Abstract
The longhorn beetle Dorysthenes paradoxus (Faldermann, 1833) (Coleoptera: Cerambycidae) is not only a serious agricultural pest but also a traditionally edible insect in China. However, no genetic information on this species has been acquired. In the present study, we report the mitochondrial genome (mitogenome) of Do. paradoxus, as the first complete mitogenome of Prioninae. The circular mitogenome of 15,922 bp encodes 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), and two ribosomal RNAs (rRNAs), and it contains an A+T-rich region. This mitogenome exhibits the lowest A+T content (71.13%) but harbors the largest AT skew (0.116) among the completely sequenced Cerambycidae species. Eleven of the 13 PCGs have a typical ATN start codon, whereas COI and ND1 are tentatively designated by AAT and TTG, respectively. Only 4 of the 13 PCGs harbor a complete termination codon, and the remaining 9 possess incomplete termination codons (T or TA). Apart from tRNASer(AGN), the other 21 tRNAs can fold into a typical clover-leaf secondary structures. The Do. paradoxus A+T-rich region contains two poly-T stretches and a tandem repeat that comprises two 47-bp-long copies. Both Bayesian inference and Maximum likelihood analyses confirmed the subfamily ranks of Cerambycidae ([Prioninae + Cerambycinae] + Lamiinae) and the close relationship between Philinae and Prioninae/Cerambycinae. However, the data did not support the monophyly of Prioninae and Cerambycinae. The mitogenome presented here provides basic genetic information for this economically important species.
Keywords: Dorysthenes paradoxus, edible insect, phylogenetic analysis, prionini
The beetles are the most diverse group of insects with over 360,000 described species worldwide (Sheffield et al. 2008), representing approximately 40% of all insect species described to date and approximately 25% of all animals (Hunt et al. 2007). Beetles are prominent in human culture: some are serious agricultural, forestry, and horticultural pests, and over 300 species are used as human food. The ecological and morphological diversity in Coleoptera have attracted the attention of biologists (Hunt et al. 2007), as has the phylogenetic relationship within this order (Zhang et al. 2016). esearchers have attempted to sequence the mitochondrial genome (mitogenome) as the reference sequence for target species, which is widely considered an effective molecular marker for research on phylogenetic analysis and phylogeography (Simon et al. 2006). In recent years, the number of coleopteran mitogenome sequences has increased dramatically in the GenBank database. Cerambycidae is a large family in the Coleoptera order, and more than 25,000 species of Cerambycidae have been described worldwide (Sama et al. 2010). Currently, 12 complete mitogenomes of Cerambycidae species are available in GenBank. Among these, seven are in the subfamily Lamiinae, three are in the subfamily Cerambycinae, one is in the subfamily Lepturinae, and one is in subfamily Prioninae (Table 1). Information about the subfamily Prioninae mitogenome is still limited. One partial mitogenome in Prioninae is available, but the taxonomic status of the specimen sequenced remains unidentified.
Table 1.
List of mitogenomes of Cerambycoidea. *Partial genome
| Family/subfamily/species | Accession no. |
Size (bp) |
A% | T% | C% | G% | A+T % |
AT skew |
GC skew |
References |
|---|---|---|---|---|---|---|---|---|---|---|
| Cerambycidae | ||||||||||
| Prioninae | ||||||||||
| Dorysthenes paradoxus | MG460483 | 15,922 | 39.67 | 31.45 | 17.87 | 11 | 71.13 | 0.116 | –0.238 | This study |
| Prioninae sp.* | JX220991 | 10,679 | 39.53 | 31.44 | 17.71 | 11.29 | 70.97 | 0.114 | –0.222 | |
| Cerambycinae | ||||||||||
| Aeolesthes oenochrous | NC_025243 | 15,747 | 39.44 | 31.75 | 18.3 | 10.5 | 71.19 | 0.108 | –0.271 | Chiu et al. (2016) |
| Massicus raddei | NC_023937 | 15,858 | 38.61 | 33.18 | 17.06 | 11.16 | 71.78 | 0.076 | –0.209 | Wang et al. (2016) |
| Xylotrechus grayii | NC_030782 | 15,540 | 41.35 | 33.94 | 15.03 | 9.68 | 75.29 | 0.098 | –0.216 | |
| Lamiinae | ||||||||||
| Anoplophora chinensis | NC_029230 | 15,805 | 39.48 | 38.17 | 13.57 | 8.78 | 77.65 | 0.017 | –0.214 | Li et al. (2016b) |
