Abstract
Purpose
Targeting MET in cancer is hampered by lack of diagnostics that accurately reflect high MET signaling and dependence. We hypothesized that assays reflecting MET signaling associated protein complexes could redefine tumors dependent on MET and could add additional precision beyond genomic assessments.
Experimental Design
We utilized biochemical approaches, cellular viability studies and proximity ligation assays to assess MET dependence. We examined MET signaling complexes in lung cancer patient specimens (N=406) and patient-derived xenograft models of solid tumors (N=308). We evaluated response to crizotinib in a MET-amplified cohort of patient-derived xenografts models of lung cancer (N=6) and provide a case report of a lung cancer patient harboring a Δexon14 MET splice variant.
Results
We found the interaction of MET with the adaptor protein GRB2 is necessary for oncogenic survival signaling by MET. MET:GRB2 complexes were identified only within MET-amplified patient-derived xenograft (PDX) models and patient specimens but exhibit substantial variability. Lack of MET:GRB2 complexes was associated with lack of response to MET TKI in cell lines and PDX models. Presence of MET:GRB2 complexes can further sub type tumors with Δexon14 MET splice variants. Presence of these complexes correlated with response to crizotinib in one patient with Δexon14 MET lacking MET gene amplification.
Conclusions
Proximity assays measuring MET:GRB2 signaling complexes provide novel insights into MET-mediated signaling and could complement current clinical genomics-based assay platforms.
Keywords: proximity ligation, cMET, crizotinib
Introduction
Subsets of lung cancer patients whose tumors harbor genomic alterations in receptor tyrosine kinases (RTK) can benefit from treatment with tyrosine kinase inhibitors (TKI). This includes targeting tumors with somatic activating mutations in the epidermal growth factor receptor (EGFR) or fusion kinases such as ALK, RET, ROS1, and NTRK. However, targeting tumors with more abundant wild type RTK protein, driven by gene amplification or by other mechanisms, has proven more difficult and therapeutic targeting of MET exemplifies this challenge (1). The MET Mab Lung Phase III trial (NCT01456325) highlighted difficulties in targeting MET in lung cancer. The trial used an immunohistochemistry (IHC) based diagnostic assay for MET protein expression as an eligibility requirement for enrollment yet no differences were observed when onartuzumab was added to erlotinib and the trial was halted due to futility (2). It is likely that the IHC-based enrollment criteria was insufficiently stringent and this experience reinvigorated a search to better identify patients whose tumors are truly dependent on MET signaling. In non-small cell lung cancer (NSCLC), MET is amplified in 3–4% of therapy naïve cases among both adenocarcinoma and squamous cell carcinoma histologies (3,4). Responses to MET TKI in patients with high MET copy number have been reported and prospective clinical trials are testing this approach (5,6). The Δexon14 splice variant, which disrupts CBL binding resulting in reduced MET degradation and elevated MET protein abundance, occurs in approximately 4% of lung adenocarcinoma and is found in up to 25% of lung sarcomatoid carcinomas (4,7,8). Patients with MET Δexon14 splice variants can respond to MET TKI and these mutations frequently co-occur with MET amplification in advanced disease and typically express high levels of MET protein by IHC (9) (10). MET activation can also occur through stromal signaling driven by its ligand hepatocyte growth factor (HGF) which can be detected by phosphorylated MET.
Based on these observations, clinical trials are now using MET copy number, Δexon14 MET alterations, and/or measurements of phosphorylated MET to enrich for patients likely to benefit from MET-directed therapies (11). There are multiple phase II trials using type I MET TKI. For example, NCT02414139 is evaluating capmantinib in pretreated NSCLC with three enrollment arms based on MET gene copy number (<4, 4≥6, and >6) in addition to a fourth arm for_METΔexon14 mutation (+) patients. Patients with MET amplification or METΔexon14 mutations are eligible for the crizotinib arm in NCI-MATCH (NCT02465060) and METROS (NCI2499614) is enrolling patients with MET amplification to receive single agent crizotinib. Newer agents such as tepotinib and savolitinib are also being evaluated in METΔexon14 mutation (+) patients (NCT02864992, NCT02897479). Type II MET TKI (such as cabozantinib, glesatinib, and merestinib) typically have a broader target profile and have recently been shown to retain activity upon acquisition of resistance to type I MET TKI (12,13). These agents are currently being evaluated either as single agents or in combination regimens. Recently, cabozantinib (either alone or in combination with erlotinib) was shown to have benefit relative to erlotinib alone in a phase II evaluation of advanced EGFR wildtype NSCLC in the 2nd and 3rd line setting (14). Beyond targeting MET as primary therapy, additional studies are targeting MET associated with acquired resistance to kinase inhibitors. MET is up-regulated as a bypass track mechanism to drive resistance to EGFR TKI through genomic amplification and HGF can mediate resistance to kinase inhibitors (15,16).
Despite these observations, it remains difficult to determine a priori what level of MET protein abundance, produced either through gene amplification or Δexon14 MET splice variants, predicts MET dependence and responsiveness to MET TKI. One approach defined the cutoff for MET gene amplification by exploiting the mutually exclusive nature of more than one driver gene aberration (17). Here we present an alternative approach of characterizing tumor cells for MET signaling-associated protein complexes. We and others have previously shown that EGFR signaling complexes can be utilized to identify active EGFR activity and stratify overall survival of NSCLC patients treated with erlotinib (18–20). Thus, we hypothesized that MET dependent tumors require sufficient activated MET protein to drive downstream signaling pathways and this requires the formation of key protein complexes that can be visualized and measured in tumors. We experimentally determined key adaptor proteins necessary for MET signaling in lung cancer cells and developed proximity ligation assays (PLA) that visualize these complexes in cancer cell lines, patient derived xenograft (PDX) models, and large cohorts of lung cancer tumor tissues (20). We provide evidence for the predictive capacity of MET:GRB2 signaling complexes and reveal unexpected discordance from genomic inferences. These findings have important implications and applicability for ongoing efforts to target MET by redefining oncogenic signaling driven by MET gene amplification and/or MET Δexon14 splice variants and determining the functional role of MET in acquired resistance to targeted agents.
Materials & Methods
Cell lines & Reagents
Sources of cell lines are as previously described (21). All cell lines have been maintained in a central bank at Moffitt Cancer Center since 2008, authenticated by STR analysis (ACTG Inc, Wheeling, IL) as of September 2010, and are routinely tested and negative for mycoplasma (PlasmoTest, InvivoGen, San Diego, CA). Experiments were conducted <15 passages post thaw. Viability assays were conducted with Cell Titer Glo assay kits (Promega) and read on Spectramax M5 plate reader. For siRNA-mediated knockdown, On-Target Smartpools (Dharmacon) were used and transfections performed with Lipofectamine RNAiMax (Invitrogen).
Xenografts and single mouse trial
Patient-derived xenograft models were created from surgically-resected lung cancers at Oncotest GmBH as described (22). Briefly, following implantation into nude mice (passage 1, P1), the tumor xenografts were passaged until establishment of stable growth patterns and stocks of early passage xenografts were frozen in liquid nitrogen. For drug treatments, serially-passaged tumors were implanted into left and right flanks in 2 mice (4 tumors per model). Crizotinib (100mg/kg) was administered twice weekly for up to 2 weeks once tumors reached ≥100mm3. Tumor measurements were performed twice weekly as described (22). Excised tumors were fixed in 10% neutral buffered formalin and embedded into paraffin. All experiments were conducted in concordance with the German animal welfare act.
MET/CEN7 FISH and Δexon14 MET mutation testing
Fluorescent in situ hybridization for MET/CEN7 was performed as described at University Hospital Göttingen and University of Colorado (3,23). Sarcomatoid specimens harboring Δexon14 MET were previously described (24). Δexon14 MET in the case reported here was identified through comprehensive genome profiling by Foundation Medicine.
