRAF, KRAS, TCF7L2, FBXW7 and/or CTNNB1 (detected in any polyp per patient) in correlation to guideline-based risk group classification, we have found that in the no-risk group oncogenic mutations were found in 48.0% of cases, whereas 38.1% of low-risk patients and 68.6% of high-risk patients were positive for either BRAF, KRAS, TCF7L2, FBXW7 and/or CTNNB1 mutations. The variant allele frequency in relation to risk groups and polyp location (right vs left colon) is shown in Figure 3. As demonstrated, KRAS mutations are more frequent in the left colon in contrast to CTNNB1 mutations that are almost exclusively found in patients with right sided polyps. Mutations in TCF7L2 were only found in high risk patients with polyps in the left colon. However, both transcription-associated genes (TCF7L2 and FBXW7) were detected in only few patients, in general.
Figure 3.
Patients with colonic polyps, classified as no-, low- or high-risk candidates, according to current guidelines4,5 are associated with distinct molecular patterns of oncogenic mutations, stratified according to colonic location of the respective polyp (yellow = right colon, blue = left colon).
Discussion
The aim of any surveillance strategy after polyp detection and resection is to reduce the incidence and mortality of CRC.4,21 Following the initial index colonoscopy, surveillance intervals for each patient must be specified. So far, clinical, endoscopic and histopathological data define the respective time line for each patient. Nevertheless, CRC in between two screening colonoscopies occur in about 3%.22 Previously reported data assume one out of 27 CRCs as interval CRC, which were more frequently located in the right colon.23 However, location of polyps so far has not been regarded important in common surveillance guidelines. Furthermore, a characterisation of colonic polyps in a more molecular field is not recommended yet. Nevertheless, a more detailed characterisation of polyps (and patients) could help to better identify high-risk patients/polyps in addition of established risk group classification.4,5
CRC develops from adenomas through a sequence of genetic events. The APC/Wnt/β-catenin pathway plays a major role in CRC carcinogenesis in both sporadic and hereditary CRC and APC mutations were reported to be found in approximately 30–70% of sporadic adenomas and sporadic CRCs.24 Within the GENESIS study, APC mutations were found in only 5.4% of adenomas, which is in contrast to these data. Nevertheless, a few studies have addressed a broader mutational landscape with subsequent prevalence of oncogene mutations in colorectal lesions.25–27 In particular, the role of KRAS oncogenic mutations in colorectal polyps remains unclear. As shown in Figure 3, hyperplastic polyps without any grade of dysplasia may already harbour these relevant oncogenic mutations, mainly in the left colon. In contrast, there was a trend that BRAF mutations were more often found in right-sided polyps, confirming previous data.28 As shown in Figure 2 and in more detail in Figure 3, even small hyperplastic polyps without any grade of dysplasia (no-risk patients) harbour oncogenic BRAF mutations. Previously published data reported, that BRAF mutation were strongly linked to HP and SSA.27 Nevertheless, BRAF mutations are too common in serrated polyps to be used as a marker of a high-risk situation. BRAF mutations are early events in the serrated pathway, and by itself not a driver mutation for progression to CRC. Therefore, BRAF mutations may be a good marker for determining that a CRC likely arose from the serrated pathway,29 but it is not a good marker by itself for predicting which polyps will go on to cancer. However, such an association needs further prove in long-term follow-up studies to clarify the real impact of BRAF mutations on carcinogenesis of CRC. In terms of distinguishing between SSA/P and HP in a clinical field, the results of a recently published study show, that the prevalence of SSA/Ps in diminutive hyperplastic-appearing polyps in the rectosigmoid is low (2.1–6%), which supports the safety and feasibility of a ‘do not resect’ policy for diminutive hyperplastic-appearing rectosigmoid polyps.30 Nevertheless, molecular markers for distinguishing both SSA/P and HP are still lacking.
Of interest, activating mutations for Wnt signaling pathway (CTNNB1) were found significantly associated with adenomas, as well as with right-sided polyps. As previously reported, ß-catenin expression is significantly associated with an early relapse after endoscopic polypectomy. Conclusively, CTNNB1 might be suitable as a risk marker after polypectomy, but also need further clarification. Moreover, we identified significant mutations in transcription-associated genes, such as TCF7L2 and FBXW7. It has been reported that TCF7L2 mutations are leading to increased CRC cell growth.31 Others recently mentioned that mutations in the FBXW7 gene were more common in younger patients (≤45 years) suffering from CRC. Hence, apart from oncogenes and ß-catenin expression, the presence of one of these transcription-associated genes might also imply a higher risk for interval lesions after polypectomy.