| Anoplophora glabripennis | NC_008221 | 15,774 | 39.61 | 38.7 | 13.07 | 8.59 | 78.31 | 0.012 | –0.207 | Fang et al. (2016) |
| Apriona swainsoni | NC_033872 | 15,412 | 38.9 | 35.81 | 15.99 | 9.29 | 74.71 | 0.041 | –0.265 | |
| Batocera lineolata | NC_022671 | 15,418 | 38.75 | 35.73 | 16.21 | 9.31 | 74.48 | 0.040 | –0.271 | Wang et al. (2012) |
| Monochamus alternatus | NC_024652 | 15,874 | 39.76 | 39.26 | 12.3 | 8.68 | 79.02 | 0.006 | –0.173 | Li et al. (2016a) |
| Psacothea hilaris | NC_013070 | 15,856 | 38.76 | 37.88 | 14.17 | 9.2 | 76.63 | 0.011 | –0.213 | Kim et al. (2009) |
| Thyestilla gebleri | NC_034752 | 15,505 | 38.5 | 35.56 | 16.64 | 9.28 | 74.07 | 0.040 | –0.284 | |
| Lepturinae | ||||||||||
| Rhagium mordax* | JX412743 | 10,676 | 41.14 | 36.76 | 12.84 | 9.25 | 77.9 | 0.056 | –0.194 | |
| Chrysomelidae | ||||||||||
| Galeruca daurica | NC_027114 | 16,615 | 40.51 | 37.63 | 13.11 | 8.73 | 78.15 | 0.048 | –0.251 | Zhou et al. (2016) |
| Diabrotica barberi | KF669870 | 16,366 | 41.26 | 37.91 | 12.29 | 8.54 | 79.17 | 0.042 | –0.220 | Coates (2014) |
| Diabrotica virgifera | KF658070 | 16,650 | 41.56 | 37.97 | 12.09 | 8.38 | 79.53 | 0.045 | –0.221 | Coates (2014) |
| Crioceris duodecimpunctata | AF467886 | 15,880 | 40.14 | 36.74 | 13.44 | 9.67 | 76.89 | 0.044 | –0.195 | Stewart and Beckenbach (2003) |
| Vesperidae | ||||||||||
| Philinae | ||||||||||
| Spiniphilus spinicornis | NC_029515 | 15,707 | 38.58 | 30.94 | 19.35 | 11.13 | 69.52 | 0.11 | –0.369 | Nie et al. (2017) |
The longhorn beetle Dorysthenes paradoxus (Faldermann, 1833) (Coleoptera: Cerambycidae) belongs to the insect subfamily Prioninae, within the family Cerambycidae. This species is widely distributed in China. In general, this species is considered a serious agricultural pest that attacks the underground stems of gramineous crops, such as maize, sorghum, rice, sugarcane, and bluegrass (Yan et al. 1997). In the Shanxi and Liaoning Provinces of China, the adult beetle has been traditionally accepted and used as an edible insect with a long history of use. In the sample collection site used in this study, each year on the day that autumn begins, many people like to capture the adults of the beetle after a rain. In recent years, the price of this beetle has soared. However, little attention has been paid to biological studies of this beetle, although its life cycle, habits, and morphological characteristics of four developmental stages and the digestive tract have been investigated (Yan et al. 1997, Hao et al. 1999). In the GenBank database, only four nucleotide sequences for three mitochondrial DNA fragments encoding partial COI (GU130426 and KJ159158), lrRNA (GU130418), and srRNA (GU130410) genes are available. Even though the genetic background is in high demand, no genetic information has been acquired.
With the aim to provide more useful information for studies of the population structure and genetic diversity of Do. paradoxus, in the present study, the complete mitogenome sequence of this economically important species was determined. This genome is the first mitogenome in the subfamily Prioninae, so we performed a phylogenetic analysis based on the available complete mitogenome sequences to provide insight into the phylogenetic relationships within the family Cerambycidae.
Materials and Methods
Specimen and DNA Extraction
Adult beetles were captured at Beiqiuzhang village (N35°32′42.05″; E111°27′39.42″), Gujiang town, Jiangxian county, Shanxi Province, China on 8 August 2015. The freshly collected specimens were preserved immediately in 95% ethanol. The specimens were identified as Do. paradoxus based on the morphology by Prof. Hong Fang (Department of Entomology, College of Plant Protection, Shenyang Agricultural University, China). Total genomic DNA was extracted from the leg muscle tissue of an adult individual using the Trace DNA Extraction Method (Wang et al. 1997). The partial leg was immersed in the extraction buffer (10 mM/liter Tris–HCl, pH 8.0, 10 mM/liter EDTA, pH 8.0, 100 mM/liter NaCl, 2% SDS, 0.039 M/liter DTT, and 100 mg/liter Proteinase K) and then treated with the traditional phenol/chloroform method. In our laboratory, this method has been confirmed to be suitable to extract total genomic DNA from animal, insect, plant, and microorganism samples.