Immunoprecipitation and Immunobloting
Immunoprecipitations were performed from 1.5mg cell lysates using 10ul of MET antibody/sample (clone D1C2) (Cell Signaling) as described (20). Antibodies for immunoblots were used and obtained as follows. pMETY1234/5, MET, phospho-AKTS473, phospho-AKTT308 AKT, phospho-MEK, MEK, PARP1, phospho-ERK, ERK (1:1000, Cell Signaling); GRB2 (1:500, BD); GAB1 (1:500, Santa Cruz); β-actin (1:10,000, Sigma).
Immunohistochemistry
Tissue microarrays were previously described (20). All specimens were collected through an institutionally-approved protocol with the consent of the patient and in accordance with federal ethical guidelines. Five micron sections were stained using the SP44 clone antibody on a Ventana Discovery instrument according to manufacturer’s instructions. For patient-derived xenografts, staining was performed with MET clone D1C2 (Cell Signaling) and phospho-MET clone D26 (Cell Signaling).
Proximity ligation assays
Proximity ligation assays were conducted and scored as described (20) using Duolink Far Red amplification kits (Sigma) with antibodies to MET clone D1C2 (Cell Signaling) diluted 1:300 and GRB2 clone 81 (BD) diluted 1:50. For tissue specimens, images were acquired at 200X using a PM2000 (HistoRX) epiflourescent microscope equipped with an automated stage.
Computed Tomography (CT)
Coronal and axial images were obtained on a standard-definition multi-slice 64-detector row scanner (Siemens Medical Solutions, Erlangen, Germany). Slices of 2.5 mm thickness through the chest and abdomen were collected after the administration of non-ionic iohexol intravenous contrast (Omnipaque; GE healthcare).
Patient treatment
The patient was treated with oral crizotinib 200 mg twice daily continuously, using commercial supply of crizotinib obtained through her insurance. An initial dose reduction was selected based upon her frailty and body surface area.
Statistical Analyses
For 2×2 contingency tables relating phospho-MET and MET:GRB2 complexes to MET inhibitor sensitivity, 2-tailed Fisher’s exact tests were utilized. In order to asses if association existed between MET:GRB2 complexes and the PDX response to crizotinib, we first calculated average tumor volume change (%) for each PDX models and then regressed the average % change in tumor volume on proportion of fields scored as “high” (2+/3+).
Results
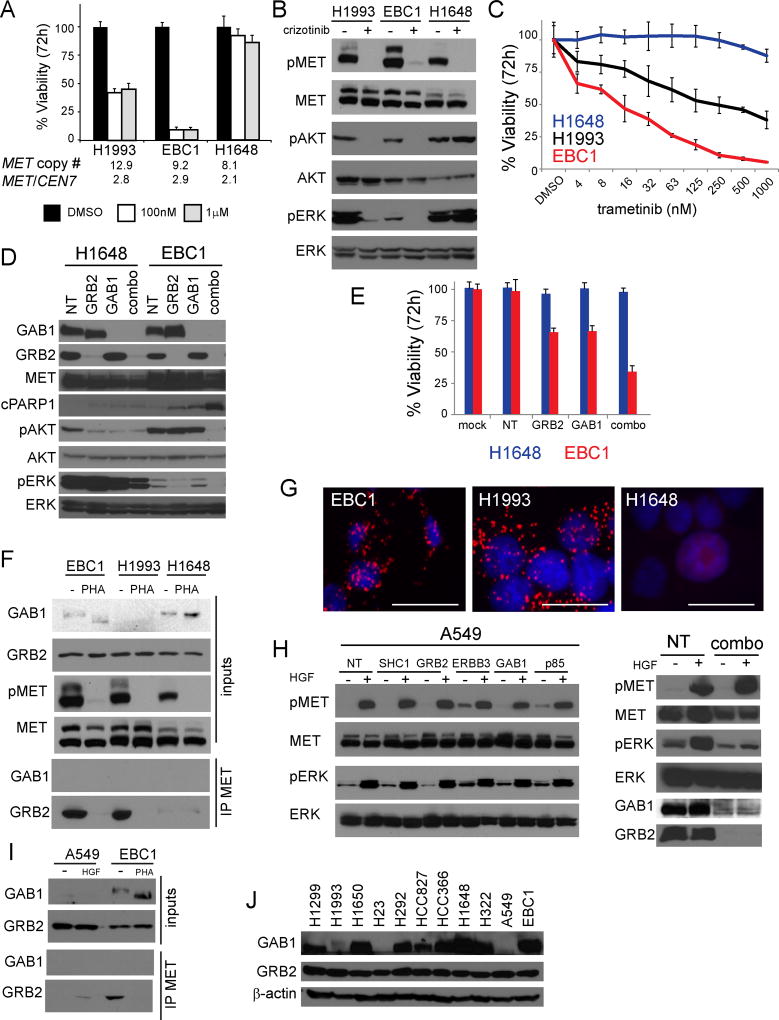
With the goal to develop assays for MET signaling-associated complexes, we first identified relevant adaptor molecules required for oncogenic MET signaling in lung cancer cells. Using three MET-amplified lung cancer cell lines with a range of sensitivities to MET kinase inhibition, we assessed viability and downstream signaling. We observed H1993 and EBC1 exhibited sensitivity to crizotinib which correlated with reductions in phosphorylated ERK (pERK) and AKT (pAKT), while H1648 were resistant and maintained phosphorylation of ERK and AKT (Fig 1A, B) (25,26). This decoupling of downstream effectors by MET TKI indicated a reliance on MAP kinase signaling, a well described feature of oncogenic drivers (27), so we next treated cells with the MEK inhibitor trametinib. We found that both H1993 and EBC1 were more sensitive to MEK inhibition than H1648, suggesting that MET dependency is mediated through the MAP kinase (MAPK) pathway (Fig 1C). Conversely, PI3K pathway inhibition with BKM120 resulted in similar dose curves in both MET TKI sensitive (H1993, EBC1) and resistant cell lines (H1648), indicating that the MET-addicted lines are not uniquely dependent on PI3K signaling (Sup Fig 1A). We next assessed downstream signaling and viability after siRNA-mediated knockdown of adaptors involved in MET signaling to MAPK. Individual knockdown of GRB2 substantially reduced pERK, while individual knockdown of GAB1 had no effect (Fig. 1D). Dual knockdown of GRB2 and GAB1 was sufficient to completely abrogate both pAKT and pERK in EBC1 cells and resulted in increased PARP cleavage consistent with induction of cell death. In H1648 cells, we observed a decrease in pAKT with individual or combination knockdown of GAB1 and GRB2 and minimal effects on pERK, which was markedly higher in the H1648 as compared to EBC1. In MET TKI sensitive EBC1 cells, knockdown of either GAB1 or GRB2 resulted in nearly 40% reduction in viability at 72hrs, while the combination resulted in a 70% reduction (Fig. 1E). In MET TKI-resistant H1648 cells, however, viability was unaffected by GRB2 or GAB1 knockdown (Fig. 1E), suggesting GAB1 and GRB2 are not involved in survival signaling in these cells. Similar experiments conducted in the MET-amplified H1993 lung adenocarcinoma cell line yielded inconclusive results due to poor transfection/knockdown efficiency and low GAB1 protein expression levels (Sup Fig. 1B). Thus, GRB2 is critical to couple MET to downstream MAPK signaling.
Figure 1. MET:GRB2 complexes are correlated with sensitivity to MET tyrosine kinase inhibitors.
