Abstract
Background: Insulin resistance disrupts metabolic processes and leads to various chronic disease states such as diabetes and metabolic syndrome (MetS). However, the mechanism linking insulin resistance with cardiometabolic disease pathophysiology is still unclear. One possibility may be through circulating microRNAs (c-miRs), which can alter gene expression in target tissues. Our goal was to assess the relationship of c-miRs with insulin sensitivity, as measured by the gold standard, hyperinsulinemic-euglycemic clamp technique.
Methods: Eighty-one nondiabetic, sedentary, and weight-stable patients across a wide range of insulin sensitivities were studied. Measurements were taken for blood pressure, anthropometric data, fasting glucose and lipids, and insulin sensitivity measured by clamp. After an initial screening array to identify candidate miRs in plasma, all samples were assessed for relationships between these c-miRs and insulin sensitivity, as well as associated metabolic factors.
Results: miR-16 and miR-107 were positively associated with insulin sensitivity (R2 = 0.09, P = 0.0074 and R2 = 0.08, P = 0.0417, respectively) and remained so after adjustment with body mass index (BMI). After adjusting for BMI, miR-33, -150, and -222 were additionally found to be related to insulin sensitivity. Regarding metabolic risk factors, miR-16 was associated with waist circumference (r = −0.25), triglycerides (r = −0.28), and high-density lipoprotein (r = 0.22), while miR-33 was inversely associated with systolic blood pressure (r = −0.29). No significant relationships were found between any candidate c-miRs and BMI, diastolic blood pressure, or fasting glucose.
Conclusions: Our results show that relative levels of circulating miR-16, -107, -33, -150, and -222 are associated with insulin sensitivity and metabolic risk factors, and suggest that multiple miRs may act in concert to produce insulin resistance and the clustering of associated traits that comprise the MetS. Therefore, miRs may have potential as novel therapeutic targets or agents in cardiometabolic disease.
Keywords: : plasma, circulating, microRNA, insulin sensitivity, insulin resistance, metabolism
Introduction
Insulin resistance is one of the major contributors to the development of Type 2 diabetes mellitus (T2DM), as well as other metabolic disease states, such as metabolic syndrome (MetS) and cardiovascular disease.1 The development of insulin resistance precedes diagnosis of these diseases, and its mechanism is still unclear. However, interventions that increase insulin sensitivity in patients with prediabetes or MetS, such as exercise, weight loss, or thiazolidinedione medications, have been shown to prevent or delay progression to T2DM and improve metabolic risk factors. Thus, understanding the pathophysiology underlying insulin sensitivity could inform the development of better therapies for patients with cardiometabolic disease.
The role of microRNA (miRNA; miR) in the pathophysiology of diabetes and cardiovascular disease has been of emerging interest in recent years, but has not been fully explored. miRNAs are short, noncoding RNA strands (about 22 nucleotides long) that base-pair with mRNA to posttranscriptionally modify gene expression. They are found in all tissues, but recent studies have suggested that they can also be secreted from cells and enter the bloodstream.
Multiple mechanisms are operative to protect circulating miRs against degradation, including packaging into microvesicles,2 association with proteins like Argonaute2 (AGO2) or nucleophosmin 1 (NPM1), or binding to high-density lipoprotein (HDL).3 Studies have also shown that secreted miR can then specifically influence gene expression in remote recipient cells2,4 and that levels of certain circulating miR species are associated with various conditions, such as cancer,5,6 cardiovascular disease,7 and diabetes.8,9
In animal models, manipulation of miRs in tissues has been shown to affect whole-body metabolism and insulin sensitivity.10,11 However, much less is known about the relationship between levels of circulating miRs (c-miRs) and insulin sensitivity. In humans, most studies used screening methodology to profile patterns of c-miR differences between metabolic disease and disease-free states (i.e., patients with diabetes, MetS, or polycystic ovary syndrome vs. controls),9,12–16 rather than specifically looking at c-miR in relation to insulin sensitivity. This is an important distinction because decreases in insulin sensitivity precede development of these disease states. In addition, studies that did assess insulin sensitivity often used the homeostatic model assessment for insulin resistance (IR)17–19 instead of the gold standard, hyperinsulinemic-euglycemic clamp technique.
Thus, to better elucidate the relationship between circulating miRs and insulin sensitivity prior to the development of metabolic disease states, we first used a screening approach to identify candidate c-miRs in nondiabetic, human plasma samples, then individually evaluated each of the miRs for association with insulin sensitivity, as measured by clamp. The goal of this study was to identify miRs that may be related to human insulin sensitivity and associated MetS risk factors.
Materials and Methods
Participants and sample collection/processing
Participants were sequentially recruited for metabolic characterization at the University of Alabama at Birmingham's (UAB) Clinical Research Unit from 2004 to 2011. Participants had body mass index (BMI) >21 kg/m2, did not engage in regular exercise, and were weight stable for at least 3 months prior (±3% of body weight). Participants were excluded if they had BMI >50 kg/m2, evidence of T2DM, cardiovascular, renal, thyroid, or hepatic disease, or if they were taking any medications that could affect body composition, lipid, or carbohydrate metabolism. All protocols were approved by the UAB Institutional Review Board, and written consent was obtained from all participants.