PCR Amplification and Sequencing
The entire mitogenome of Do. paradoxus was amplified in 14 overlapping fragments (Table 2). In the first step, four fragments were successfully amplified with universal insect PCR primers (Simon et al. 2006). Then, seven species specific primer pairs (Dp1-14) were designed, based on the initial sequences of the four obtained fragments and three mitochondrial DNA fragments available in the GenBank database (COI, lrRNA, srRNA). Finally, we designed three primer pairs (Dp15-20) to fill the remaining gaps corresponding to the sequences from GU130426, GU130418, and GU130410. PCR amplifications were done using TaKaRa LA Taq or EX Taq polymerase (Takara Biotechnology Co. Ltd., Dalian, China), under the following procedure: 94°C for 2 min, followed by 35 cycles of 1 min at 94°C and 1 min at 50–55°C, with a subsequent 10 min final extension at 72°C. All amplified products were sequenced firstly using upstream and downstream primers from both directions and later by primer walking. Sanger sequencing was performed at SinoGenoMax Co., Ltd., Beijing, China.
Table 2.
Primers used for PCR amplification
| Fragment | Primer | Sequence(5ʹ–3ʹ) | Target | Position |
|---|---|---|---|---|
| Universal primers (Simon et al. 2006) | ||||
| F1 | TK-J3790 | CATTAgATgACTgAAAgCAAgTA | tRNA Lys | 3,703–3,725 |
| F1 | A6-N4552 | ATgTCCWgCAATYATATTWgC | ATP6 | 4,481–4,501 |
| F2 | C3-J4792 | gTTgATTATAgACCWTgRCC | COIII | 4,695–4,715 |
| F2 | C3-N5460 | TCAACAAAATgTCARTAYCA | COIII | 5,389–5,408 |
| F3 | N5-J7572 | AAAgggAATTTgAgCTCTTTTWgT | ND5 | 7,395–7,418 |
| F3 | N4-N8727 | AAATCTTTRATTgCTTATTCWTC | ND4 | 8,570–8,592 |
| F4 | CB-J10933 | gTTCTACCTTgAggNCAAATRTC | Cytb | 10,728–10,750 |
| F4 | CB-N11526 | TTCTACTggTCgRgCTCCAATYCA | Cytb | 11,343–11,363 |
| Species specific primers (This study) | ||||
| F5 | Dp2-F | gAATgACTACAAAAACTTCCACCTgC | COI | 2,887–2,912 |
| F5 | Dp3-R | TTAAggggTgATATTTgTgggATT | ATP8 | 3,845–3,868 |
| F6 | Dp4-F | AACCCTTgCTTTACCCTTATgATT | ATP6 | 4,303–4,326 |
| F6 | Dp5-R | ATTgAAAAACCTAATAGTAATAAA | COIII | 4,794–4,817 |
| F7 | Dp6-F | CCTCgTTCTTTgTggCAACTggAT | COIII | 5,255–5,278 |
| F7 | Dp7-R | TTAgggTTggTTTCTTATTgTTTggT | ND5 | 7,618–7,643 |
| F8 | Dp8-F | TAAACCAgAATAATAAgAACCAT | ND5 | 8,170–8,192 |
| F8 | Dp9-R | gCATTATCAACggCgAATCCTCC | Cytb | 10,830–10,852 |
| F9 | Dp10-F | AATCCTATTTTgATCgTTggTAg | Cytb | 11,300–11,322 |
| F9 | Dp11-R | AACTTTTTTTTCATTTTTACACTT | 16SrRNA | 12,875–12,898 |
| F10 | Dp12-F | TTCTCATCCAACCATTCATTCCAg | 16SrRNA | 13,071–13,094 |
| F10 | Dp13-R | AAAAggggAgATgggTTACAAT | 12SrRNA | 14,133–14,158 |
| F11 | Dp14-F | TTCAgAATAATAgggTATCTAATCC | 12SrRNA | 14,380–14,404 |
| F11 | Dp1-R | TCCTCTTTCTTgTCTgATAATgTggg | COI | 2,174–2,199 |
| F12 | Dp15-F | ggAggTggAgATCCTATTCTAT | COI | 2,080–2,101 |
| F12 | Dp16-R | ggACgCTCTATCTTggAgTAAT | COII | 3,039–3,060 |
| F13 | Dp17-F | AATCCTATTTTgATCgTTggTAg | Cytb | 11,300–11,322 |
| F13 | Dp18-R | TATATATTTTTgTTTATgggggTA | 16SrRNA | 13,264–1,3287 |