A) MET-amplified NSCLC cell lines analyzed by Cell Titer Glo 72 hr after crizotinib treatment. Error bars represent standard deviation from triplicate wells of a representative experiment. B) Immunoblot analysis of downstream signaling events after crizotinib (3h 1µM); pAKT = S473 residue. C) 72hr titration of trametinib in MET-amplified NSCLC cell lines analyzed by Cell Titer Glo 72 hr. Error bars represent standard deviation from triplicate wells of a representative experiment. D) Immunoblot analysis of downstream signaling events after siRNA-mediated adaptor knockdown (72hr); pAKT = S473 residue. E) H1648 cells (MET amplified, MET TKI resistant) and EBC1 cells (MET-amplified, MET TKI sensitive) were analyzed by Cell Titer Glo 72 hr after knockdown of adaptors. Error bars represent standard deviation from triplicate wells of a representative experiment. F) Endogenous co-immunoprecipitation in MET-amplified NSCLC cell lines +/− PHA665752 (3h 1µM). G) Proximity ligation assays for MET:GRB2 complexes visualized at 200X magnification. Scale bar represents 25µm. H) Immunoblot analysis of HGF-induced activation of ERK in A549 (5’ 50ng/ml) with single knockdown of adaptors (left) or combination knockdown (right). I) Endogenous co-immunoprecipitation of HGF-stimulated A549 cells (5’ 50ng/ml) and MET-amplified EBC1 cell lines +/− PHA665752 (3h 1µM). J) Immunoblot analysis of GAB1 and GRB2 expression levels in a panel of 11 NSCLC cell lines.
We next assessed if GRB2 was in complex with MET in the MET-amplified cells sensitive to MET TKI. Using endogenous co-immunoprecipitation, we could readily identify GRB2 in association with MET in EBC1 and H1993, while the interaction was minimally detectable in H1648 cells (Fig. 1F). The MET:GRB2 interaction was abrogated by treatment with the MET inhibitor PHA665752 (PHA), while MET:GAB1 was not detectable under these conditions, We also observed a PHA-induced molecular weight shift of GAB1, which is likely due to loss of GAB1 phosphorylation (Fig S1C). We next constructed MET:GRB2 proximity ligation assays (PLA) to enable expansion of this analysis to larger cohorts of tissues and work with needle biopsy material from formalin fixed paraffin embedded material (20). We confirmed MET:GRB2 signaling complexes by PLA in EBC1 and H1993, but not H1648 (Fig 1G). To determine the role of each adaptor in the context of ligand-induced signaling, we used A549 cells that lack MET amplification, but can be stimulated by HGF. Knockdown of individual adaptors did not affect HGF-induced increases in pERK (Fig 1H, Sup 1D) and dual knockdown of GAB1 and GRB2 was required to prevent HGF-induced increases in pERK (Fig 1H). Finally, we used endogenous co-immunoprecipitation to compare levels of MET:GRB2 in EBC1 cells with non-MET-amplified A549 cells exposed to HGF. While MET:GRB2 complexes are induced by HGF in A549, the magnitude of this was markedly less compared to that observed in the EBC1 cells (Fig. 1I). In multiple experiments we were unable to detect MET:GAB1 and observed striking variation in GAB1 expression levels, a pattern that was confirmed by comparing expression levels across a panel of NSCLC cell lines (Fig 1J). Collectively, these data indicate a critical role of MET:GRB2 complexes in MET-addicted cells.
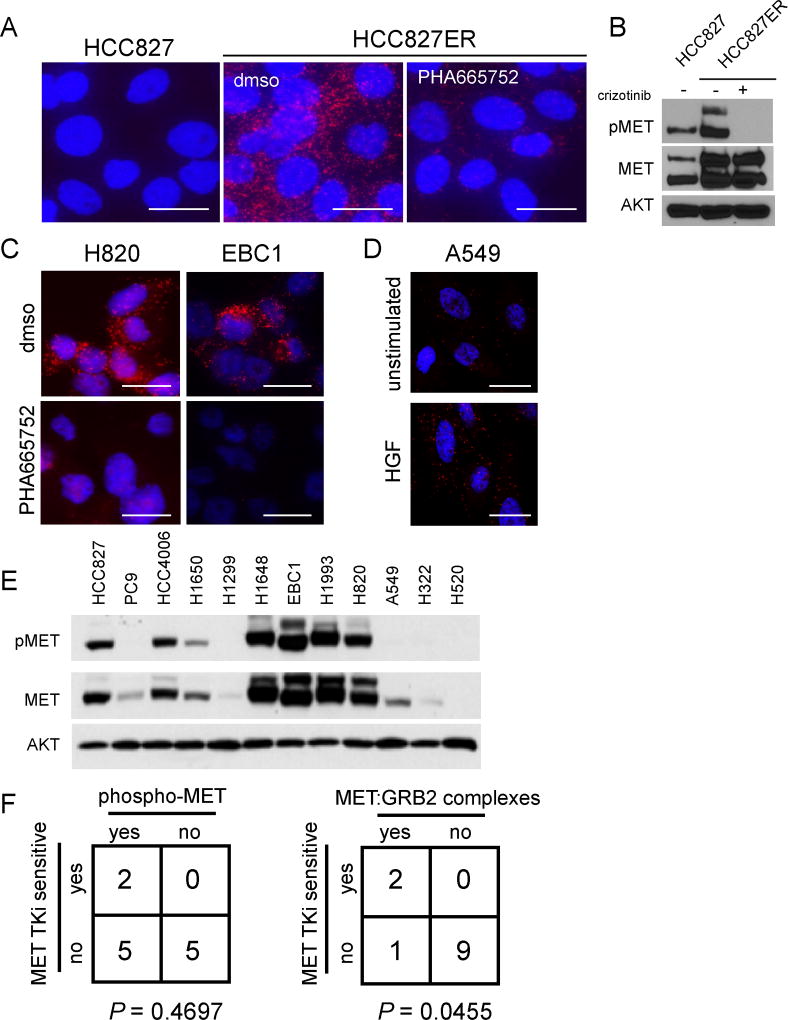
To expand assessment of MET:GRB2 PLA to predict MET signaling and sensitivity to MET TKI, we assessed MET:GRB2 complexes using PLA in additional model cell lines. We utilized HCC827 cells and the erlotinib-resistant derivative HCC827ER as a model for acquired resistance. The parental HCC827 line harbors an activating EGFR mutation and is highly sensitive to single agent EGFR TKI, while the HCC827ER derivative acquires resistance through MET amplification, yet is sensitive to dual EGFR and MET TKI (15). Using MET:GRB2 PLA, we observed complexes in the MET-amplified resistant cells, but not the parental cells. Furthermore, these complexes were substantially reduced upon treatment with PHA, indicating that our PLA reflects the actual signaling complex and not an artifact of more abundant MET protein because PHA treatment does not reduce total MET protein levels. (Fig. 2A,B). Similarly, we also observed MET:GRB2 complexes in NCI-H820 cells that have activating EGFR mutation and MET amplification and are sensitive to dual EGFR and MET TKI (28). MET TKI (PHA) reduced abundance of MET:GRB2 complexes H820 and EBC1 (Fig. 2C). HGF induced an increase in MET:GRB2 complexes (Fig. 2D), as similarly observed by IP-western blotting (Fig. 1I), but the abundance of ligand-induced signaling complexes is markedly lower than basal levels in MET-amplified cells, suggesting that the magnitude of signaling complexes is different between ligand-mediated vs. constitutively-active MET signaling. We extended our analysis to a panel of twelve NSCLC cancer cell lines in order to further explore the relationship between phosphorylated MET, presence of MET:GRB2 complexes and sensitivity patterns to MET TKI. We found that phospho-MET was detectable in multiple non-MET-amplified cell lines (Fig. 2E). We observed an association between sensitivity to MET TKI (PHA665752) and MET:GRB2 complexes detected by PLA (P = 0.0455), but not phosphorylated MET detected via immunoblotting (P = 0.4697) (Fig 2F, Supp table 1). The single discordant cell line in this analysis was the EGFR-amplified NCI-H820, which does not respond to single agent MET TKI. Thus, MET-driven lung cancer cells have abundant MET:GRB2 complexes, which maintain downstream signaling and impart sensitivity to MET TKI.
