Abstract
Osteocytes are deeply embedded in the mineralized matrix of bone and are nonproliferative, making them a challenge to isolate and maintain using traditional in vitro culture methods without sacrificing their inimitable phenotype. We studied the synergistic effects of two microenvironmental factors that are vital in retaining, ex vivo, the phenotype of primary human osteocytes: hypoxia and three-dimensional (3D) cellular network. To recapitulate the lacunocanalicular structure of bone tissue, we assembled and cultured primary human osteocytic cells with biphasic calcium phosphate microbeads in a microfluidic perfusion culture device. The 3D cellular network was constructed by the following: (1) the inhibited proliferation of cells entrapped by microbeads, biomimetically resembling lacunae, and (2) the connection of neighboring cells by dendrites through the mineralized, canaliculi-like interstitial spaces between the microbeads. We found that hypoxia synergistically and remarkably upregulated the mature osteocytic gene expressions of the 3D-networked cells, SOST (encoding sclerostin) and FGF23 (encoding fibroblast growth factor 23), by several orders of magnitude in comparison to those observed from two-dimensional and normoxic culture controls. Intriguingly, hypoxia facilitated the self-assembly of a nonproliferating, osteoblastic monolayer on the surface of the 3D-networked cells, replicating the osteoblastic endosteal cell layer found at the interface between native bone and bone marrow tissues. Our ability to replicate, with hypoxia, the strong expressions of these mature osteocytic markers, SOST and FGF23, is important since these (1) could not be significantly produced in vitro and (2) are new important targets for treating bone diseases. Our findings are therefore expected to facilitate ex vivo studies of human bone diseases using primary human bone cells and enable high-throughput evaluation of potential bone-targeting therapies with clinical relevance.
Keywords: : primary human osteocytes, hypoxia, bone tissue engineering, microfluidics, sclerostin, 3D culture
Introduction
Osteocytes are the most abundant cell type in bone, making up about 90–95% of bone cells. They are the terminally differentiated cells of the osteogenic lineage and serve as master regulators in bone remodeling.1–3 However, osteocytes are the least characterized and understood among the bone cells due to their location in bone and three-dimensional (3D) networked cellular structure. They are deeply embedded in mineralized extracellular matrix and are nonproliferative, making them a challenge to isolate and maintain using traditional in vitro culture methods without sacrificing their inimitable phenotype. On the other hand, animal models are not only costly but also have limited relevance to human diseases, as they do not recapitulate key biological processes seen in humans. This is particularly true for animal models being unable to accurately predict the efficacy and safety of drug outcomes in humans.4–8
There are several osteocytic murine cell line models such as the MLO-A5,9 MLO-Y4,10,11 and IDG-SW312 and a human preosteocytic cell line, HOB-01-C1.13 Most of these cell lines, however, do not faithfully recapitulate mature osteocytic phenotypes. In fact, cell lines, in general, are under clinical scrutiny since immortalizing human cells into cell lines by gene transfection perturbs the cells' gene expression profiles and cellular physiology and, hence, renders cell lines incapable of capturing the heterogeneity of primary cells and tissues.14–17
Specifically, osteocytic cell lines do not adequately express the key markers of mature osteocytes—SOST and FGF23. SOST encodes sclerostin, which inhibits osteoblasts' bone-forming functions. FGF23 encodes fibroblast growth factor 23, which plays a critical role in phosphate homeostasis. These mature osteocytic gene expressions are gaining great importance in targeted drug therapy, namely, sclerostin antibody therapy is emerging as a potential treatment of osteoporosis and debilitating osteolytic lesions that form in patients of multiple myeloma and other bone metastatic cancers.18–22 In addition, elevated FGF23 levels have been reported in patients with multiple myeloma and other cancers, and are being explored as a potential therapeutic target in reducing tumor burden.23,24
For these reasons, it is imperative to develop more predictive, 3D tissue-engineered models to bridge the gap of inconsistency between preclinical trials and actual human outcomes.25,26 Specifically, it is vital to be able to recapitulate SOST and FGF23 gene expressions, ideally using primary human osteocytes. However, to isolate and make use of primary human osteocytes, it is vital to maintain these cells ex vivo by providing a physiologically relevant microenvironment.
We hypothesize that there are two critical physiological elements that will favor the ex vivo maintenance of primary human osteocyte phenotype: (1) oxygen tension and (2) 3D cellular network. Since osteocytes reside deep within heavily mineralized bone, impeded oxygen diffusion leads to a hypoxic environment. It has been shown that osteocytes reside in oxygen tensions below 5% oxygen and express hypoxic markers ORP150 and HIF1α.27,28 Also, osteogenic cell lines have been used to show that oxygen tension plays an important role in the transition of osteoblasts to osteocytes and maintenance of osteogenic activity.27,29
We and others have demonstrated that osteogenic differentiation is facilitated in 3D culture systems.30–34 For example, we showed that the murine cell line MLO-A5 and primary murine osteocytes could be assembled with 20–25 μm microbeads and cultured in a 3D perfusion device to replicate the following: (1) the 3D cellular network of osteocytes in the lacunocanalicular structure of human bone tissue and (2) their expression of SOST and FGF23.33,34 In this biomimetic approach, the proliferation of cells entrapped by microbeads was mitigated, while neighboring cells became interconnected by dendrites to form a 3D cellular network and underwent significant osteocytic differentiation. Most recently, we used this 3D tissue model approach to successfully support the osteocytic differentiation of primary human osteoblastic cells (article under review).35
The goal of this study was to assess the importance of hypoxia as another critical factor for the ex vivo maintenance of physiologically relevant osteocytes. The specific aims of this article were to (1) construct 3D bone tissue models using primary human osteocytic cells under different oxygen tensions and (2) evaluate and compare the hypoxic effects on osteocytic differentiation and proliferation in 3D as wells as two-dimensional (2D) cultures.
Materials and Methods
Isolation of primary human osteocytic and osteoblastic cells
Discarded bone samples were collected with consent from patient orthopedic surgeries, in accordance with the protocol (Pro5059) that was approved by the Institution Review Board (IRB) of Hackensack University Medical Center. For this study, bone samples from the knee were used from three different patients for experiments (Table 1). After retrieval from surgery, the bone sample was promptly washed in Hank's balanced salt solution (HBSS) without calcium and magnesium and then rinsed in α-MEM supplemented with 10% penicillin–streptomycin (P/S) for 10 min on ice. Bone samples were cleaned of any muscle, cartilage, and periosteal tissues using a scalpel, then cut into 3–5 mm bone chips, and placed in large conical tubes.