There are certain limitations of our study worth mentioning. First, we do not have follow-up data to evaluate whether the genetic changes found in this study have any influence on clinical outcome. Second, only representative polyp biopsies were analysed. A certain degree of intratumoral heterogeneity even in the benign situation cannot be excluded as described for example in CRC.32 That only biopsies were available for blinded histopathological assessment might also explain why the number of SSA/P was rather low (n = 6 in the present study). We were not able to identify reliable markers that might help to reliably distinguish HPs from SSA/Ps. Second, in particular in serrated adenomas methylation markers (e.g. CIMP)18 are considered to be important, but could not be addressed in this study. Last not least, not only changes in the molecular setup in colonic mucosa pave the way to CRC. Recent data also suggest shifts in the faecal microbiota being associated with adenomatous polyps33 and CRC.34 Finally, work over the last decade has proposed that epigenetic changes such as DNA methylation, histone modification of protein coding genes might also play an important role in CRC development.35 Hence, CRC develops through a series of events resulting in the individual risk for CRC formation.
In summary, we can conclude that we were able to show that assessment of the mutational landscape of resected polyps/poly biopsy can easily be integrated in the workflow of current colonoscopy practice. There are distinct genetic patterns related to size and location of polyps and the clinician can appreciate this additional information to better estimate a patient's individual risk. In line with that, identifying polyps with an altered genetic (high risk) profile that traditionally would be classified as low risk would result in an earlier surveillance colonoscopy to prevent advanced interval polyps. This might help prevent morbidity and mortality due to CRC. In contrast persons classically classified as high risk that do not harbour (high risk) genetic alterations can be monitored as the low risk group. To evaluate the robustness of genetic risk prediction, we will conduct a prospective, multicentre, non-randomised cohort study (GENESIS II).
Supplementary Material
Acknowledgements
For excellent technical assistance, the authors thank Uwe Oelmüller, Daniel Grölz and Tomasz Krenz from Qiagen, Hilden, Germany, the whole endoscopy team from Ulm University, Germany, and the Outpatient Clinic for Gastroenterology in Dornstadt, Germany, and also Magdalena Bienek-Ziolkowski and Rosina Sing from the Biobank, Department of Internal Medicine I, Ulm University, Germany. Thanks are also given to the tissue bank of the Technical University of Munich and its associated Medical School for the PAXgene tissue workflow. The following author contributions were made: concept and design of the study: AWB, AM; generation, collection, assembly, analysis and/or interpretation of data: AWB, DS, KR, LP, CL, LL, ND, KFB, JSH, MQ, AK, EZ, FO, TS, AM; drafting or revision of the manuscript: AWB, LP, AK, EZ, FO, TS, AM; approval of the final version of the manuscript: AM.
Declaration of conflicting interests
The authors declare no conflict of interest.
Ethics approval
This article reports a registered clinical trial: www.clinicaltrials.gov number, NCT02595645.
Funding
No third-party funding. Material support by QIAGEN, Hilden, Germany.
Informed consent
Participation to the study was voluntary. Written, informed consent was obtained from each patient included in this study.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136: E359–E386. [DOI] [PubMed] [Google Scholar]
- 2.Nishihara R, Wu K, Lochhead P, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med 2013; 369: 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkin WS, Edwards R, Kralj-Hans I, et al. Once-only flexible sigmoidoscopy screening in prevention of colorectal cancer: A multicentre randomised controlled trial. Lancet 2010; 375: 1624–1633. [DOI] [PubMed] [Google Scholar]
- 4.Hassan C, Quintero E, Dumonceau JM, et al. Post-polypectomy colonoscopy surveillance: European Society of Gastrointestinal Endoscopy (ESGE) guideline. Endoscopy 2013; 45: 842–851. [DOI] [PubMed] [Google Scholar]
- 5.Lieberman DA, Rex DK, Winawer SJ, et al. Guidelines for colonoscopy surveillance after screening and polypectomy: A consensus update by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology 2012; 143: 844–857. [DOI] [PubMed] [Google Scholar]
- 6.Atkin WS, Morson BC, Cuzick J. Long-term risk of colorectal cancer after excision of rectosigmoid adenomas. N Engl J Med 1992; 326: 658–662. [DOI] [PubMed] [Google Scholar]
- 7.Brenner H, Hoffmeister M, Stegmaier C, et al. Risk of progression of advanced adenomas to colorectal cancer by age and sex: Estimates based on 840,149 screening colonoscopies. Gut 2007; 56: 1585–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winawer SJ, Zauber AG, O'Brien MJ, et al. Randomized comparison of surveillance intervals after colonoscopic removal of newly diagnosed adenomatous polyps. The National Polyp Study Workgroup. The N Engl J Med 1993; 328: 901–906. [DOI] [PubMed] [Google Scholar]