Clinical measurements
Anthropometric measures of height, weight, and waist circumference (WC) were recorded, and standardized blood pressure measurements were obtained. Serum and plasma (in EDTA tubes) were obtained after an overnight fast and stored at −80°C until use. Lipids, glucose, and insulin levels were determined using a conventional lipid panel colorimetric assay (Stanbio Laboratory, Boerne, TX), glucose analyzer (YSI 2300; Yellow Springs Instruments, Yellow Springs, OH), and immunofluorescence (TOSOH A1A-II analyzer; TOSOH Corp., South San Francisco, CA), respectively.
Insulin sensitivity, assessed as the glucose disposal rate (GDR), was measured using the hyperinsulinemic-euglycemic clamp technique as described previously.20,21 Briefly, participants were given a primed-continuous infusion of insulin (Humulin; Eli Lilly, Indianapolis, IN) at 200 mU/m2/min to maximally stimulate glucose uptake and suppress hepatic glucose production, and serum glucose was clamped at 5.0 mmol/L for at least 3 hr. Maximal glucose uptake was determined as the mean glucose infusion rate over the last three 20-min intervals, and the GDR was calculated based on the glucose infusion rate after adjustments for glucose pool size and normalization per kilogram of lean body mass as assessed by dual-energy X-ray absorptiometry.
MiR microarray screening
For screening purposes, 15 participants with the highest GDR (insulin sensitive [IS]) and 15 with the lowest GDR (IR) were chosen to assess for differential c-miR expression based on insulin sensitivity. Total RNA (including miRs) was extracted from 200 μL of plasma samples of these individuals using the miRNeasy Serum/Plasma Kit (QIAGEN) according to the manufacturer's protocol. Within each group, participants were divided into three pooled samples of five individuals each, which were then reverse-transcribed with the miScript II RT Kit (QIAGEN) into cDNA using the miScript HiSpec Buffer for mature miR profiling. miRNA microarrays were performed with diluted pooled samples using a quantitative, real-time polymerase chain reaction (qrt-PCR; StepOnePlus Real-Time PCR System, Applied Biosystems) on the Human Diabetes miScript miRNA PCR Array (QIAGEN; MIHS-115Z).
Analysis of the miR microarray data was performed with miScript miRNA PCR Array Data Analysis software (SABiosciences/QIAGEN). Only one invariant miR (miR-490) emerged from this analysis and was found to be stably expressed across subsequent individual assays, so it was used as the reference gene for the remainder of the study. For screening purposes, samples were determined to be candidate miRs if they had a greater than fourfold difference between the two groups, P value <0.30. Additional miRs that did not meet criteria and/or were not included in the array were added as candidate genes if they had the potential to be related to insulin sensitivity based on previous literature (see Results section for details).
Candidate miR evaluation
A subset of 81 participants were chosen for this study based on their GDR, (with an effort to include an equal number of whites and blacks, males and females), the availability of the plasma sample, and the absence of overt hemolysis. For validation, levels of candidate miRs were individually assessed in each of the 81 plasma samples. RNA extraction and reverse transcription to cDNA were the same as described above. cDNA samples were diluted 1:10 for determining relative expression using qrt-PCR with the miScript SYBR Green PCR Kit (QIAGEN) in duplicate. Primers used for evaluation of individual mature miR expressions were as follows (All from QIAGEN, Cat. No. 218300, miScript Primer Assays): mi-33, mi-490, mi-150, RNU6, mi-16, mi-140, mi-133a, mi-199a, mi-218, mi-200a, SNORD61, mi-34a, mi-107, and mi-222. All data were normalized to miR-490 and subsequent analyses used–ΔCt values for relative expression.
Statistical analyses
Descriptive data are presented as mean ± standard deviation, with comparisons made through Student's t-test or nonparametric equivalents, as appropriate. miR-107 and miR-133a were transformed by adding a constant and square-rooting the values, and triglycerides (TG), HDL, and fasting glucose were log-transformed for normality for subsequent analyses. Simple linear regressions were conducted between insulin sensitivity and relative expression levels of circulating candidate miR (-ΔCt), along with an adjusted multiple linear regression model to include BMI, with standardized β coefficients reported for the miR and BMI. Pearson's correlations were also determined between circulating miRs and relevant metabolic risk factors (measures of obesity, blood pressure, lipids, and fasting glucose). All statistical analyses were performed with the SAS 9.4 statistical software package (SAS Institute, Inc., Cary, NC).
Results
A total of 81 samples were used for this study, across a wide range of insulin sensitivity, as measured by the hyperinsulinemic-euglycemic clamp technique. The majority of patients were female (67.9%), and there was comparable representation of African Americans and European Americans (51.9% AA; 45.7% EA; 2.7% other). Patient characteristics are depicted in Table 1.
Table 1.
Patient Clinical Characteristics
| Age (years) | 39.0 ± 10.3 |
| Sex | 55 female, 26 male |
| Race | 1 Asian, 42 Black, 37 White, 1 Hispanic |
| Body mass index (kg/m2) | 31.4 ± 5.2 |
| WC (cm) | 97.4 ± 14.1 |
| Systolic blood pressure (mmHg) | 116.8 ± 14.1 |
| Diastolic blood pressure (mmHg) | 68.1 ± 9.5 |
| Triglycerides (mg/dL) | 110.0 ± 58.6 |
| HDL-cholesterol (mg/dL) | 50.0 ± 20.5 |
| GDR (mg/min/kg LBM) | 14.4 ± 4.7 |
| Fasting glucose (mg/dL) (n = 80) | 92.9 ± 10.9 |
| Fasting insulin (μU/mL) (n = 48) | 19.3 ± 12.6 |
| Total body fat (%) | 41.1 ± 10.0 |
| Total fat mass (kg) (n = 71) | 34.9 ± 11.7 |
Data are presented as mean ± standard deviation, or n if categorical (n = 81 unless otherwise noted). GDR is adjusted for LBM.