Table 1.
Patient Information
| Patient | Gender | Age | Health status |
|---|---|---|---|
| 1 | Male | 64 | Osteoarthritis |
| 2 | Male | 63 | Osteoarthritis |
| 3 | Female | 59 | Osteoarthritis |
The bone chips were subjected to a series of digestions as described previously,36 with the modifications to optimize human bone cell isolation as implemented recently by Sun et al.35 Each digestion took place in 8 mL of digestive solution at 37°C for 20 min with shaking. Digestions 1–3, 5, and 7 were in 0.25 mg/mL collagenase type 1A in α-MEM. Digestions 4 and 6 were in 5 mM EDTA with 1% bovine serum albumin (BSA) in Dulbecco's phosphate-buffered saline (D-PBS; without calcium and magnesium), pH 7.4. After each digestion, the digestive supernatant was removed and the bone chips were washed 3 × with 5 mL HBSS.
For osteoblastic cell isolation, the supernatant from the fourth digestion was collected, centrifuged at 360 g for 8 min, and the cell pellet was resuspended in the medium. These cells were plated in a six-well tissue culture plate without collagen coating and allowed to proliferate for 14 days before characterization.
After the seventh digestion, the bone chips were plated onto collagen-coated six-well tissue culture plates. Osteocytic cells were allowed to migrate out of the bone chips for 8 days, after which the cells were harvested for experiments or used for further characterizations.
As shown in Supplementary Figure S1 (Supplementary Data are available online at www.liebertpub.com/tea), isolated osteoblastic cells were characterized by probing for the alkaline phosphatase (ALP) activity, whereas osteocytic cells were characterized by probing for sclerostin, as described below. For each sample, cells were counted under the microscope in at least 10 random fields of view (each with an area of 0.6 mm2) using a magnification of 100 × in triplicate wells to calculate percentage of osteoblasts and osteocytes.
General cell culture
All primary cells were cultured in α-MEM supplemented with 10% fetal bovine serum and 1% P/S. All cultures were maintained at 37°C in a sterile, humidified, 5% CO2 incubator. Cells and bone chips were washed with phosphate-buffered saline (PBS) and the medium was replaced every 3–4 days. For experiments using the differentiation medium, 3 mM β-glycerophosphate and 50 μg/mL l-ascorbic acid were added to the base medium. For hypoxia conditions, cultures were maintained in a Heracell Vios 160i incubator (Thermo Fisher) and culture medium was preincubated to 1% oxygen level for 24 h before exposing to cells.
Engineering of 3D human bone tissue in perfusion culture
A microfluidic culture device was made with polydimethylsiloxane (PDMS) with eight culture chambers, as described previously.33,34 Soft lithography was used to create 200 μm thick hexagonal patterns of 6 × 12 mm with a central 3 mm diameter culture chamber. Each culture chamber was secured with a 200 μm thick Microweb filter membrane (Millipore). The PDMS was then bonded to a glass slide. The device was sterilized by washing all chambers and microfluidic channels with 70% isopropyl alcohol.
In the central culture chamber, the 3D human bone tissue was reconstructed using biphasic calcium phosphate (BCP) microbeads (CaP Biomaterials). BCP microbeads were sieved to a size of 20–25 μm diameter, sterilized under UV light for 20 min, and then coated with type 1 collagen for 1 h at 37°C and washed 3 × in PBS. Primary human osteocytic cells were harvested, counted, and combined with prepared BCP microbeads in a 1:1 ratio. To each culture chamber, 3D tissues were constructed by seeding a mixture of 100,000 cells and 100,000 beads.
Perfusion was initiated by connecting the inlet with polyethylene tubing that served the differentiation medium to the central culture chamber with a syringe pump (KD Scientific), at a rate of 1 μL/min. Effluent medium was collected by polyethylene tubing connecting the outlets to a collection vial (Fig. 1A). Parallel culture devices were prepared and placed in hypoxic (1% oxygen) or normoxic (20% oxygen) culture. Simultaneously, 2D cultures were set up by seeding 10,000 cells into triplicate wells of a 24-well plate for hypoxic and normoxic culture, to serve as static controls. Tissues were allowed to grow for 14 days and then harvested for further characterization.
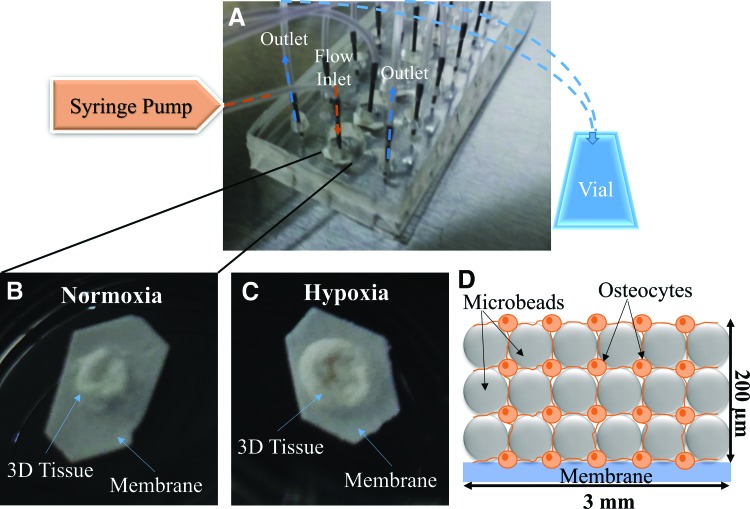
FIG. 1.