- 9.Lorentzen JA, Grzyb K, De Angelis PM, et al. Oncogene mutations in colorectal polyps identified in the Norwegian Colorectal Cancer Prevention (NORCCAP) Screening Study. Clin Med Insights Pathol 2016; 9: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker K, Zhang Y, Jin C, et al. Proximal versus distal hyperplastic polyps of the colorectum: Different lesions or a biological spectrum? J Clin Pathol 2004; 57: 1089–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qumseya BJ, Coe S, Wallace MB. The effect of polyp location and patient gender on the presence of dysplasia in colonic polyps. Clin Transl Gastroenterol 2012; 3: e20–e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corley DA, Levin TR, Doubeni CA. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med 2014; 370: 2541–2541. [DOI] [PubMed] [Google Scholar]
- 13.Rex DK, Ahnen DJ, Baron JA, et al. Serrated lesions of the colorectum: Review and recommendations from an expert panel. Am J Gastroenterol 2012; 107: 1315–1329. quiz 4, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glatz K, Pritt B, Glatz D, et al. A multinational, Internet-based assessment of observer variability in the diagnosis of serrated colorectal polyps. Am J Clin Pathol 2007; 127: 938–945. [DOI] [PubMed] [Google Scholar]
- 15.Gibson JA, Odze RD. Pathology of premalignant colorectal neoplasia. Dig Endosc 2016; 28: 312–323. [DOI] [PubMed] [Google Scholar]
- 16.Kahi CJ. How does the serrated polyp pathway alter CRC screening and surveillance? Dig Dis Sci 2015; 60: 773–780. [DOI] [PubMed] [Google Scholar]
- 17.Fearon ER, Carethers JM. Molecular subtyping of colorectal cancer: Time to explore both intertumoral and intratumoral heterogeneity to evaluate patient outcome. Gastroenterology 2015; 148: 10–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 2006; 38: 787–793. [DOI] [PubMed] [Google Scholar]
- 19.Snover DC, Ahnen, DJ, Burt RW, et al. Serrated polyps of the colon and rectum and serrated polyposis. In: Bosman FT, Carneiro F, Hruban RH and Theise ND (eds) WHO classification of tumours of the digestive system. 4th ed. Lyon: IARC, 2010, pp.160–165.
- 20.Shihab HA, Gough J, Cooper DN, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat 2013; 34: 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atkin WS, Valori R, Kuipers EJ, et al. European guidelines for quality assurance in colorectal cancer screening and diagnosis. First edition–colonoscopic surveillance following adenoma removal. Endoscopy 2012; 44: S151–S163. [DOI] [PubMed] [Google Scholar]
- 22.le Clercq CM, Bouwens MW, Rondagh EJ, et al. Postcolonoscopy colorectal cancers are preventable: A population-based study. Gut 2014; 63: 957–963. [DOI] [PubMed] [Google Scholar]
- 23.Singh S, Singh PP, Murad MH, et al. Prevalence, risk factors, and outcomes of interval colorectal cancers: A systematic review and meta-analysis. Am J Gastroenterol 2014; 109: 1375–1389. [DOI] [PubMed] [Google Scholar]
- 24.De Filippo C, Luceri C, Caderni G, et al. Mutations of the APC gene in human sporadic colorectal cancers. Scand J Gastroenterol 2002; 37: 1048–1053. [DOI] [PubMed] [Google Scholar]
- 25.Yan HH, Lai JC, Ho SL, et al. RNF43 germline and somatic mutation in serrated neoplasia pathway and its association with BRAF mutation. Gut Epub ahead of print 2016. doi: 10.1136/gutjnl-2016-311849. [DOI] [PubMed] [Google Scholar]
- 26.Sakai E, Fukuyo M, Ohata K, et al. Genetic and epigenetic aberrations occurring in colorectal tumors associated with serrated pathway. Int J Cancer 2016; 138: 1634–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rau TT, Atreya R, Aust D, et al. Inflammatory response in serrated precursor lesions of the colon classified according to WHO entities, clinical parameters and phenotype-genotype correlation. J Pathol Clin Res 2016; 2: 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawada T, Yamamoto E, Yamano HO, et al. Assessment of epigenetic alterations in early colorectal lesions containing BRAF mutations. Oncotarget 2016; 7: 35106–35118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan TL, Zhao W, Leung SY, et al. BRAF and KRAS mutations in colorectal hyperplastic polyps and serrated adenomas. Cancer Res 2003; 63: 4878–4881. [PubMed] [Google Scholar]
- 30.Ponugoti P, Lin J, Odze R, et al. Prevalence of sessile serrated adenoma/polyp in hyperplastic-appearing diminutive rectosigmoid polyps. Gastrointest Endosc 2017; 85: 622–627. [DOI] [PubMed] [Google Scholar]
- 31.Tang W, Dodge M, Gundapaneni D, et al. A genome-wide RNAi screen for Wnt/beta-catenin pathway components identifies unexpected roles for TCF transcription factors in cancer. Proc Natl Acad Sci U S A 2008; 105: 9697–9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horst D, Chen J, Morikawa T, et al. Differential WNT activity in colorectal cancer confers limited tumorigenic potential and is regulated by MAPK signaling. Cancer Res 2012; 72: 1547–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hale VL, Chen J, Johnson S, et al. Shifts in the fecal microbiota associated with adenomatous polyps. Cancer Epidemiol Biomarkers Prev 2017; 26: 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peuker K, Muff S, Wang J, et al. Epithelial calcineurin controls microbiota-dependent intestinal tumor development. Nat Med 2016; 22: 506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao Q, Chen L, Liu J, et al. Comprehensive analysis of epigenetic pattern of long noncoding RNA loci in colorectal cancer. Gene 2016; 595: 9–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.