GDR, glucose disposal rate; LBM, lean body mass; WC, waist circumference; HDL, high-density lipoprotein.
MiRs and insulin sensitivity
Based on the screening array criteria above, five candidate miRs were chosen for further analysis from the microarray: miR-133a, -200a, -34a, SNORD61, and RNU6-2. Eight other candidate miRNAs, miR-150, -199a, -140, -16, -107, -222, -218, and -33, were additionally included for this study, as they have previously been shown to have differential expression in diabetic patients,10,22,23 and/or a potential mechanistic relationship to insulin sensitivity.24–29 Thus, a total of 13 candidate miRs were chosen for further analysis in each of the 81 samples. Four of these miRs were excluded from our final results (miR-218, -200a, SNORD61, and RNU6-2) due to having a majority of samples under the detectable limit (Ct values >35).
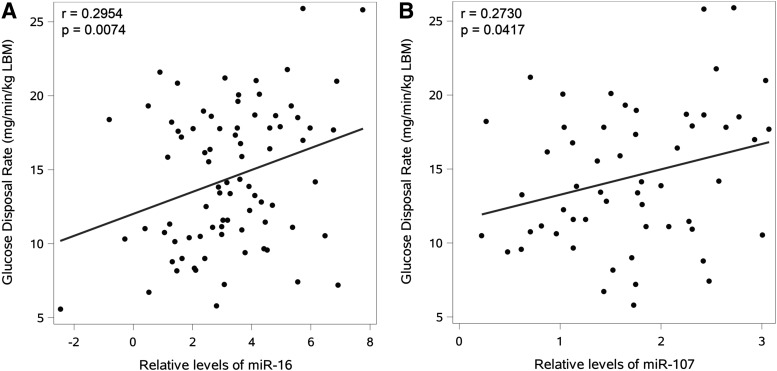
Simple linear regressions of candidate miRs with GDR (Table 2) showed that miR-16 and miR-107 were positively associated with insulin sensitivity (Fig. 1). After adjustment for BMI, miR-16 and -107 were still significantly associated with GDR, and miR-33, -150, and -222 were additionally found to be related to GDR. BMI was a significant covariate for all models involving these c-miRs (Table 2). No significant differences between gender were noted.
Table 2.
Linear Regressions of Candidate miRs with Insulin Sensitivity
| Unadjusted model | Adjusted model | ||||||
|---|---|---|---|---|---|---|---|
| miR | n | R2 | P | miR, partial β | BMI, partial β | Model, R2 | Model, P value |
| miR-16 | 81 | 0.0873 | 0.0074** | 0.25* | −0.39** | 0.2183 | <0.0001** |
| miR-33 | 52 | 0.0526 | 0.1018 | 0.26* | −0.36** | 0.1814 | 0.0028** |
| miR-34a | 44 | 0.0005 | 0.8851 | 0.05 | −0.34* | 0.0693 | 0.0865 |
| miR-107 | 56 | 0.0746 | 0.0417* | 0.32** | −0.43** | 0.2310 | 0.0004** |
| miR-133a | 73 | 0.0177 | 0.2623 | 0.13 | −0.39** | 0.1496 | 0.0013** |
| miR-140 | 66 | 0.0163 | 0.3072 | 0.12 | −0.41** | 0.1559 | 0.0018** |
| miR-150 | 81 | 0.0334 | 0.1025 | 0.21* | −0.43** | 0.1994 | <0.0001** |
| miR-199a | 67 | 0.0436 | 0.0898 | 0.22 | −0.40** | 0.1807 | 0.0006** |
| miR-222 | 81 | 0.0321 | 0.1097 | 0.20* | −0.43** | 0.1962 | <0.0001** |
Insulin sensitivity was measured with a hyperinsulinemic-euglycemic clamp technique for GDR adjusted for LBM. Unadjusted model: simple linear regression between candidate miRs and insulin sensitivity. Adjusted model: addition of BMI as a covariate, with standardized β estimates and adjusted R2 reported. miR-107 and miR-133a were transformed with a constant and square-rooted for normality.
Statistically significant values are highlighted in bold.
= P < 0.05.
= P < 0.01.
BMI, body mass index.
FIG. 1.
Relationship of miR-16 and miR-107 with insulin sensitivity. Both miR-16 (A) and miR-107 (B) were positively associated with insulin sensitivity, as measured by glucose disposal rate (GDR), adjusted for lean body mass (LBM). miR-107 was transformed with a constant and square-rooted for normality.
Candidate miRs and risk factors for metabolic disease
Candidate miRs were then tested for associations with common risk factors for cardiometabolic disease, including BMI, WC, systolic and diastolic blood pressure (SBP, DBP), circulating TG, HDL-cholesterol, and fasting glucose (Table 3). miR-16 was negatively associated with WC and TG and positively associated with HDL. miR-33 was negatively associated with SBP. No significant relationships were found between any candidate miRs with BMI, DBP, or fasting glucose. All of the candidate miRs (that were detectable) were found to be highly correlated with each other (Table 4).