Microfluidic perfusion device for constructing 3D-engineered tissue. (A) Actual device, which contains tissues in central chamber with fluid flowing into one inlet fed with a syringe pump and two outlets emptying into collection vials. Tissues were constructed using biphasic calcium phosphate microbeads and primary human osteocytes in a 1:1 ratio. Tissues were harvested after 14 days of culture in (B) normoxia and (C) hypoxia. (D) An idealized illustration of the spatial distribution of cells and microbeads in the tissue constructs (not drawn to scale). 3D, three dimensional. Color images available online at www.liebertpub.com/tea
RNA isolation, reverse transcription, and polymerase chain reaction
Total RNA was isolated using the Purelink RNA Mini kit (Ambion), following the manufacturer's instructions. Complementary DNA (cDNA) was prepared by reverse transcription using 1 μg of the total RNA isolated. Reverse transcription took place in a 30 μL reaction with 16.5 μL of 61 ng/μL RNA, 3 μL of 0.5 mg/mL oligoDT, 1 μL RNasin ribonuclease inhibitor, 6 μL 5 × reaction buffer, 1.5 μL 10 mM dNTP, and 2 μL reverse transcriptase enzyme (Promega). The reaction mixture was incubated in a 37°C water bath for 1.5 h. The reaction was stopped by incubating at 95°C for 3 min and then immediately placing on ice.
For real-time polymerase chain reaction (RT-PCR) assays, 20 μL reaction mixtures were prepared using 2 μL cDNA, 1 μL of 20 × Taqman primer (Table 2), 10 μL of 2 × Taqman master mix, and 7 μL DEPC-treated water. The quantitative RT-PCR fast assay was carried out on a StepOnePlus (Applied Biosystems, CA). The following amplification cycle was repeated 40 times: 95°C, 20 s; 90°C, 1 s; 60°C, 20 s.
Table 2.
Taqman Primers used in Real-Time Polymerase Chain Reaction
| Gene | Primer (ThermoFisher) | Amplicon length (bp) |
|---|---|---|
| ALPL | Hs01029144_m1 | 79 |
| DMP1 | Hs01009390_m1 | 132 |
| FGF23 | Hs00221003_m1 | 64 |
| SOST | Hs00228830_m1 | 81 |
Gene expression was normalized to 18S and fold changes in relative expression were determined using the ΔCt method. The following gene expressions were analyzed: ALPL (encoding ALP, an osteoblast marker), DMP1 (encoding dentin matrix acidic phosphoprotein 1, a preosteocyte marker), FGF23 (encoding fibroblast growth factor 23, a mature osteocyte marker), and SOST (encoding sclerostin, a mature osteocyte marker).
Histology
After 14 days, 3D tissues were harvested and fixed in 4% paraformaldehyde (PFA). Fixed samples were sent to the Histology Core Facility at the New Jersey Medical School of Rutgers University for paraffin embedding and sectioning. Each sample was cut into 10 μm thick sections and placed on slides. Sections were stained with hematoxylin and eosin (H&E; Sigma-Aldrich) and visualized under a microscope to examine cell distribution and morphology.
To characterize the cells, immunostaining of sclerostin, ALP, and HIF1α was done on parallel sections. After deparaffinization and rehydration, sections were subjected to heat-induced antigen retrieval in EDTA buffer, pH 8.5 (Sigma-Aldrich). Sections were permeabilized with 0.1% Triton X-100 for 10 min and blocked with 3% BSA (w/v) for 1 h at RT. Rabbit anti-human sclerostin (1:20), rabbit anti-human ALP (1:20), and rabbit anti-human HIF1α (1:200; Abcam) were exposed to respective samples overnight at 4°C. The sections were incubated with a secondary stain (TRITC-conjugated goat anti-rabbit IgG, 1:150; Abcam) for 1 h at RT and counterstained with DAPI and mounted. Secondary antibody only (without primary) and rabbit IgG isotype served as negative controls. Sections were visualized under a fluorescence microscope.
2D hypoxia experiments
Primary human osteocytic cells were seeded onto 24-well plates (50,000 cells per well) and allowed to grow for 14 days in the differentiation medium in parallel hypoxic (1% oxygen level) and normoxic (20% oxygen level) cultures. All 2D experiments were conducted under static conditions and cells were washed and the medium changed every 3–4 days. On day 14, triplicate wells were used in each of the assessments described below.
Viability assay
Cell viability was assessed using a Live/Dead viability/cytotoxicity kit according to the manufacturer's instructions (Molecular Probes). Briefly, the staining solution was prepared in PBS with 6 μM Calcien-AM (live stain) and 6 μM Ethidium homodimer-1 (dead stain). After adding staining solution to each well, the plate was incubated at 37°C for 30 min. The cells were visualized under a fluorescence microscope; live cell bodies fluoresced green after being excited by blue light, whereas dead cells' nuclei fluoresced red after being excited by green light. Live and dead cells were counted in at least 10 random fields of view (each with an area of 0.6 mm2) in triplicate wells under the microscope at 100 × magnification to calculate percent viability of cells.
F-actin staining
Samples were stained with ActinRed 555 according to the manufacturer's instructions (Thermo Fisher). Cells were fixed with 4% PFA for 10 min and permeabilized using 0.1% Triton X-100 for 10 min. After adding staining solution to each well, the plate was incubated for 30 min at room temperature. The cells were then counterstained with DAPI and visualized under a fluorescence microscope; the cytoskeleton of the cell fluoresced red, whereas the cell nucleus fluoresced blue. For calculating cell densities, nuclei were counted in at least 10 random fields of view in triplicate wells as described above.
Mineralization
After samples were fixed with 4% PFA for 10 min, Alizarin Red S staining and quantification assay (ScienCell) were used according to the manufacturer's instructions. Briefly, stain was added to each well, incubated for 15 min, washed 3 × , and observed under the microscope; mineralized zones stained red. The stain was extracted and quantified by measuring absorbance at 405 nm.
ALP activity staining
Samples were prepared for staining using an ALP activity staining kit according to the manufacturer's instructions (Takara). Briefly, samples were fixed with 4% PFA for 10 min. After adding the ALP substrate solution to each well, the plate was incubated at 37°C for 30 min. The reaction was stopped by washing 3 × with distilled water. Cells were visualized under the microscope; positive cells were stained purple. For quantification, the stain was extracted using dimethyl sulfoxide for 15 min at 37°C. Absorbance was read at 490 nm.
Sclerostin immunostaining
For the detection of sclerostin, samples were treated as described above for histology immunostaining for sclerostin. Cells were visualized under a fluorescence microscope; sclerostin-positive cells fluoresced red and the cell nuclei fluoresced blue.