Figure 2. MET:GRB2 complexes reflect signaling activity and MET TKI sensitivity in NSCLC cell lines.
A) in situ PLA was used to analyze MET:GRB2 complexes in HCC827 and erlotinib-resistant, MET-amplified derivative HCC827ER +/− MET TKI B) Immunoblot showing phospho- and total MET protein levels in HCC827 and HCC827ER cell lines. C) MET:GRB2 complexes in MET-amplified NSCLC cell lines H820 and EBC1 +/− MET TKI (3h 1µM), and D) A549 cells +/− HGF (5’ 100ng/ml). Data shown are representative of three independent experiments. E) Immunoblot showing phospho- and total MET protein levels in a panel of NSCLC cell lines. F) Chi-square analysis of relationship between sensitivity to PHA665,752 and presence of phospho-MET (left) or MET:GRB2 complexes (right) in NSCLC cell lines. P-values shown are derived from a two-tailed Fisher’s exact test.
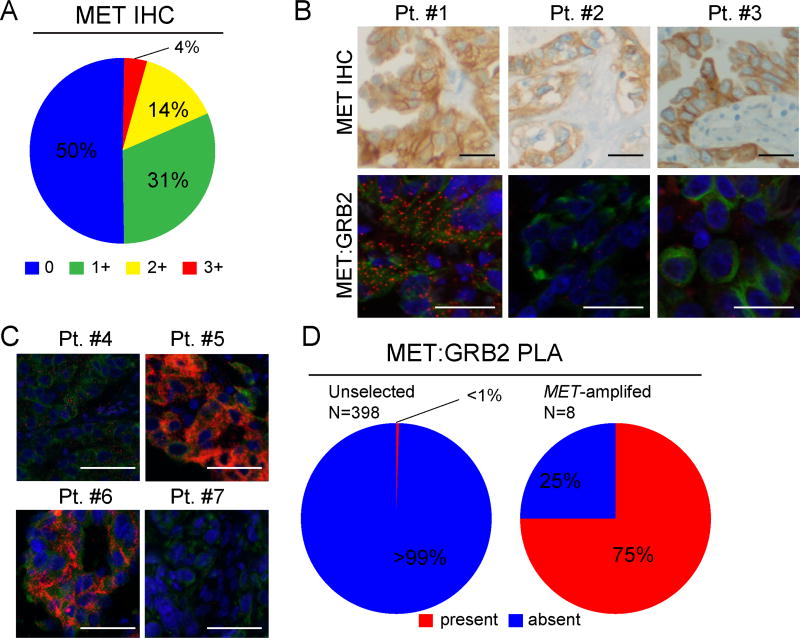
These complexes are detectable by PLA, which facilitates analysis of patient tissue specimens to identify MET-driven tumors. We next surveyed multiple cohorts of NSCLC and other solid tumors using tissue microarrays, surgical resections and biopsy specimens in order to determine the prevalence of MET:GRB2 complexes in human tumor tissues. Because MET has been pursued as a clinical target in multiple cancers, we initially examined a series of 308 PDX tissue microarrays, which included duplicate spots of a variety of solid tumors (Fig. S2). Surprisingly, we found very low prevalence of MET:GRB2 complexes across the cohort of tumors, with only 2 clear cell renal models and 4 lung models having abundant foci. No activating mutations were identified in MET in these models. This low prevalence was markedly lower than previously observed using EGFR:GRB2 PLA in the same PDX cohort (20). We next surveyed multiple tissue microarrays from predominantly early stage NSCLC patients. We concurrently performed MET IHC and MET:GRB2 PLA to assess the relationship between MET protein expression and presence of MET signaling complexes. Using a 0–3+ scoring algorithm, we scored 398 patients as: 50% “0”, 31% “1+”, 15% “2+” and 4% 3+ (Fig 3A). No specimens lacking MET expression by IHC contained MET:GRB2 signaling complexes and only three of 78 that expressed high levels of MET protein (as measured by 2+ or 3+) had detectable MET:GRB2 complexes (Fig. 3B,D). Thus, elevated MET protein expression is necessary, but not sufficient for signaling complex formation. We then evaluated MET:GRB2 complexes in MET-amplified NSCLC specimens from biopsies and surgical resections with FISH-verified mean MET/CEN7 ratios ranging from (3.7 to 7.4). Consistent with results from our cell line studies, we found that 75% (6/8) of these specimens had detectable MET:GRB2 complexes (Fig. 3C,D). Notably, the abundance of these signaling complexes varied considerably across and within specimens and no correlation between average MET copy number and abundance was identified.
Figure 3. MET:GRB2 complexes are infrequent in human NSCLC patient specimens.
A) Enumeration of IHC scores for MET (N=398). B Representative images comparing similar regions of patient FFPE tumor specimens. Upper panels show MET protein expression as detected by SP44 antibody. Lower panels show MET:GRB2 complexes in red detected by in situ PLA, with cytokeratin in green acquired at 200X. C) Representative images from a second cohort of patients with MET high level amplification, defined MET/CEN7 > 5 detected by FISH. D) Frequencies of detectable MET:GRB2 complexes in human NSCLC cohorts. Scale bars are 25µm (A-C,E) and 50µm (F).