Table 3.
Correlation of Candidate miRs with Metabolic Parameters Related to Insulin Sensitivity
| miR | n | BMI | WC | SBP | DBP | TG | HDL | Fasting glucose |
|---|---|---|---|---|---|---|---|---|
| miR-16 | 81 | −0.11 | −0.25* | −0.15 | −0.11 | −0.28* | 0.22* | 0.01a |
| miR-33 | 52 | 0.08 | −0.05 | −0.29* | −0.19 | −0.20 | 0.21 | 0.02 |
| miR-34a | 44 | 0.09 | 0.19 | 0.16 | 0.18 | −0.07 | 0.05 | 0.08 |
| miR-107 | 56 | 0.11 | −0.005 | −0.13 | −0.08 | −0.23 | 0.09 | −0.05 |
| miR-133a | 73 | −0.01 | −0.13 | −0.20 | −0.11 | −0.14 | 0.10 | 0.18a |
| miR-140 | 66 | −0.01 | −0.09 | −0.02 | 0.04 | −0.21 | 0.08 | 0.07a |
| miR-150 | 81 | 0.07 | −0.05 | −0.01 | 0.01 | −0.15 | 0.08 | 0.12a |
| miR-199a | 67 | 0.03 | −0.05 | −0.08 | −0.07 | −0.22 | 0.09 | 0.08a |
| miR-222 | 81 | 0.06 | −0.05 | −0.09 | −0.09 | −0.19 | 0.13 | 0.11a |
Pearson's correlation coefficients are reported for each metabolic parameter as individually assessed with each miRNA. Circulating triglycerides, HDL, and fasting glucose were log-transformed for normality, although this generally did not affect results, and miR-107 and miR-133a were transformed with a constant and square-rooted for normality.
Statistically significant values are highlighted in bold.
n–1.
P < 0.05.
BMI (kg/m2); WC (cm); SBP, systolic blood pressure (mmHg); DBP, diastolic blood pressure (mmHg); TG, triglycerides (mg/dL); HDL cholesterol (mg/dL).
Table 4.
Correlation of Relative Expression of miRs (−ΔCt) With Each Other
| miR-33 | miR-34a | miR-107 | miR-133a | miR-140 | miR-150 | miR-199a | miR-222 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | r | n | r | n | R | n | r | n | r | n | r | n | r | n | r | |
| miR-16 | 52 | 0.49* | 44 | 0.51* | 56 | 0.63** | 73 | 0.47** | 66 | 0.76** | 81 | 0.78** | 67 | 0.71** | 81 | 0.78** |
| miR-33 | - | - | 41 | 0.80** | 50 | 0.88** | 52 | 0.73** | 51 | 0.84** | 52 | 0.56** | 52 | 0.92** | 52 | 0.87** |
| miR-34a | - | - | 43 | 0.76** | 44 | 0.65** | 44 | 0.82** | 44 | 0.65** | 43 | 0.81** | 44 | 0.82** | ||
| miR-107 | - | - | 54 | 0.73** | 55 | 0.93** | 56 | 0.75** | 56 | 0.95** | 56 | 0.95** | ||||
| miR-133a | - | - | 62 | 0.76** | 73 | 0.61** | 62 | 0.76** | 73 | 0.71 | ||||||
| miR-140 | - | - | 66 | 0.82** | 64 | 0.96** | 66 | 0.97** | ||||||||
| miR-150 | - | - | 67 | 0.79** | 81 | 0.84** | ||||||||||
| miR-199a | - | - | 67 | 0.97** | ||||||||||||
| miR-222 | - | - | ||||||||||||||
As previously, miR-107 and miR-133a were transformed with a constant and square-rooted for normality.
P < 0.001.
P < 0.0001.
r ≥ 0.80 is represented in bold.
Discussion
Using miRNA microarrays, we screened pooled samples of human plasma from IS and IR participants to identify candidate miRNA species for further study. Thirteen specific miRNAs were identified for validation in a larger number of individual plasma samples from patients who had been metabolically characterized for their degree of insulin sensitivity and MetS traits. From these assays, we discovered that circulating levels of miR-16 and miR-107 were positively associated with insulin sensitivity, and after controlling for BMI, three other miRNAs (miR-150, -222, and -33) were also significantly associated with insulin sensitivity. miR-16 was also found to be variably related to traits that comprise the MetS, including TG levels, HDL cholesterol, and WC, and miR-33 was related to systolic blood pressure.