| F14 | Dp19-F | TgTTACgACTTATTTCACCTTggg | 12SrRNA | 13,986–14,009 |
| F14 | Dp20-R | gTTTgTgCCAgCAGTTgCggTTA | 12SrRNA | 145,49–14,571 |
| For amplification and sequencing of the A+T-rich region | ||||
| Dp21-F | TAAAAAGAAGATCAAAATCCCAG | AT-rich | 14,977–14,999 | |
| Dp23-R | CTATAAgTTATTAAAAACTAAT | AT-rich | 15,566–15,587 | |
| Dp24-F | ATTAgTTTTTAAAAACTTATAg | AT-rich | 15,566–15,587 | |
| Dp22-R | TTgATACTTTAggAggTggTTT | tRNA Gln | 85–106 | |
| N = [A, C, G, T]; R = [A, G]; Y = [C, T]; W = [A, T] | ||||
When we attempted to sequence the AT-rich region, the occurrence of a polyT(12) at positions 15,132–15,143 and a polyA(21) at positions 15,841–15,861 made their internal sequencing results unreadable. To address this issue, several primers based on the possible sequence deduced by the indefinite sequencing results of primers Dp21-F and Dp22-R were used. Fortunately, primer Dp23-F and its reverse complement primer Dp24-R, paired with Dp21-F and Dp22-R, respectively, were suitable for amplification and sequencing which allowed us to cover the remaining gaps.
Sequence Annotation and Genomic Analysis
The partial mitogenome of Prioninae sp. (Coleoptera: Cerambycidae) is available in GenBank, so we used it as the reference sequence for mapping the amplified fragments of Do. paradoxus. The overlapping sequences were manually checked and assembled into a complete mitogenome via the alignment of neighboring fragments using the Clustal X program (Thompson et al. 1997). Protein-coding genes (PCGs) were identified by comparison with sequences from other Cerambycidae species. The 5ʹ ends of PCGs were assumed to be at the first legitimate in-frame start codon ATN in an open reading frame (ORF) that was not located within an upstream gene encoded on the same strand. A truncated stop codon (T or TA) adjacent to the beginning of the downstream gene was designated the termination codon (Wolstenholme 1992). The transfer RNA (tRNA) genes and their secondary structures were identified using the MITOS web server (Bernt et al. 2013). The large rRNA (lrRNA) gene was annotated to extend to the boundaries of the flanking tRNALeu(CUN) and tRNAVal. The 3ʹ end of the small rRNA (srRNA) gene was annotated to be adjacent to the start of tRNAVal, whereas the 5ʹ end was determined by comparing orthologous sequences of other known mitogenomes. The entire A+T-rich region was subjected to a search for the tandem repeats using Tandem Repeats Finder program (Benson 1999). Composition skew was used to describe the base composition of nucleotide sequences, with the relative number of As to Ts (AT skew = [A – T]/[A + T]) and Gs to Cs (GC skew = [G – C]/[G + C]) (Perna and Kocher 1995). Overlapping regions and intergenic spacers between genes were counted manually. Circular genome maps were generated by OrganellarGenomeDRAW (Lohse et al. 2013) and revised by hand. The sequence data were deposited in GenBank under accession MG460483.