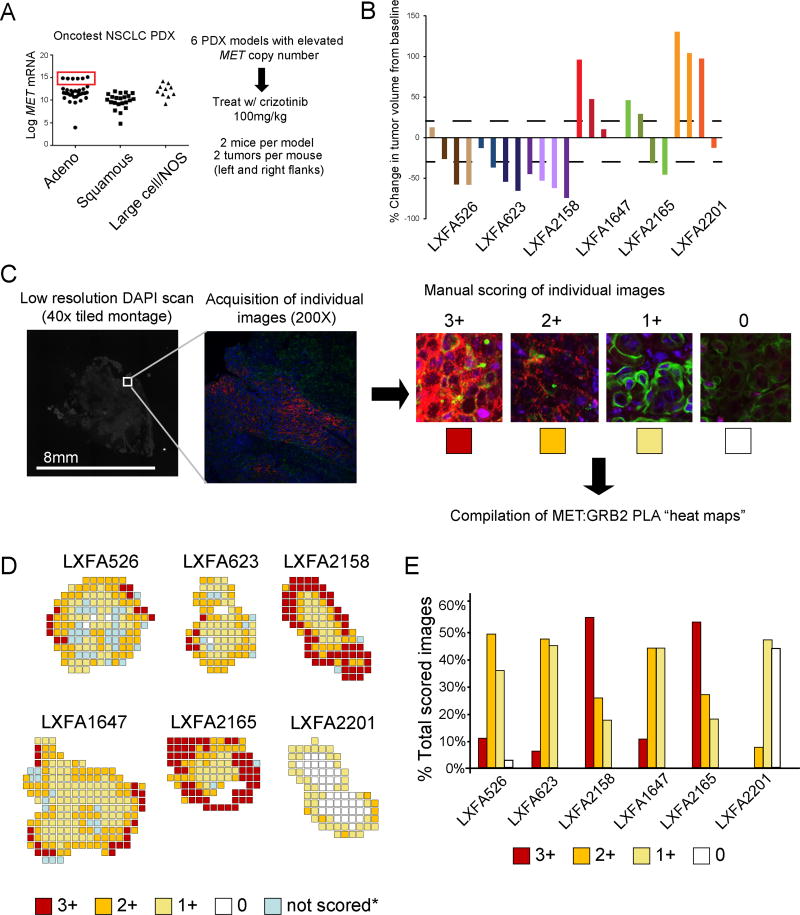
To address the relationship between MET:GRB2 complexes and response to MET TKI we performed a “single mouse trial” in six MET-amplified PDX models of lung adenocarcinoma, which is consistent with ongoing clinical trial patient enrichment strategies. These PDX models lacked ALK translocations and had between 4–14 copies of MET with similar mRNA expression levels, which were approximately 3-logs higher than other available lung adenocarcinoma models as measured by Affymetrix array (Fig. 4A). No mutations in MET, EGFR or KRAS were detected and mutational burden ranged from 829 to 1440 (Supp. Table 2). After serial passage, we established duplicate bilateral xenografts into female NMRI nu/nu mice and initiated treatment with crizotinib. We observed variable responses to crizotinib both between and within these PDX models and one model (LXFA2201) exhibited innate resistance to crizotinib in three of four implanted tumors (Fig 4B). To characterize MET signaling activity in these models, we utilized tumors from an untreated animal obtained at Day 0 to mimic pre-treatment biopsies. We performed H&E staining, IHC for total MET and phosphorylated METTyr1234/5 (pMET) and also evaluated MET:GRB2 complexes by PLA. In all six models we found uniformly high intensity MET staining, while pMET was also readily detectable but less intense (Fig. S3). Using MET:GRB2 PLA on whole tissue sections, we were able to analyze the entire tumor through a series of tiled images which were manually scored to generate geographical “heat maps” of MET:GRB2 intensity (Fig. 4C). We observed extensive heterogeneity in signaling complexes both within and between specimens (Fig 4D, Fig, S4). In the MET TKI resistant model (LXFA2201), we observed a markedly lower percentage of fields scored as 2+ or 3+ (Fig. 3E), suggesting less MET dependence. A regression model indicated a negative association between the proportion of high (2+/3+) MET:GRB2 PLA and percent change in tumor volume with clear segregation of the LXFA2201 model (Fig. S5). Although we were underpowered to reach statistical significance (P=0.08) due to the small number MET-amplified PDX models available, such an approach will likely have increased utility in larger cohorts.
Figure 4. MET:GRB2 signaling complexes correlate with response to crizotinib in vivo.
A) A subset of adenocarcinoma PDX models exhibit MET overexpression due to copy number variation. These PDX models (N=6) were used to test initial response to crizotinib. B) Waterfall plots showing individual tumor responses in response to crizotinib (duration 6–11 days). Two tumors per mouse, two mice each (N=4, per PDX model). Identical colors signify that tumors are from same mouse. C) Schematic for construction of PLA heatmaps with examples of scoring bins. Each 2048x2048 image was manual scored in order to compile color-coded heat maps D) MET:GRB2 PLA heat maps from the “time 0” untreated PDX in (B) used to mimic the pretreatment biopsy. Individual images were mapped back to location within whole tumor. E) Distribution of PLA scores for each model, shown as a percentage of total number images scored for each tumor.
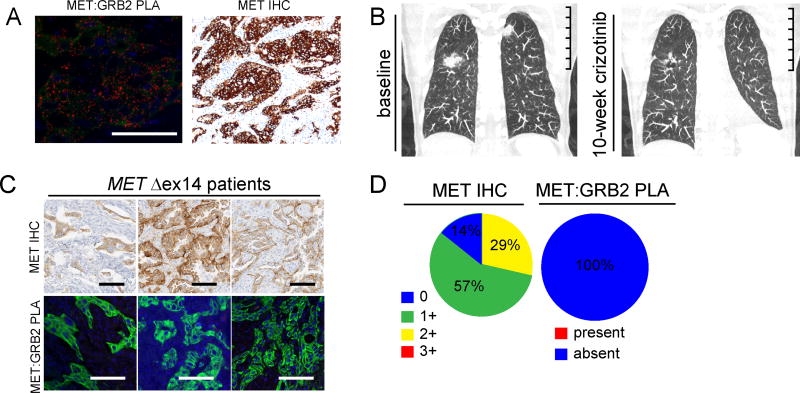
Based on responses to MET TKI in patients harboring MET Δexon14 mutations we analyzed patient tissues and cellular models of this genetic lesion (9,29,30). We identified an untreated 74-year-old female former smoker with stage IV lung adenocarcinoma with bilateral lung metastases harboring a c. 2888–70_2976>18 MET Δexon14 splice site variant (MAF of 14%) without evidence of MET copy number gain. A pretreatment biopsy revealed abundant MET:GRB2 complexes by PLA and high MET abundance by IHC (H-score 270) (Fig. 5A). She began first-line treatment with crizotinib, and had significant improvement of cancer-related inanition and dyspnea by week 2. Follow-up 10-week CT scan showed interval shrinkage in lung metastases (Fig 5B). We expanded this analysis to seven early-stage MET Δexon14 tumors (six sarcomatoid carcinoma and one adenocarcinoma), all of which had received surgery as the primary treatment modality. None of the seven had received neo-adjuvant chemotherapy and all were naïve to targeted therapy regimens (Sup table 3). Surprisingly, all seven specimens lacked detectable MET:GRB2 complexes. Since some early-stage MET Δexon14 tumors have been shown to lack high levels of MET protein expression (29), we hypothesized that the absence of detectable MET:GRB2 complexes in our MET Δexon14 cohort was due to insufficient expression of MET. Subsequent MET IHC analysis failed to detect high levels of MET protein in all seven surgically-resected specimens (Fig 5C,D). Thus, similar to the study by Awad and colleagues (29), not all lung cancers harboring MET Δexon14 mutations express high levels of MET protein.
Figure 5. MET:GRB2 signaling complexes in METΔex14 specimens.
A) Representative micrographs of baseline tumor showing MET:GRB2 PLA (left) and MET IHC (right). Scale bars 50µm. B) Baseline and 10-week coronal CT scan from patient (MET c.2888-70_2976>18 exon 14 splice variant) following treatment with crizotinib. Imaging shows shrinkage in lung metastases. Scale bar is 10cm. C) Example images of MET:GRB2 PLA and MET IHC in METΔex14 expressing lung adenocarcinoma and sarcomatoid tumors. Images shown are taken from same region from sequential slides; scale bars in both sets represent 100 µm. D) Summation of IHC and MET:GRB2 PLA analysis.