Our data showing a positive association of circulating miR-16 with insulin sensitivity in humans seem to correspond with observations regarding miR-16 and tissue-specific metabolism and insulin response in rodent models. Lee et al. showed that miR-16 levels were significantly decreased in insulin resistant skeletal muscle both in vitro and in vivo, and found that miR-16 helps control skeletal muscle insulin-stimulated protein synthesis through regulation of production of mTOR and p70S6K1, as well as increasing autophagy through regulation of Bcl-2 synthesis.30 miR-16 was also found to be significantly downregulated in insulin-sensitive tissues of high sucrose diet-induced insulin resistant mice, with concomitant increase in PHLPP expression (a protein involved with regulation of both inflammation and insulin resistance) and that overexpression of miR-16 in L6 myoblasts improved insulin sensitivity of the cells through upregulation of GLUT4 and MEF2A expression.27
In vitro, both high glucose and high insulin conditions resulted in reduced miR-16 levels in L6 cells,31 and high glucose conditions also result in increased Myc expression and decreased miR-16 expression in colon cancer cells, with subsequent upregulation of miR-16 target genes Myb and VEGFR2 and proliferation of cells.32
In terms of circulating miR-16, circulating adipocyte-derived exosomal miR-16 has been shown to decrease after gastric bypass surgery in humans, concomitant with improvements in insulin sensitivity,33 and colorectal cancer patients with higher blood glucose had significantly lower levels of serum miR-16.32 However, our results are a discrepant with one study based on Framingham Offspring and POOL cohorts that showed that miR-16 was positively associated with fasting insulin levels.19 Interestingly, miR-16 has also been implicated in the cross talk between insulin resistant skeletal muscle and pancreatic β cells through modulation of Ptch1.34 Of note, miR-16 has at least 27 target genes in silico that participate in the insulin signaling pathway, including insulin-like growth factor 1 receptor, and the insulin receptor itself.33 These data from the literature, taken together with our findings, suggest that miR-16 may be involved during the development of insulin resistance and the chronic inflammation underlying cardiometabolic disease.
It should also be noted that some studies have used miR-16 as a constitutive miRNA for data normalization to examine alterations in other circulating miRs.35 However, this has been refuted by other studies,36 and our data show that expression of miR-16 in plasma is variable and quantitatively related to metabolic traits. In fact, we found that miR-490 was the most stable miR across all samples, even more so than the spike-in control (data not shown). Thus, future studies might consider using miR-490 as a potential endogenous control for qrt-PCR studies on miR analyses conducted on human plasma samples.
Our data linking plasma miR-107 and insulin sensitivity in humans are consistent with the observation that circulating levels of miR-107 increased after 12 weeks of endurance training.37 However, our data are at odds with previous reports in preclinical model systems. miR-107 has been shown to be upregulated by glucose in a dose-dependent manner in pancreatic β cells in vitro,38 and it is also upregulated in the livers of obese mice.10 Injection of miR-107 into either the liver or fat pads in mice caused a rise in blood glucose and insulin levels, with decreased glucose tolerance and insulin sensitivity10,39 through the actions of caveolin-1. In fact, based on these and similar findings, AstraZeneca has developed RG-125 (AZD4076), an N-acetylgalactosamine (GalNAc)-conjugated anti-miR-103/107, to treat NASH in patients with T2DM or prediabetes, which is currently in Phase I clinical trials.40
miR-107 also seems to promote lipid accumulation in liver cells through targeting of fatty acid synthase41 and inhibition of mitochondrial β-oxidation by decreasing levels of hydroxyacyl-CoA dehydrogenase (HADHA).39 Interestingly, the latter was rescued by the endoplasmic reticulum (ER) stress inhibitor 4-BPA, and HADHA overexpression itself rescued miR-107-induced lipid accumulation. Given that this group also noticed the same glucose and insulin response with miR-107 injection, along with increased hepatic ER stress markers BiP and CHOP, miR-107 may be a link between oxidative stress and insulin resistance.
It is interesting to note that miR-107 may also promote angiogenesis in tissues under hypoxic conditions. One study showed that miR-107 is increased in the ischemic region of the brain after an ischemic stroke in rats and downregulated DICER1, which normally helps process precursor miR, leading to increased expression of vascular endothelial growth factor and angiogenesis.42 Meanwhile, a different study showed that circulating miR-107 was downregulated after ischemic stroke in rats.43 This compels the speculation that hypoxic stressors, such as under insulin resistant conditions, may also promote tissue uptake of circulating miR-107.
The combined data point to an interesting possibility to explain the discrepancy between the current human data and preclinical studies. Perhaps under insulin resistant/oxidative stress states, circulating miR-107 levels might decrease because they are being delivered to cells and tissues of need to compensate for impaired substrate metabolism and insulin action at those sites. This would explain a sort of counterbalance, featuring an inverse regulation of miRNA levels in plasma versus tissues. Regardless, given these studies, it seems quite possible that circulating levels of certain miRNAs may not reflect their expression levels (and subsequent biological effects) in target tissues.
Our results regarding the positive association of miR-150 with insulin sensitivity correspond well with Ying et al.'s study, which showed that miR-150 plays a critical role in preventing the pro-inflammatory activation of adipose tissue and systemic insulin resistance by regulating B cell-dependent interactions through targeting Elk1, Etf1, and Myb.26 However, this is in conflict with our previous data, which showed that whole-body miR-150 knockout mice had lower body weight, decreased expression of inflammatory cytokines in the adipose tissue (e.g., IL-6, MCP-1, tumor necrosis factor alpha (TNFα)), and improved glucose tolerance and insulin sensitivity after a high fat diet compared with controls.24 In addition, the data regarding circulating miR-150 and patients with T2DM also seem controversial in terms of the direction of association.23,8 Thus further studies are needed to clarify miR-150's role in insulin resistance and development of diabetes.