Statistical analysis
When determining statistically significant differences, Student's t-test was used when comparing two groups, whereas analysis of variance was used for multiple comparisons. A p ≤ 0.1 was considered a significant difference. All experiments were done in triplicate, except 3D experiments for gene expression analysis were done in duplicate. Data are reported as mean ± standard deviation.
Results
3D tissue engineered under hypoxia resembles in vivo bone more precisely
Human primary osteocytic cells isolated, as shown in Supplementary Figure S1B, were assembled with BCP microbeads and cultured to create 3D tissues. After 14 days of 3D culture, engineered tissues were harvested from the microfluidic perfusion device (Fig. 1A). Representative images of the tissues cultured under normoxia and hypoxia are shown in Figure 1B and C, respectively. The anticipated 3D-networked structure and spatial distribution of the cells assembled with the microbeads are illustrated in Figure 1D.
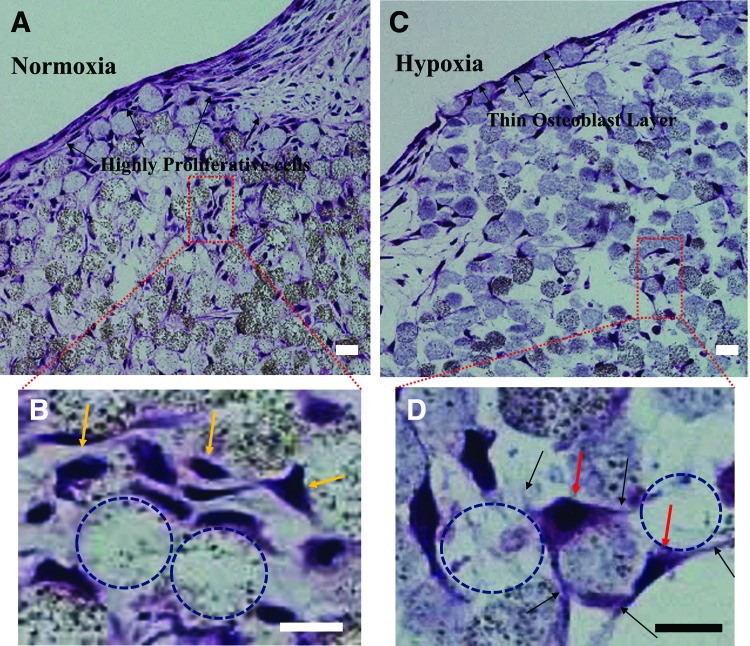
After sectioning and staining with H&E (Fig. 2), the normoxic tissue appeared very dense with the presence of proliferative cells, especially at the tissue surface (Fig. 2A). Cells entrapped between microbeads were overall less proliferative than on the surface; however, some areas in between microbeads still seemed to proliferate in the normoxic tissue (Fig. 2B). The hypoxic tissue contained virtually nonproliferative cells, which resulted in a self-assembled, thin layer of cells on the surface of the 3D tissue construct, closely mimicking the endosteum found in native bone (Fig. 2C). Furthermore, cells within the hypoxic tissue also retained a virtually nonproliferative state and distributed themselves in spaces in between the microbeads (Fig. 2D). This hypoxic structure mimics more precisely the in vivo lacunae with dendritic processes extending to neighboring osteocytes, very much like canaliculi in vivo.
FIG. 2.
Histology sections of 3D-engineered tissues harvested after 14 days and stained with hematoxylin and eosin. (A) Normoxia tissue with proliferative cells observed on the tissue surface and (B) in between the microbeads (yellow arrows). (C) Hypoxia tissue with a thin layer of cells, resembling the osteoblastic endosteum observed in vivo. (D) Nonproliferative cell bodies (red arrows) with dendritic connections (black arrows) present between microbeads. Voids left by pulled out microbeads are indicated using dashed circles. Scale bar = 20 μm. Color images available online at www.liebertpub.com/tea
3D hypoxic culture upregulates the expression of osteocyte-specific genes
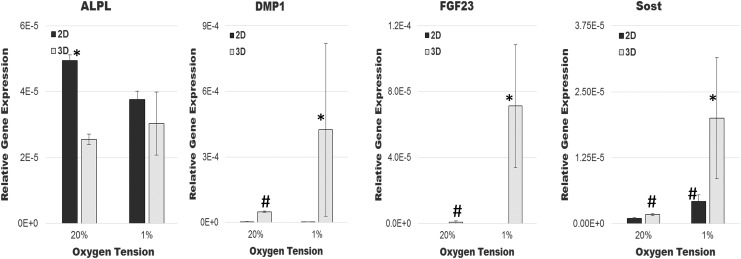
Osteoblast-specific gene expression of ALPL, osteoblast-to-osteocyte transition gene expression of DMP1, and mature osteocyte-specific gene expression of FGF23 and SOST were examined after 14 days of culturing primary human osteocytic cells under different culture conditions (Fig. 3). Osteoblastic ALPL expression was highest in traditional 2D normoxic culture and was expressed significantly less under all other culture conditions. Gene expression of the transition gene, DMP1, was lowest in 2D cultures with upregulation in 3D cultures.
FIG. 3.
Relative gene expressions of primary human osteocytic cells cultured in 3D (gray bars) or 2D (black bars) and under normoxia (20% oxygen) or hypoxia (1% oxygen) after 14 days. *p ≤ 0.1 compared to all other cultures. #p ≤ 0.05 compared to 2D normoxia. 2D, two dimensional.
The expressions of mature osteocyte-specific genes SOST and FGF23 were remarkably high in hypoxic 3D cultures, in comparison to other culture conditions. Specifically, SOST was expressed at low levels in traditional 2D normoxic cultures. There was a significant increase in expression in 3D normoxic and 2D hypoxic cultures. Remarkably, a synergistic upregulation of SOST was observed in 3D hypoxic culture. Likewise, little to no FGF23 was observed in 2D cultures, but an increase was observed in 3D normoxic cultures, with a synergistic upregulation of FGF23 under 3D hypoxic cultures.