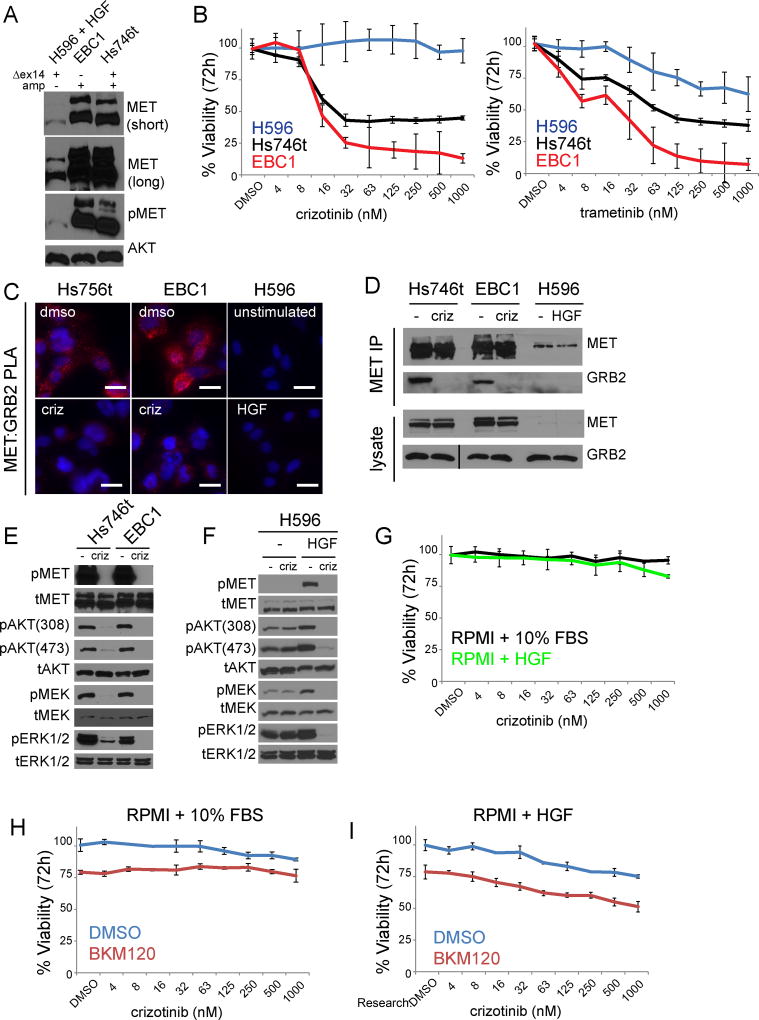
A review of multiple case reports describing responses to single agent MET TKI in patients harboring MET Δexon14 mutations revealed that co-existence of MET amplification is frequent (9–11,29–31). Furthermore, when MET IHC is reported, it is uniformly high among responders. Thus, we extended our analysis further using two MET Δexon14 cellular models: the lung adenosquamous line H596 which harbors a MET Δexon14 mutation and the gastric line Hs746t which has both MET Δexon14 mutation and MET gene amplification. While both cell lines express a smaller MET protein than MET wildtype cell lines, the expression levels of total and phosphorylated MET are markedly lower in H596, relative to Hs746t (Fig 6A). In viability assays the H596 was resistant to crizotinib, while Hs746t was sensitive, and H596 was more resistant to MEK inhibition than both Hs746t and EBC1, suggesting differential reliance on the MAP kinase pathway for proliferation (Fig 6B). By PLA and co-immunoprecipitation, Hs746t have abundant MET:GRB2 complexes (similar to EBC1), which are abrogated by crizotinib. We were unable to detect MET:GRB2 interactions in H596 cells either under basal conditions or upon HGF stimulation (Fig 6C,D). Under basal conditions Hs746t and EBC1 cells exhibited loss of downstream signaling, measured by phosphorylated ERK and AKT, in response to crizotinib, while no such changes were observed in H596 cells (Fig 6E,F). H596 cells exhibited disruption of downstream signaling only in the presence of HGF, but this had no effect on cellular viability in response to crizotinib (Fig 6F,G). One reason for the lack of sensitivity in H596 could be the co-existence of a PIK3CA mutation (32). However, we did not observe maintenance of PI3K signaling (as assessed by phosphorylated AKTT308) due to the presence of the PIK3CA mutation in this cell line, which has been reported to results in a synergistic enhancement in sensitivity to dual MET/PI3K inhibition (24). Consistent with this, we found that addition of the PI3K inhibitor BKM120 resulted in only additive effects when combine with crizotinib in both full serum media and HGF-only media (Fig. 6H,I). Thus, HGF stimulation transiently activates downstream signaling, but does not induce a switch to MET-mediated proliferation in H596 cells. We conclude that high level protein expression is required for constitutive MET:GRB2 complex formation and TKI sensitivity in Δexon14 mutations, but is not universally present in all MET Δexon14 tumor tissues.
Figure 6. METΔexon14 mutation alone is not sufficient to impart sensitivity to MET TKI.
A) Immunoblot analysis of equal amounts of lysate (50µg) METΔex14 cell lines run on 6% acrylaminde gel. Total AKT serves as loading control. B) MET-amplified and METΔex14 expressing NSCLC cell lines analyzed by Cell Titer Glo 72 hr after crizotinib (left) and trametinib (right) treatment. Error bars represent standard deviation from triplicate wells of a representative experiment. C) PLA analysis of MET-amplified and METΔex14 expressing NSCLC cell lines, +/− 1µM crizotinib for 3hrs (Hs746t, EBC1) or +/− 100ng/ml HGF for 5 minutes (H596). Images are representative of three independent experiments, scale bars represent 20µm. D) Endogenous co-immunoprecipitation in MET-amplified NSCLC and METΔex14 expressing cell lines +/− crizotinib (3h 1µM). Line in total GRB2 blot represents second “crop box”, which was required due to a tear in acrylamide gel producing an artificial molecular weight shift. Image is the same exposure. E) Immunoblot showing downstream signaling in wt and ex14 mutant MET amplified lines, +/− crizotinib (3h 1µM). F Immunoblot showing downstream signaling in non-amplified MET ex14 mutant H596 cells +/− crizotinib (3h 1 µM) cultured in either RPMI1640 + 10% FBS or serum-free RPMI1640, then stimulated with HGF (100ng/ml, 5 minutes). G) H596 cells analyzed by Cell Titer Glo 72 hr after crizotinib cultured in either RPMI1640 + 10% FBS or serum-free RPMI1640 + 100ng/ml HGF. Each curve separately normalized to DMSO. Error bars represent standard deviation from triplicate wells of a representative experiment. H,I) H596 cells analyzed by Cell Titer Glo 72 hr after crizotinib cultured +/− 500nM BKM120 in either RPMI1640 + 10% FBS (H) or serum-free RPMI1640 + 100ng/ml HGF (I). Each curve normalized to the “double DMSO” in order to visualize the effect of BKM120 in the absence of crizotinib. Error bars represent standard deviation from triplicate wells of a representative experiment.
Discussion
We demonstrate that MET:GRB2 protein complex abundance predicts MET survival signaling and correlates with sensitivity to MET TKI in MET-driven cellular models of lung cancer. The low frequency of MET:GRB2 complexes in lung cancer tumor specimens suggests that MET is likely not acting as an oncogenic driver in most circumstance and is consistent with low response rates observed in unselected patients with single agent MET inhibition. Moreover, our data demonstrate that some tumors with high levels of MET gene amplification or harboring Δexon14 splice variants are insufficient to produce MET signaling-associated protein complexes and therefore do not to impart sensitivity to MET TKI.
Only within subsets of MET-amplified cell line models, PDX models, or human tumor tissues did we observe clear evidence of MET:GRB2 complexes. We failed to observed strong MET:GRB2 complexes upon HGF stimulation suggesting that ligand stimulation provides a weak stimulus as compared to gene amplification. Furthermore, we observed large variability in the abundance of MET:GRB2 complexes even among similar high MET copy number samples. Our data from MET-amplified cell lines, patient specimens and crizotinib-treated MET amplified PDX models suggests that even among MET amplified subsets, responses to MET TKI will vary considerably. Our cell line and PDX models, although limited in number, show a correlation between presence of complexes and sensitivity to MET TKI. Despite presence of MET alterations used to predict MET therapy sensitivity, through either copy number gain (H1648) or exon 14 mutations (H596), none of these cell line models exhibited MET:GRB2 complexes and were resistant to MET TKI. Our PDX model with overt progression (LXFA2201) had markedly lower abundance of MET:GRB2 complexes, yet was indistinguishable from other models by MET copy number, protein expression, and phosphorylated MET. These observations are consistent with early clinical experience using crizotinib to treat MET-amplified lung cancer patients and raise the possibility of using PLA to better discern patients with MET amplification receiving benefit from MET targeting agents.