Our findings that miR-16 and miR-33 were related to both insulin sensitivity and other metabolic factors corroborates with other studies that have shown that relative expression of miRs linked to insulin sensitivity seems to be related to other metabolic risk factors.17 In addition, there is evidence that certain miR involved in diabetes may also play a role in the development of cardiovascular disease and other diabetic complications.44,45 This suggests that miRs may be involved with the interaction between insulin resistance and development of additional traits that characterize cardiometabolic disease.
However, we also noticed that all of our candidate miRs seemed to be mutually correlated, despite the fact that they were not all related to other metabolic risk factors. The fact that multiple miRs were associated with insulin resistance and that each miR in isolation was only modestly correlated with GDR may indicate that multiple miRs act in concert to regulate pathways that in combination produce insulin resistance. This concept is supported by studies such as Zhou et al., which showed that synergistic knockout of three miRs in skeletal muscle cells significantly improved insulin-stimulated glucose uptake.46 In addition, overlapping subsets of miRs may act in concert to determine which MetS traits emerge from a background of insulin resistance.
Alternatively, it is possible that another underlying confounding factor explains the co-association of these miRs. For example, miR-16, -126, -146a, and -223, which have all previously been associated with differential expression in diabetic patients, have also been shown to be upregulated after patients with rheumatoid arthritis were treated with an anti-TNFα drug, suggesting that they may play a role in immune function and anti-inflammatory processes.47 To this point, overexpression of miR-16 in macrophages has been shown to decrease secretion of TNF-α, IL-6, and IFN-β.27 In addition, many of these c-miRs have also been shown to be differentially expressed in various other disease states, such as cancer5,6 and tissue injury,48,49 so one should be cautious in interpreting single, or even combinations of, miRs as being solely biomarkers for insulin sensitivity per se.
One of the strong features of the current study is the use of hyperinsulinemic glucose clamps to quantify insulin sensitivity in our volunteers. To our knowledge, this is the first study that associates individual circulating miRs in adult humans with insulin sensitivity as measured by this gold standard.
There were a few limitations in our study, however. First, it is important to mention that this is a study that is based on associations and, therefore, should be viewed as hypothesis generating, rather than suggesting a strict cause and effect directionality between the c-miRs and insulin sensitivity. Second, we used banked, frozen plasma samples, some of which had been subjected to multiple freeze thaws before use. While some studies have shown that some miRs seem to be fairly stable over multiple freeze–thaw cycles,5,6 others have suggested that some miRNA species may vary in their stability under these conditions.50 In addition, while our data show a remarkable constancy of miR-490 levels across all samples, the use of miR-490 as a reference gene does not seem to have been used in previous reports relating to diabetes or insulin sensitivity. However, as it was the only invariant miR that fit the criteria in our screening arrays and was the most stable miR across our sample population, we felt that it was the best one to use as a reference for our study. Future studies are needed to verify the generalizability of miR-490 as a potential endogenous control for qrt-PCR studies on miR analyses conducted on human plasma samples.
It is also unclear whether different degrees of hemolysis in human blood samples could affect the data.51,52 For example, miR-16 is present in red blood cells and miR-107 in basophils,53,54 and these cells could have been disrupted during processing to release falsely to elevate the levels of these miRs in the samples. In fact, one study showed that levels of both of these miRs may be affected by hemolysis.54 However, efforts were made to exclude samples with overt hemolysis before miR extraction in our study based on visual inspection. In addition, given the use of high-dose insulin clamps in our study, our findings are more reflective of the relationship between c-miRs and peripheral insulin action and cannot isolate the effect of hepatic insulin action.
Finally, it is also important to note that the R2 values in our study were relatively modest, indicating that each single miRNA can explain only a small degree of individual variation in insulin sensitivity and other traits.
In conclusion, our study implicates a cluster of miRNA species as having a potential role in human insulin resistance. Plasma levels of miR-16, -107, -150, -33, and -222 were found to be significantly associated with insulin sensitivity, and miR-16 and miR-33 were also associated with MetS traits. These miRNAs could serve as biomarkers in humans, or more importantly, act as pathophysiological mediators in diseases characterized by insulin resistance. Additionally, given that miRNAs can be transported in blood and delivered to specific tissues where they can impact biological functions2, the miRNAs we have identified, or others, may have potential as pharmacological agents that could be introduced into the bloodstream for targeted delivery to metabolically relevant tissues. miRNAs could therefore constitute a novel therapeutic approach for the treatment and prevention of cardiometabolic disease. At the same time, it is important to point out that for individual miRs, the R2s were modest, indicating that each miR can only explain a small portion of the variance in insulin sensitivity.
Thus, subsets of miRs may act in concert in the pathogenesis of insulin resistance, and it may be more efficacious to use multiple miRs together when considering miRs as potential biomarkers and/or therapeutic agents.
Acknowledgments
The authors thank Dana Golson, RN, CDE, Robert Petri, Armando Enriquez, MT, and Tracie Thomas, RN for their hard work in acquiring the clinical data and plasma samples that were used for this study. The authors also thank Wei Zhang, MD, PhD, Ling Tian, PhD, and Nanlan Luo, for their advice on qrt-PCR, Xiangqin Cui, PhD for her advice regarding the screening arrays, Guanlan Xu, PhD, Chu-Fang (Herman) Chou, PhD, and Gang Liu, PhD, for their advice with miR assays and analysis, and Peng Li, PhD and Suzanne Judd, PhD, for their statistical advice.