Spatial distribution of osteoblasts and osteocytes is recapitulated in 3D bone tissue engineered under hypoxia
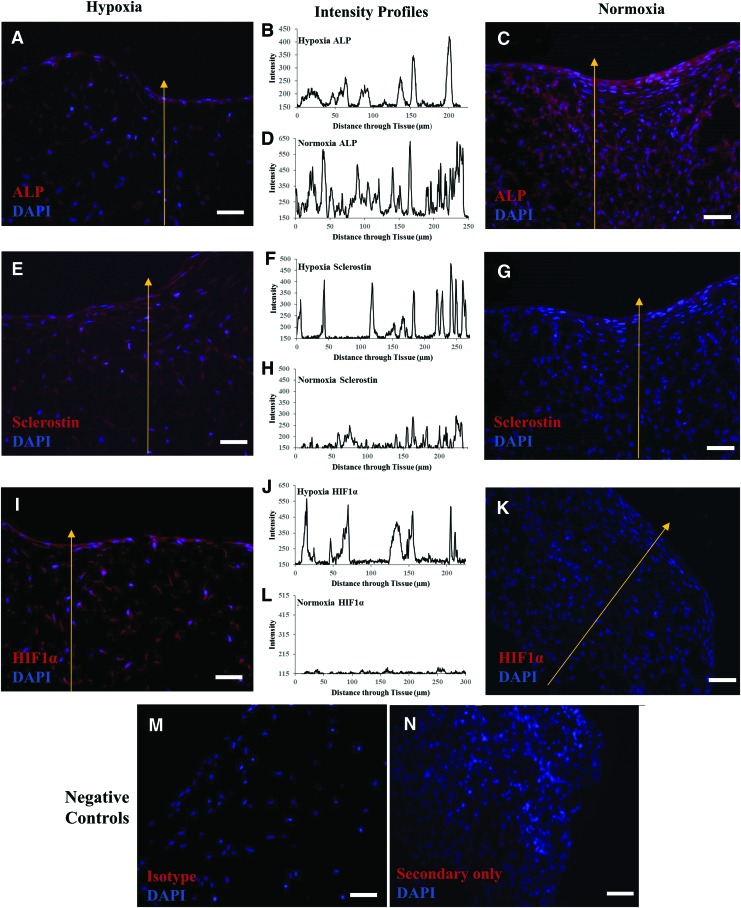
The replication of osteocyte-like phenotype in hypoxic 3D tissue was further verified by the immunostaining of histology sections (Fig. 4), probing for sclerostin (an osteocyte-specific marker), ALP (an osteoblast-specific marker), and HIF1α (a hypoxic marker). For each section stained, a fluorescence intensity profile was quantitated along a path from the interior to the surface of the tissue (indicated with a yellow arrow in Fig. 4).
FIG. 4.
Immunostained sections of 3D-engineered bone tissues cultured in normoxia (right) or hypoxia (left). Intensity profiles (center) were quantitated through each tissue along the indicated yellow arrow, starting from the interior of the tissue to the top surface. Parallel tissue sections were separately stained and quantitated for the presence of (A–D) ALP, (E–H) sclerostin, or (I–L) HIF1α. Negative controls were stained with (M) Isotype and (N) Secondary antibody only. All sections were counterstained with DAPI. Scale bar = 50 μm. ALP, alkaline phosphatase. Color images available online at www.liebertpub.com/tea
The thin layer of cells on the surface of the hypoxic tissue stained highly positive for ALP (Fig. 4A, B), whereas its expression diminished in the interior region of the hypoxic tissue, as shown by peaks decreasing in intensity further into the tissue. The maximum intensity for ALP was found to correspond to the surface layer of cells (Fig. 4B). The cells in the hypoxic tissue also stained highly positive for sclerostin throughout the tissue with uniform intensity (Fig. 4E, F). The results suggest that the hypoxic tissue consisted of (1) 3D-networked osteocytic cells in its interior and (2) a self-assembled osteoblastic cell layer, similar to the endosteal layer in native bone. It was confirmed that the cells throughout the hypoxic tissue experienced hypoxia as evident from the observed positive staining of HIF1α (Fig. 4I) and uniform peaks in the intensity profile (Fig. 4J).
The 3D normoxic tissue was very dense with proliferative cells that stained highly positive for ALP throughout the tissue (Fig. 4C). The ALP expression level was consistent throughout the tissue, from the surface to the interior (Fig. 4D). There was very little sclerostin staining in the cells of the normoxic tissue (Fig. 4G), with low intensity (Fig. 4H). This suggested that the normoxic tissue consisted mostly of osteoblastic cells and some osteocytic cells. The absence of HIF1α staining in the normoxic tissue confirmed that the tissue interior was thoroughly oxygenated (Fig. 4K, L).
Hypoxia maintains human primary osteocyte phenotype longer in ex vivo 2D cultures
To evaluate the effectiveness of hypoxic culture in maintaining the osteocytic phenotype ex vivo, we cultured isolated human primary osteocytic cells in 2D under hypoxia and normoxia. As seen in Figure 5, after growing for 14 days, cells were stained for viability, morphology (F-actin), ALP activity, sclerostin expression, and mineralization (alizarin red). As seen in Figure 5A, cell viability was unaffected in hypoxia (98.72% viability), which was similar to that observed in normoxia (98.25% viability).
FIG. 5.
Phenotypic characterization of primary human osteocytes cultured in 2D cultures for 14 days under normoxia (images on right) and hypoxia (images on left). (A) Quantification of total, live, dead cells, and % viability of cells in normoxia and hypoxia. *p ≤ 0.001 compared to total cell density in hypoxia. (B, C) Representative images showing live (green) and dead (red) cells. (D, E) F-actin staining (red) and DAPI (blue). (F) Quantitation of ALP activity in normoxia and hypoxia. *p ≤ 0.001 compared to hypoxia. (G, H) ALP activity staining (purple). (I, J) Sclerostin immunostaining (red) and DAPI (blue). (K) Quantitation of Alizarin Red staining in normoxia and hypoxia. (L, M) Alizarin red staining showing mineralization node (red). Scale bar = 50 μm. Color images available online at www.liebertpub.com/tea
There were, on average, 903 ± 99 cells/mm2 in hypoxic culture (Fig. 5A). Cells remained viable and healthy for 14 days in hypoxic culture, as evident by the majority of cells staining positive for the live stain and little to no dead cells (Fig. 5B). The hypoxic cells exhibited a distinct morphology as indicated by the cytoskeleton staining (Fig. 5D). There was very little ALP activity in hypoxia (Fig. 5F), which is evident by little to no staining in Figure 5G. However, some cells stained positive for sclerostin (Fig. 5I). Alizarin Red staining was high in hypoxia (Fig. 5K) and random nodes of mineralization were observed throughout the hypoxic culture as seen in Figure 5L.