Although we identified MET:GRB2 complexes in one advanced-stage patient who responded to crizotinib, we were originally surprised by the lack of observable MET:GRB2 complexes in the additional tumor samples and cells with Δexon14 MET splice variants. We believe this reflects insufficient abundance of MET protein necessary to form signaling complexes, as our analysis of patient specimens harboring Δexon14 MET mutations revealed only 1 of 7 specimens would have met the criteria for positivity in the MetMab lung trial. Similar observations have been reported demonstrating lower abundance of MET protein by IHC in early stage lung cancers with MET Δexon14 splice variants (29). We also observed this in the H596 cell line, which was derived from a surgically-resected lesion harboring a MET Δexon14 mutation but lacking MET gene amplification. Although the H596 cell line harbors an additional mutation in PIK3CA that has been reported to provide a bypass survival signal (24), our data show minimal effects of combined MET and PI3K inhibition. Mutations in PIK3CA are known to co-occur with other oncogene drivers and have been found in responders to both MET and EGFR TKI, suggesting that in some cases they are not sufficient to provide bypass survival signaling (32–34). To the best of our knowledge, all reported MET Δexon14 patients with clinical responses to MET TKI were advanced stage and (when reported) exhibited high levels of MET protein by IHC (9,29,30). Metastatic cells may gain additional fitness through mechanisms that further increase MET protein level, either through MET copy gain, which co-exists with MET Δexon14 mutations in up to 50% or through other transcriptional or epigenetic mechanisms that augment MET protein abundance. It will be important to determine if MET:GRB2 complexes correlate with duration of response in patients with MET Δexon14 mutations and future studies will address this.
While nucleic acid-based methodologies are now well-established in clinical practice, emerging methods to characterize protein function in clinical specimens could redefine genomic subclasses and provide additional precision to predictive medicine. Engagement of adaptors is necessary for activation of downstream signaling by RTK and diagnostic methodologies to quantify these signaling complexes represents a promising new strategy for patient enrichment. Moreover, this approach might have broad applicability to targeting other oncogenic RTKs, such as fibroblast growth factor receptors (FGFR), where early clinical data indicates limited responses despite enrichment through gene copy number assessment (35). Recent methodological advancements have enabled the adaptation of assays measuring protein complexes to CLIA-compliant automated stainers used in clinical pathology labs worldwide (36). We envision that this approach can be combined with existing assay technologies in prospective correlative studies in order to clinically validate the detection of protein complexes and their association with response to targeted therapies.
Supplementary Material
Translational Relevance.
Small molecule inhibition of MET has been evaluated in multiple solid malignancies on the basis of preclinical studies. Despite setbacks, multiple trials are ongoing and current patient stratification strategies are using MET copy number alterations and presence of Δexon14 MET splice variants for enrollment. Here, we utilize proximity ligation assays to define MET activity in lung cancer. We show that MET:GRB2 signaling-associated complexes are predictive of response to MET kinase inhibitors and offer a novel approach to annotate MET signaling activity in tumors with MET gene amplification or Δexon14 MET splice variants. These assays could augment patient stratification strategies in ongoing clinical trials investigating MET-directed targeted therapies.
Acknowledgments
Grant support: This work was supported by the Florida Department of Health through the Bankhead-Coley program (5BC07, awarded to E.B.H.)
References
- 1.Haura EB, Smith MA. Signaling Control by Epidermal Growth Factor Receptor and MET: Rationale for Cotargeting Strategies in Lung Cancer. Journal of Clinical Oncology. 2013;31(32):4148–50. doi: 10.1200/jco.2013.50.8234. [DOI] [PubMed] [Google Scholar]
- 2.Spigel D, Edelman M, O'Byrne K, Paz-Ares L, Shames D, Yu W, et al. Onartuzumab plus erlotinib versus erlotinib in previously treated stage IIIb or IV NSCLC: Results from the pivotal phase III randomized, multicenter, placebo-controlled METLung (OAM4971g) global trial. Journal of Clinical Oncology, 2014 ASCO Annual Meeting Abstracts. 2014;32 (No 15_suppl (May 20 Supplement)) [Google Scholar]
- 3.Schildhaus H-U, Schultheis AM, Rüschoff J, Binot E, Merkelbach-Bruse S, Fassunke J, et al. MET Amplification Status in Therapy-Naïve Adeno- and Squamous Cell Carcinomas of the Lung. Clinical Cancer Research. 2015;21(4):907–15. doi: 10.1158/1078-0432.ccr-14-0450. [DOI] [PubMed] [Google Scholar]
- 4.Tong JH, Yeung SF, Chan AW, Chung LY, Chau SL, Lung RWM, et al. MET amplification and exon 14 splice site mutation define unique molecular subgroups of Non-small Cell Lung Carcinoma with poor prognosis. Clinical Cancer Research. 2016 doi: 10.1158/1078-0432.ccr-15-2061. [DOI] [PubMed] [Google Scholar]
- 5.Caparica R, Yen CT, Coudry R, Ignatius Ou S-H, Varella-Garcia M, Camidge DR, et al. Responses to crizotinib can occur in high level MET-amplified non-small cell lung cancer independent of MET exon 14 alterations: A brief report. Journal of Thoracic Oncology. 2016 doi: 10.1016/j.jtho.2016.09.116. [DOI] [PubMed] [Google Scholar]
- 6.Ou S-HI, Kwak EL, Siwak-Tapp C, Dy J, Bergethon K, Clark JW, et al. Activity of Crizotinib (PF02341066), a Dual Mesenchymal-Epithelial Transition (MET) and Anaplastic Lymphoma Kinase (ALK) Inhibitor, in a Non-small Cell Lung Cancer Patient with De Novo MET Amplification. Journal of Thoracic Oncology. 2011;6(5):942–6. doi: 10.1097/JTO.0b013e31821528d3. [DOI] [PubMed] [Google Scholar]
- 7.The Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma PC, Jagadeeswaran R, Jagadeesh S, Tretiakova MS, Nallasura V, Fox EA, et al. Functional Expression and Mutations of c-Met and Its Therapeutic Inhibition with SU11274 and Small Interfering RNA in Non–Small Cell Lung Cancer. Cancer Research. 2005;65(4):1479–88. doi: 10.1158/0008-5472.can-04-2650. [DOI] [PubMed] [Google Scholar]
- 9.Paik PK, Drilon A, Fan P-D, Yu H, Rekhtman N, Ginsberg MS, et al. Response to MET Inhibitors in Patients with Stage IV Lung Adenocarcinomas Harboring MET Mutations Causing Exon 14 Skipping. Cancer Discovery. 2015;5(8):842–9. doi: 10.1158/2159-8290.cd-14-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schrock AB, Frampton GM, Suh J, Chalmers ZR, Rosenzweig M, Erlich RL, et al. Characterization of 298 Patients with Lung Cancer Harboring MET Exon 14 Skipping Alterations. Journal of Thoracic Oncology. 2016;11(9):1493–502. doi: 10.1016/j.jtho.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Reungwetwattana T, Liang Y, Zhu V, Ou S-HI. The race to target <em>MET</em> exon 14 skipping alterations in non-small cell lung cancer: The Why, the How, the Who, the Unknown, and the Inevitable. Lung Cancer. 2017;103:27–37. doi: 10.1016/j.lungcan.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 12.Bahcall M, Sim T, Paweletz CP, Patel JD, Alden RS, Kuang Y, et al. Acquired <em>MET</em><sup>D1228V</sup> Mutation and Resistance to MET Inhibition in Lung Cancer. Cancer Discovery. 2016;6(12):1334–41. doi: 10.1158/2159-8290.CD-16-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li A-N, Yang J, Zhang X-C, Zhang Z, Su J, Gou L-Y, et al. Acquired MET Y1248H and D1246N mutations mediate resistance to MET inhibitors in non-small cell lung cancer. Clinical Cancer Research. 2017 doi: 10.1158/1078-0432.ccr-16-3273. [DOI] [PubMed] [Google Scholar]