Funding
This work was supported by the National Institutes of Health (R01DK038765), UAB Medical Scientist Training Program (T32GM008361), Pre-Doctoral Training in Obesity-Related Research (T32HL105349), and the Merit Review program of the Department of Veterans Affairs, as well as the UAB Diabetes Research Center (P30DK079626) and Nutrition Obesity Research Center (P30DK056336).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.McFarlane SI, Banerji M, Sowers JR. Insulin resistance and cardiovascular disease. J Clin Endocrinol Metab 2001;86:713–718 [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Liu D, Chen X, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell 2010;39:133–144 [DOI] [PubMed] [Google Scholar]
- 3.Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011;13:423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, Liang H, Zhang J, et al. Secreted microRNAs: A new form of intercellular communication. Trends Cell Biol 2012;22:125–132 [DOI] [PubMed] [Google Scholar]
- 5.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513–10518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997–1006 [DOI] [PubMed] [Google Scholar]
- 7.Navickas R, Gal D, Laucevicius A, et al. Identifying circulating microRNAs as biomarkers of cardiovascular disease: A systematic review. Cardiovasc Res 2016;111:322–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zampetaki A, Kiechl S, Drozdov I, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 2010;107:810–817 [DOI] [PubMed] [Google Scholar]
- 9.Wu L, Dai X, Zhan J, et al. Profiling peripheral microRNAs in obesity and type 2 diabetes mellitus. APMIS 2015;123:580–585 [DOI] [PubMed] [Google Scholar]
- 10.Trajkovski M, Hausser J, Soutschek J, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011;474:649–653 [DOI] [PubMed] [Google Scholar]
- 11.Jordan SD, Kruger M, Willmes DM, et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol 2011;13:434–446 [DOI] [PubMed] [Google Scholar]
- 12.Seyhan AA, Nunez Lopez YO, Xie H, et al. Pancreas-enriched miRNAs are altered in the circulation of subjects with diabetes: A pilot cross-sectional study. Sci Rep 2016;6:31479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang L, Huang J, Chen Y, et al. Identification of several circulating microRNAs from a genome-wide circulating microRNA expression profile as potential biomarkers for impaired glucose metabolism in polycystic ovarian syndrome. Endocrine 2016;53:280–290 [DOI] [PubMed] [Google Scholar]
- 14.Pek SL, Sum CF, Lin MX, et al. Circulating and visceral adipose miR-100 is down-regulated in patients with obesity and Type 2 diabetes. Mol Cell Endocrinol 2016;427:112–123 [DOI] [PubMed] [Google Scholar]
- 15.He QF, Wang LX, Zhong JM, et al. Circulating microRNA-21 is downregulated in patients with metabolic syndrome. Biomed Environ Sci 2016;29:385–389 [DOI] [PubMed] [Google Scholar]
- 16.Higuchi C, Nakatsuka A, Eguchi J, et al. Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabolism 2015;64:489–497 [DOI] [PubMed] [Google Scholar]
- 17.Wang R, Hong J, Cao Y, et al. Elevated circulating microRNA-122 is associated with obesity and insulin resistance in young adults. Eur J Endocrinol 2015;172:291–300 [DOI] [PubMed] [Google Scholar]
- 18.Willeit P, Skroblin P, Moschen AR, et al. Circulating microRNA-122 is associated with the risk of new-onset metabolic syndrome and type 2 diabetes. Diabetes 2017;66:347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah R, Murthy V, Pacold M, et al. Extracellular RNAs are associated with insulin resistance and metabolic phenotypes. Diabetes Care 2017;40:546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garvey WT, Kwon S, Zheng D, et al. Effects of insulin resistance and type 2 diabetes on lipoprotein subclass particle size and concentration determined by nuclear magnetic resonance. Diabetes 2003;52:453–462 [DOI] [PubMed] [Google Scholar]
- 21.Garvey WT, Olefsky JM, Griffin J, et al. The effect of insulin treatment on insulin secretion and insulin action in type II diabetes mellitus. Diabetes 1985;34:222–234 [DOI] [PubMed] [Google Scholar]
- 22.Ortega FJ, Mercader JM, Moreno-Navarrete JM, et al. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care 2014;37:1375–1383 [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Sundquist J, Zoller B, et al. Determination of 14 circulating microRNAs in Swedes and Iraqis with and without diabetes mellitus type 2. PLoS One 2014;9:e86792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo N, Garvey WT, Wang D-Z, et al. MicroRNA-150 Regulates lipid metabolism and inflammatory response. J Metab Syndr 2013;2 DOI: 10.4172/2167-0943.1000131 [DOI] [Google Scholar]
- 25.Yan ST, Li CL, Tian H, et al. MiR-199a is overexpressed in plasma of type 2 diabetes patients which contributes to type 2 diabetes by targeting GLUT4. Mol Cell Biochem 2014;397:45–51 [DOI] [PubMed] [Google Scholar]
- 26.Ying W, Tseng A, Chang RC, et al. MiR-150 regulates obesity-associated insulin resistance by controlling B cell functions. Sci Rep 2016;6:20176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talari M, Kapadia B, Kain V, et al. MicroRNA-16 modulates macrophage polarization leading to improved insulin sensitivity in myoblasts. Biochimie 2015;119:16–26 [DOI] [PubMed] [Google Scholar]