When cultured in normoxia for 14 days, cells remained viable and highly proliferative. There were, on average, 1970 ± 282 cells/mm2 in normoxic culture (Fig. 5A, C). The normoxic cells grew without any particular orientation as the cytoskeleton staining reveals a random mesh-like appearance (Fig. 5E). The ALP activity was significantly higher in normoxia (Fig. 5F) and observed throughout the culture (Fig. 5H), whereas sclerostin protein expression was undetected using immunostaining (Fig. 5J). Alizarin red staining was lower in normoxia (Fig. 5K) and nodes of mineralization were not observed in culture (Fig. 5M).
Discussion
In this study, we used a combination of oxygen tension and 3D culture systems to enhance the expression of vital markers in primary human osteocytic cells ex vivo. Cell lines were previously used to show that oxygen tension plays a role in the differentiation of osteoblasts to osteocytes.27,29 We not only confirmed these results using MLO-A5 cell lines in 2D cultures (data not shown) but also validated this claim, for the first time, using isolated primary human osteocytic cells in 2D and 3D.
As an enhancement to our recently reported35 methodology of isolating osteoblastic cells from human bone chips, we were able to isolate more osteocytic cells in this study. As our previous study reported, the optimal numbers of osteoblastic cells (81–95%) could be obtained by collecting cells from the fourth digestion and proliferating them over 14 days. Note that the majority of these cells stained positive for the ALP activity (Supplementary Fig. S1A). More osteocytic cells were obtained by allowing them to migrate out from the bone chips after seven digestions. The majority of cells that migrated out of the bone chips over an 8-day period were identified to be osteocytic (86.4% ± 5.9%), as evident by sclerostin-positive staining (Supplementary Fig. S1B) with a very small population of osteoblastic cells (12.9% ± 5.6%), which stained weakly positive for the ALP activity (Supplementary Fig. S1C). Since osteocytes are nonproliferative, it was procedurally important to harvest and characterize these cells by 8 days. This kept them from dedifferentiating, as the proliferative nature of osteoblastic cells could take over the culture.
When using osteoblastic cells for the 3D tissue construction under normoxia, we observed previously35 that the proliferation of the cells entrapped by the 20–25 μm BCP microbeads was mitigated. At the same time, the cells differentiated significantly to become osteocytic with dendritic connections. This approach was used as a key mechanism for producing the 3D-networked cellular structure of osteocytes in the lacunocanalicular structure of bone. Similarly, in this study, the osteocytic cells entrapped by the microbeads in both normoxic and hypoxic tissue samples (Fig. 2A, C) formed the 3D cellular network. In addition, the gene expression profiles of primary human osteocytic cells cultured in 3D-engineered tissues showed a significant increase in osteocytic markers (SOST, FGF23, and DMP1) with a decrease in osteoblastic marker (ALPL), in comparison to 2D culture samples (Fig. 3). The gene expression results were consistent with our previous studies on 3D-reconstructed bone tissues using MLO-A5,33 primary murine osteocytes,34 and primary human osteoblastic cells.35
Despite these general similarities in normoxia, there were two profound effects of hypoxia on the 3D tissue development. First, very remarkably, cells on the 3D tissue surface did not proliferate significantly under hypoxia, resulting in the formation of an osteoblastic monolayer (Fig. 2C). The monolayer was determined to be osteoblastic since it was stained strongly by the ALP staining (maximum peak in Fig. 4B), whereas the cells inside the 3D structure exhibited diminished intensity. Therefore, this layer resembled the osteoblastic endosteal layer observed at the interface region between native bone and bone marrow tissues. In contrast, cells located on the surface of the normoxic 3D tissue were able to proliferate freely to form several layers of osteoblastic cells (Figs. 2A and 4C). This hypoxia-induced self-assembly of the endosteum-like layer is particularly noteworthy, as it has not been observed in any other culture system reported to date.30–32,37,38
Second, hypoxia significantly enhanced the gene expressions of SOST and FGF23, while decreasing that of ALPL. Taking culture conditions separately, replacing traditional normoxic 2D cultures with hypoxia significantly increased SOST expression by fourfold, while introducing the 3D configuration to the traditional 2D normoxic cultures significantly increased SOST by twofold (Fig. 3). However, the synergistic effect of combining hypoxia with 3D culture was profound, as reflected by the 20-fold increase in SOST expression in hypoxic 3D cultures. Likewise, the 90-fold increase of FGF23 was observed in hypoxic 3D cultures. These gene expression results were consistent with the histological immunostaining of tissue sections that showed cells within the 3D structure were stained more strongly by sclerostin under hypoxia (Fig. 4E, F) than in normoxia (Fig. 4G, H). ALPL gene expression of 3D hypoxic culture was significantly lower compared to 2D normoxic culture. Although there was no detectable significant difference in 3D hypoxic culture and 3D normoxic culture when only looking at gene expression, there was an observable and quantifiable difference in protein expression as seen in the immunostaining of the 3D sections, where the intensity of the ALP staining was uniformly higher in normoxia (Fig. 4A–D).
In addition, the comparison of the 2D culture results showed a more osteocyte-like phenotype under hypoxia (Fig. 5). Cells exhibited a distinct morphology in hypoxia, as indicated by the cytoskeleton staining (Fig. 5D), whereas cells in normoxia seemed to proliferate with no particular orientation. The detection of some sclerostin activity, the presence of mineralization, and the absence of ALP activity, further indicated a more osteocyte-like phenotype in hypoxic 2D cultures. Also, there were, on average, 903 ± 99 cells/mm2 in hypoxic culture, which was significantly less (p < 0.001) than in normoxic culture (1970 ± 282 cells/mm2). This showed normoxic cultures were more proliferative and thus, more osteoblast like. Taken together, these results suggest that hypoxia retains osteocytic phenotype ex vivo, whereas normoxia allows the rapid dedifferentiation of osteocytes back to an osteoblastic phenotype.