- 14.Neal JW, Dahlberg SE, Wakelee HA, Aisner SC, Bowden M, Huang Y, et al. Erlotinib, cabozantinib, or erlotinib plus cabozantinib as second-line or third-line treatment of patients with <em>EGFR</em> wild-type advanced non-small-cell lung cancer (ECOG-ACRIN 1512): a randomised, controlled, open-label, multicentre, phase 2 trial. The Lancet Oncology. 17(12):1661–71. doi: 10.1016/s1470-2045(16)30561-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–43. doi: 10.1126/science.1141478. doi 1141478. [DOI] [PubMed] [Google Scholar]
- 16.Harbinski F, Craig VJ, Sanghavi S, Jeffery D, Liu L, Sheppard KA, et al. Rescue screens with secreted proteins reveal compensatory potential of receptor tyrosine kinases in driving cancer growth. Cancer Discovery. 2012;2(10):948–59. doi: 10.1158/2159-8290.CD-12-0237. [DOI] [PubMed] [Google Scholar]
- 17.Noonan SA, Berry L, Lu X, Gao D, Barón AE, Chesnut P, et al. Identifying the Appropriate FISH Criteria for Defining MET Copy Number-Driven Lung Adenocarcinoma through Oncogene Overlap Analysis. Journal of Thoracic Oncology. 2016;11(8):1293–304. doi: 10.1016/j.jtho.2016.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaishnavi A, Schubert L, Rix U, Marek LA, Le AT, Keysar S, et al. EGFR mediates responses to small molecule drugs targeting oncogenic fusion kinases. Cancer Research. 2017 doi: 10.1158/0008-5472.can-17-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toki MI, Carvajal-Hausdorf DE, Altan M, McLaughlin J, Henick B, Schalper KA, et al. EGFR-GRB2 Protein Colocalization Is a Prognostic Factor Unrelated to Overall EGFR Expression or EGFR Mutation in Lung Adenocarcinoma. Journal of Thoracic Oncology. 2016;11(11):1901–11. doi: 10.1016/j.jtho.2016.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith MA, Hall R, Fisher K, Haake SM, Khalil F, Schabath M, et al. Annotation of human cancers with EGFR signaling-associated complexes using proximity ligation assays. Science Signaling. 2015;8(r4) doi: 10.1126/scisignal.2005906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Bennett K, Stukalov A, Fang B, Zhang G, Yoshida T, et al. Perturbation of the mutated EGFR interactome identifies vulnerabilities and resistance mechanisms. Molecular Systems Biology. 2013;9:705. doi: 10.1038/msb.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krumbach R, Schüler J, Hofmann M, Giesemann T, Fiebig H-H, Beckers T. Primary resistance to cetuximab in a panel of patient-derived tumour xenograft models: Activation of MET as one mechanism for drug resistance. European Journal of Cancer. 2011;47(8):1231–43. doi: 10.1016/j.ejca.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 23.Zucali PA, Ruiz MG, Giovannetti E, Destro A, Varella-Garcia M, Floor K, et al. Role of cMET expression in non-small-cell lung cancer patients treated with EGFR tyrosine kinase inhibitors. Annals of Oncology. 2008;19(9):1605–12. doi: 10.1093/annonc/mdn240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Jia Y, Stoopler MB, Shen Y, Cheng H, Chen J, et al. Next-Generation Sequencing of Pulmonary Sarcomatoid Carcinoma Reveals High Frequency of Actionable MET Gene Mutations. Journal of Clinical Oncology. 2016;34(8):794–802. doi: 10.1200/JCO.2015.62.0674. [DOI] [PubMed] [Google Scholar]
- 25.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–307. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y-W, Staal B, Essenburg C, Lewis S, Kaufman D, Vande Woude GF. Strengthening Context-Dependent Anticancer Effects on Non–Small Cell Lung Carcinoma by Inhibition of Both MET and EGFR. Molecular Cancer Therapeutics. 2013;12(8):1429–41. doi: 10.1158/1535-7163.MCT-13-0016. [DOI] [PubMed] [Google Scholar]
- 27.Hrustanovic G, Olivas V, Pazarentzos E, Tulpule A, Asthana S, Blakely CM, et al. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nat Med. 2015;21(9):1038–47. doi: 10.1038/nm.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bean J, Brennan C, Shih J-Y, Riely G, Viale A, Wang L, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proceedings of the National Academy of Sciences. 2007;104(52):20932–7. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Awad MM, Oxnard GR, Jackman DM, Savukoski DO, Hall D, Shivdasani P, et al. MET Exon 14 Mutations in Non–Small-Cell Lung Cancer Are Associated With Advanced Age and Stage-Dependent MET Genomic Amplification and c-Met Overexpression. Journal of Clinical Oncology. 2016;34(7):721–30. doi: 10.1200/JCO.2015.63.4600. [DOI] [PubMed] [Google Scholar]
- 30.Frampton GM, Ali SM, Rosenzweig M, Chmielecki J, Lu X, Bauer TM, et al. Activation of MET via Diverse Exon 14 Splicing Alterations Occurs in Multiple Tumor Types and Confers Clinical Sensitivity to MET Inhibitors. Cancer Discovery. 2015;5(8):850–9. doi: 10.1158/2159-8290.CD-15-0285. [DOI] [PubMed] [Google Scholar]
- 31.Van Der Steen N, Giovannetti E, Pauwels P, Peters GJ, Hong DS, Cappuzzo F, et al. cMET Exon 14 Skipping: From the Structure to the Clinic. Journal of Thoracic Oncology. 11(9):1423–32. doi: 10.1016/j.jtho.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Jorge SE, Schulman S, Freed JA, VanderLaan PA, Rangachari D, Kobayashi SS, et al. Responses to the multitargeted MET/ALK/ROS1 inhibitor crizotinib and co-occurring mutations in lung adenocarcinomas with <em>MET</em> amplification or <em>MET</em> exon 14 skipping mutation. Lung Cancer. 2015;90(3):369–74. doi: 10.1016/j.lungcan.2015.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaft JE, Arcila ME, Paik PK, Lau C, Riely GJ, Pietanza MC, et al. Coexistence of <em>PIK3CA</em> and Other Oncogene Mutations in Lung Adenocarcinoma–Rationale for Comprehensive Mutation Profiling. Molecular Cancer Therapeutics. 2012;11(2):485–91. doi: 10.1158/1535-7163.MCT-11-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramirez M, Rajaram S, Steininger RJ, Osipchuk D, Roth MA, Morinishi LS, et al. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nature Communications. 2016;7:10690. doi: 10.1038/ncomms10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim SH, Sun J-M, Choi Y-L, Kim HR, Ahn S, Lee JY, et al. Efficacy and safety of dovitinib in pretreated patients with advanced squamous non-small cell lung cancer with FGFR1 amplification: A single-arm, phase 2 study. Cancer. 2016;122(19):3024–31. doi: 10.1002/cncr.30135. [DOI] [PubMed] [Google Scholar]
- 36.Hong R, Roberts E, Bieniarz C. In Situ Detection of Protein Complexes and Modifications by Chemical Ligation Proximity Assay. Bioconjugate Chemistry. 2016;27(7):1690–60. doi: 10.1021/acs.bioconjchem.6b00230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.