- 28.Davalos A, Goedeke L, Smibert P, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A 2011;108:9232–9237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lang H, Ai Z, You Z, et al. Characterization of miR-218/322-Stxbp1 pathway in the process of insulin secretion. J Mol Endocrinol 2015;54:65–73 [DOI] [PubMed] [Google Scholar]
- 30.Lee DE, Brown JL, Rosa ME, et al. MicroRNA-16 is downregulated during insulin resistance and controls skeletal muscle protein accretion. J Cell Biochem 2016;117:1775–1787 [DOI] [PubMed] [Google Scholar]
- 31.Bork-Jensen J, Scheele C, Christophersen DV, et al. Glucose tolerance is associated with differential expression of microRNAs in skeletal muscle: Results from studies of twins with and without type 2 diabetes. Diabetologia 2015;58:363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang IP, Tsai HL, Huang CW, et al. High blood sugar levels significantly impact the prognosis of colorectal cancer patients through down-regulation of microRNA-16 by targeting Myb and VEGFR2. Oncotarget 2016;7:18837–18850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hubal MJ, Nadler EP, Ferrante SC, et al. Circulating adipocyte-derived exosomal MicroRNAs associated with decreased insulin resistance after gastric bypass. Obesity (Silver Spring) 2017;25:102–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jalabert A, Vial G, Guay C, et al. Exosome-like vesicles released from lipid-induced insulin-resistant muscles modulate gene expression and proliferation of beta recipient cells in mice. Diabetologia 2016;59:1049–1058 [DOI] [PubMed] [Google Scholar]
- 35.Solayman MH, Langaee T, Patel A, et al. Identification of suitable endogenous normalizers for qRT-PCR analysis of plasma microRNA expression in essential hypertension. Mol Biotechnol 2016;58:179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice J, Roberts H, Rai SN, et al. Housekeeping genes for studies of plasma microRNA: A need for more precise standardization. Surgery 2015;158:1345–1351 [DOI] [PubMed] [Google Scholar]
- 37.Nielsen S, Akerstrom T, Rinnov A, et al. The miRNA plasma signature in response to acute aerobic exercise and endurance training. PLoS One 2014;9:e87308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang X, Muniappan L, Tang G, et al. Identification of glucose-regulated miRNAs from pancreatic {beta} cells reveals a role for miR-30d in insulin transcription. RNA 2009;15:287–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhatia H, Pattnaik BR, Datta M. Inhibition of mitochondrial beta-oxidation by miR-107 promotes hepatic lipid accumulation and impairs glucose tolerance in vivo. Int J Obes (Lond) 2016;40:861–869 [DOI] [PubMed] [Google Scholar]
- 40.Vienberg S, Geiger J, Madsen S, et al. MicroRNAs in metabolism. Acta Physiol (Oxf) 2017;219:346–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhatia H, Verma G, Datta M. miR-107 orchestrates ER stress induction and lipid accumulation by post-transcriptional regulation of fatty acid synthase in hepatocytes. Biochim Biophys Acta 2014;1839:334–343 [DOI] [PubMed] [Google Scholar]
- 42.Li Y, Mao L, Gao Y, et al. MicroRNA-107 contributes to post-stroke angiogenesis by targeting Dicer-1. Sci Rep 2015;5:13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke 2008;39:959–966 [DOI] [PubMed] [Google Scholar]
- 44.Slusarz A, Pulakat L. The two faces of miR-29. J Cardiovasc Med (Hagerstown) 2015;16:480–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kato M, Castro NE, Natarajan R. MicroRNAs: potential mediators and biomarkers of diabetic complications. Free Radic Biol Med 2013;64:85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou T, Meng X, Che H, et al. Regulation of insulin resistance by multiple MiRNAs via targeting the GLUT4 signalling pathway. Cell Physiol Biochem 2016;38:2063–2078 [DOI] [PubMed] [Google Scholar]
- 47.Castro-Villegas C, Perez-Sanchez C, Escudero A, et al. Circulating miRNAs as potential biomarkers of therapy effectiveness in rheumatoid arthritis patients treated with anti-TNFalpha. Arthritis Res Ther 2015;17:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian C, Li Z, Yang Z, et al. Plasma microRNA-16 is a biomarker for diagnosis, stratification, and prognosis of hyperacute cerebral infarction. PLoS One 2016;11:e0166688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Zhang Y, Liu Y, et al. Microvesicle-mediated transfer of microRNA-150 from monocytes to endothelial cells promotes angiogenesis. J Biol Chem 2013;288:23586–23596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farina NH, Wood ME, Perrapato SD, et al. Standardizing analysis of circulating microRNA: Clinical and biological relevance. J Cell Biochem 2014;115:805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blondal T, Jensby Nielsen S, Baker A, et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 2013;59:S1–S6 [DOI] [PubMed] [Google Scholar]
- 52.McDonald JS, Milosevic D, Reddi HV, et al. Analysis of circulating microRNA: Preanalytical and analytical challenges. Clin Chem 2011;57:833–840 [DOI] [PubMed] [Google Scholar]
- 53.Pritchard CC, Kroh E, Wood B, et al. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2012;5:492–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shkurnikov MY, Knyazev EN, Fomicheva KA, et al. Analysis of plasma microRNA associated with hemolysis. Bull Exp Biol Med 2016;160:748–750 [DOI] [PubMed] [Google Scholar]