Our ability to replicate, with hypoxia, the expressions of mature osteocytic markers, FGF23 and SOST, is important for targeted drug therapy studies. It has been previously reported that FGF23 is elevated in hypoxic cultures,39 which has been confirmed by this study. However, there have been conflicting reports regarding SOST expression in hypoxic cultures.40,41 Our results support that hypoxia increases SOST expression, which is agreeable with the study conducted by Chen et al.41 Importantly, in our RT-PCR analysis, we used a uniform RNA concentration and an endogenous control that is consistently expressed between normoxia and hypoxia.42 Genetos et al.,40 however, used a varying range of RNA concentration for RT-PCR analysis and an endogenous control, which has previously been shown to be affected by hypoxia.42,43 This may be the source of varying outcomes between their study and ours.
In conclusion, the results of this study show the importance of hypoxia and 3D cellular network as two key microenvironmental factors that are vital and synergistic in retaining the phenotype of primary human osteocytes. We found that hypoxia maintains osteocyte phenotype in 2D cultures ex vivo and plays a critical role in the self-assembly of 3D bone tissue ex vivo, and osteocyte-specific markers are synergistically enhanced in hypoxic 3D cultures. Our 3D-constructed tissues accurately recapitulate vital osteocytic gene expressions. This recapitulation is critical in engineering tissues that have relevant correlation to human outcomes. With the advent of new antibody therapies against SOST and FGF23 to alleviate osteoporosis and tumor burden, respectively, and the lack of accurate bone models that faithfully recapitulate osteocytic gene expressions, we have specifically shown how SOST and FGF23 expressions can be maximized using primary human bone cells, hypoxia, and 3D culture conditions. This culture system has the potential to be integrated into creating reliable tissue constructs for drug testing platforms, bone metastatic studies, and mechanistic studies.
Supplementary Material
Acknowledgments
Research in this publication was supported by grants from (1) the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number 1R21AR065032 to W.Y.L. and J.Z. and (2) the National Science Foundation (DMR 1409779) to W.Y.L. and J.Z. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health and the National Science Foundation.
We thank Luke Fritzky at the Histology Core Facility at Rutgers-New Jersey Medical School for paraffin embedding and sectioning and Larry Lyons for his help with the IRB protocol preparation for human bone sample collection.
Authors' Contributions
S.C., J.Z., and W.Y.L. designed the research. S.C. conducted the study and experimental work. C.M. and Y.K. provided the human bone samples. S.C., Q.S., J.Z., and W.Y.L. contributed to data interpretation and discussion. S.C. drafted the article. S.C., J.Z., and W.Y.L. revised the article content. All coauthors read and approved the final version of the article.
Disclosure Statement
No competing financial interests exist.
References
- 1.Bonewald L.F. The amazing osteocyte. J Bone Miner Res 26, 229, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dallas S.L., and Bonewald L.F. Dynamics of the transition from osteoblast to osteocyte. Ann N Y Acad Sci 1192, 437, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franz-Odendaal T.A., Hall B.K., and Witten P.E. Buried alive: how osteoblasts become osteocytes. Dev Dyn 235, 176, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Jucker M. The benefits and limitations of animal models for translational research in neurodegenerative diseases. Nat Med 16, 1210, 2010 [DOI] [PubMed] [Google Scholar]
- 5.Shanks N., Greek R., and Greek J. Are animal models predictive for humans? Philos Ethics Humanit Med 4, 2, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartung T. Thoughts on limitations of animal models. Park Relat Disord 14(SUPPL.2), 81, 2008 [DOI] [PubMed] [Google Scholar]
- 7.Couzin-Frankel J. Hope in a mouse. Science 346, 28, 2014 [DOI] [PubMed] [Google Scholar]
- 8.Rhrissorrakrai K., Belcastro V., Bilal E., Norel R., Poussin C., Mathis C., et al. Understanding the limits of animal models as predictors of human biology: lessons learned from the sbv IMPROVER Species Translation Challenge. Bioinformatics 31, 471, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato Y., Boskey A., Spevak L., Dallas M., Hori M., and Bonewald L.F. Establishment of an osteoid preosteocyte-like Cell MLO-A5 that spontaneously mineralizes in culture. J Bone Miner Res 16, 1622, 2001 [DOI] [PubMed] [Google Scholar]
- 10.Kato Y., Windle J.J., Koop B.A., Mundy G.R., and Bonewald L.F. Establishment of an osteocyte-like cell line, MLO-Y4. J Bone Miner Res 12, 2014, 1997 [DOI] [PubMed] [Google Scholar]
- 11.Bonewald LF. Establishment and characterization of an osteocyte-like cell line, MLO-Y4. J Bone Miner Metab 17, 61, 1999 [DOI] [PubMed] [Google Scholar]
- 12.Woo S.M., Rosser J., Dusevich V., Kalajzic I., and Bonewald L.F. Cell line IDG-SW3 replicates osteoblast-to-late-osteocyte differentiation in vitro and accelerates bone formation in vivo. J Bone Miner Res 26, 2634, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodine P.V., Vernon S.K., and Komm B.S. Establishment and hormonal regulation of a conditionally transformed preosteocytic cell line from adult human bone. Endocrinology 137, 4592, 1996 [DOI] [PubMed] [Google Scholar]
- 14.Li A., Walling J., Kotliarov Y., Center A., Steed M.E., Ahn S.J., et al. Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas. Mol Cancer Res 6, 21, 2008 [DOI] [PubMed] [Google Scholar]
- 15.Gillet J., Varma S., and Gottesman M.M. The clinical relevance of cancer cell lines. J Natl Cancer Inst 105, 452, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandberg R., and Ernberg I. Assessment of tumor characteristic gene expression in cell lines using a tissue similarity index (TSI). Proc Natl Acad Sci U S A 102, 2052, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domcke S., Sinha R., Levine D.A., Sander C., and Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun 4, 2126, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Compton J.T., and Lee F.Y. A review of osteocyte function and the emerging importance of sclerostin. J Bone Joint Surg Am 96, 1659, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke B.L. Anti-sclerostin antibodies: utility in treatment of osteoporosis. Maturitas 78, 199, 2014 [DOI] [PubMed] [Google Scholar]
- 20.Brunetti G., Oranger A., Mori G., Specchia G., Rinaldi E., Curci P., et al. Sclerostin is overexpressed by plasma cells from multiple myeloma patients. Ann N Y Acad Sci 1237, 19, 2011 [DOI] [PubMed] [Google Scholar]
- 21.Yavropoulou M.P., van Lierop A.H., Hamdy N.A.T., Rizzoli R., and Papapoulos S.E. Serum sclerostin levels in Paget's disease and prostate cancer with bone metastases with a wide range of bone turnover. Bone 51, 153, 2012 [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Fontana B., Morales-Santana S., Varsavsky M., Garcia-Martín A., Garcia-Salcedo J.A., Reyes-Garcia R., et al. Sclerostin serum levels in prostate cancer patients and their relationship with sex steroids. Osteoporos Int 25, 645, 2014 [DOI] [PubMed] [Google Scholar]
- 23.Suvannasankha A., Tompkins D.R., Edwards D.F., Katarina V., Crean C.D., Fournier P.G., et al. FGF23 is elevated in multiple myeloma and increases heparanase expression by tumor cells. Oncotarget 6, 19647, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng S., Wang J., Zhang Y., and Creighton C.J. FGF23 promotes prostate cancer progression. Oncotarget 6, 17291, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horvath P., Aulner N., Bickle M., Davies A.M., Nery E.D., Ebner D., et al. Screening out irrelevant cell-based models of disease. Nat Rev Drug Discov 15, 751, 2016 [DOI] [PubMed] [Google Scholar]
- 26.Hutmacher D.W., Horch R.E., Loessner D., Rizzi S., Sieh S., Reichert J.C., et al. Translating tissue engineering technology platforms into cancer research. J Cell Mol Med 13(8A), 1417, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirao M., Hashimoto J., Yamasaki N., Ando W., Tsuboi H., Myoui A., et al. Oxygen tension is an important mediator of the transformation of osteoblasts to osteocytes. J Bone Miner Metab 25, 266, 2007 [DOI] [PubMed] [Google Scholar]
- 28.Gross T.S., Akeno N., Clemens T.L., Komarova S., Srinivasan S., Weimer D.A., et al. Osteocytes upregulate HIF-1α in response to acute disuse and oxygen deprivation. J Appl Physiol 90, 2514, 2001 [DOI] [PubMed] [Google Scholar]
- 29.Zahm A.M., Bucaro M.A., Srinivas V., Shapiro I.M., and Adams C.S. Oxygen tension regulates preosteocyte maturation and mineralization. Bone 43, 25, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boukhechba F., Balaguer T., Michiels J.-F., Ackermann K., Quincey D., Bouler J.-M., et al. Human primary osteocyte differentiation in a 3D culture system. J Bone Miner Res 24, 1927, 2009 [DOI] [PubMed] [Google Scholar]
- 31.Kale S., Biermann S., Edwards C., Tarnowski C., Morris M., and Long M.W. Three-dimensional cellular development is essential for ex vivo formation of human bone. Nat Biotechnol 18, 954, 2000 [DOI] [PubMed] [Google Scholar]
- 32.Trojani C., Weiss P., Michiels J.F., Vinatier C., Guicheux J., Daculsi G., et al. Three-dimensional culture and differentiation of human osteogenic cells in an injectable hydroxypropylmethylcellulose hydrogel. Biomaterials 26, 5509, 2005 [DOI] [PubMed] [Google Scholar]
- 33.Gu Y., Zhang W., Sun Q., Hao Y., Zilberberg J., and Lee W.Y. Microbead-guided reconstruction of the 3D osteocyte network during microfluidic perfusion culture. J Mater Chem B 3, 3625, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Q., Gu Y., Zhang W., Dziopa L., Zilberberg J., and Lee W. Ex vivo 3D osteocyte network construction with primary murine bone cells. Bone Res 3, 15026, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Q., Choudhary S., Mannion C., Kissin Y., Zilberberg J., and Lee W.Y. Ex vivo construction of human primary 3D-Networked osteocytes. Bone. (Forthcoming 2017) [DOI] [PMC free article] [PubMed]
- 36.Stern A.R., Stern M.M., van Dyke M.E., Jähn K., Prideaux M., and Bonewald L.F. Isolation and culture of primary osteocytes from the long bones of skeletally mature and aged mice. Biotechniques 52, 361, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vazquez M., Evans B.A.J., Ralphs J.R., Riccardi D., and Mason D.J. In vitro 3D osteoblast-osteocyte co-culture model. Eur Cells Mater 24(SUPPL. 1), 7, 2012 [Google Scholar]
- 38.Pajevic P.D., Spatz J.M., Garr J., Adamson C., Misener L. Osteocyte biology and space flight. Curr Biotechnol 2, 179, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clinkenbeard E.L., Farrow E.G., Summers L.J., Cass T.A., Roberts J.L., Bayt C.A., et al. Neonatal Iron deficiency causes abnormal phosphate metabolism by elevating FGF23 in normal and ADHR mice. J Bone Miner Res 29, 361, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Genetos D.C., Toupadakis C.A., Raheja L.F., Wong A., Papanicolaou S.E., Fyhrie D.P., et al. Hypoxia decreases sclerostin expression and increases Wnt signaling in osteoblasts. J Cell Biochem 110, 457, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen D., Li Y., Zhou Z., Wu C., Xing Y., Zou X., et al. HIF-1α inhibits Wnt signaling pathway by activating Sost expression in osteoblasts. PLoS One 8, e65940, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y., Fan W., Zhu L., Zhao T., Ma L., Wu Y., et al. Effects of hypoxia on mRNA expression of housekeeping genes in rat brain tissue and primary cultured neural cells. Front Med China 2, 239, 2008 [Google Scholar]
- 43.Zhong H., and Simons J.W. Direct comparison of GAPDH, beta-actin, cyclophilin, and 28S rRNA as internal standards for quantifying RNA levels under hypoxia. Biochem Biophys Res Commun 259, 523, 1999 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.