Phylogenetic Analysis
To reconstruct the phylogenetic relationships among Chrysomeloidae species, a total of 17 available Cerambycidea mitogenomes, including 13 Cerambycinae and 4 Chrysomelidae, were used (Table 1). The mitogenome of Spiniphilus spinicornis (Lin & Bi, 2011) (Coleoptera: Vesperidae), which belongs to Philinae of Vesperidae, was also included due to its association with Prioninae of Chrysomeloidae in their adult morphology (Nie et al. 2017). One neuropteran species, Polytoechotes punctatus (Fabricius) (Neuroptera: Polystoechotidae) (Beckenbach and Stewart 2009), served as an outgroup. The nucleotide sequences of each of the 13 PCGs were translated into amino acid sequences then aligned with default settings, and these resultant alignments were retranslated into nucleotide alignments by MEGA 6.06 (Tamura et al. 2013). The alignment was done with Muscle using default settings. The sequences of 13 PCGs were then concatenated to allow a more complete analysis (Hassanin 2006). In the present study, the nucleotide sequence of ND5 in Massicus raddei (Blessig) (Coleoptera: Cerambycidae) was excluded due to probable sequencing misread. Bayesian Inference (BI) and Maximum likelihood (ML) methods were used for phylogenetic analysis. The GTR + G + I model was selected by the Akaike information criterion in MEGA 6.06. BI analysis was performed using MrBayes v3.1.2 (Huelsenbeck and Ronquist 2001), with posterior distributions estimated using Markov chain Monte Carlo (MCMC) sampling. The MCMC search was conducted for 100,000 generations, and sampling was done every 100 generations. The average standard deviation of split frequencies was below 0.01. Then, the first 25% of the trees were discarded as ‘burn-in’, and posterior probabilities were estimated for each node. For ML analysis, PhyML v3.0 (Guindon and Gascuel 2003) was used with the following conditions: equilibrium frequencies as ‘empirical’, proportion of invariable sites as ‘estimated’, number of substitution rates categorized in fours, gamma shape parameter as ‘estimated’, and starting tree as ’BIONJ’. The confidence values of the ML tree were evaluated via a bootstrap test with 100 replications. The obtained phylogenetic trees were visualized with Treeview 1.6.6 (Page 1996).
Results and Discussion
General Features of the Do. paradoxus Mitogenome
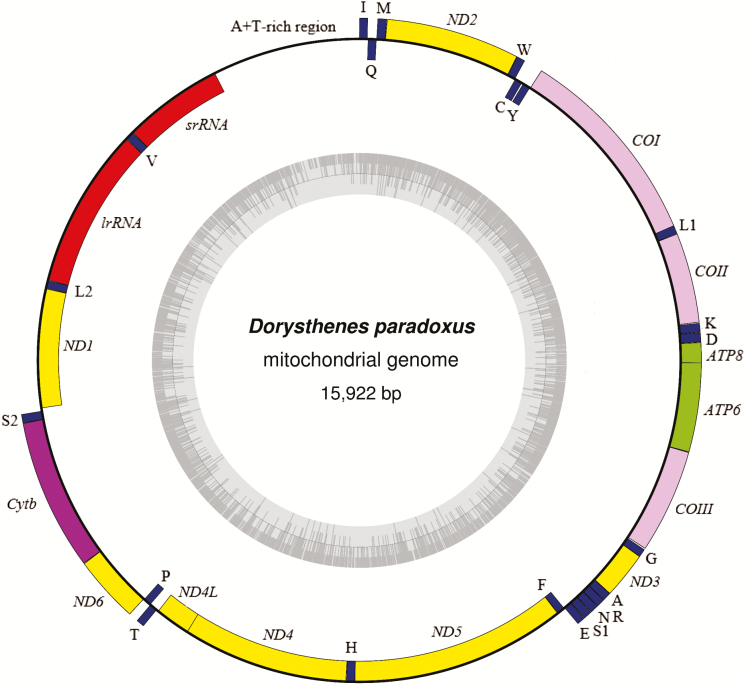
In the present study, we sequenced the entire mitogenome of Do. paradoxus by PCR amplification in 14 overlapping fragments with universal insect primers and species specific primers (Table 2). The complete mitogenome sequence of Do. paradoxus is 15,922 bp in size, which is the largest among the completely sequenced Cerambycidae insects. This mitogenome shares typical metazoan genes (Fig. 1, Table 3), including 13 PCGs, 22 tRNA genes, 2 rRNAs, and a noncoding A+T-rich region (Wolstenholme 1992). The gene arrangement and orientation of this mitogenome are identical to those of the completely sequenced coleopteran insects, with the ancestor gene order tRNAIle-tRNAGln-tRNAMet located between the A+T-rich region and ND2.
Fig. 1.
The gene map of the mitogenome of Do. paradoxus. The circle inside the GC content graph marks the 50% threshold. Genes are shown as blocks facing out if they are transcribed in the clockwise direction or facing in if they are transcribed in the counter-clockwise direction. COI, COII, and COIII refer to the cytochrome oxidase subunits, Cytb refers to cytochrome b, and ND1-6 refers to NADH dehydrogenase components. tRNA genes are denoted as one-letter symbols according to the IUPAC-IUB single-letter amino acid codes. The one-letter symbols L1, L2, S1, and S2 denote tRNALeu(CUN), tRNALeu(UUR), tRNASer(AGN), and tRNASer(UCN), respectively. lrRNA and srRNA refer to large and small rRNA, respectively.
Table 3.
Annotation of the mitogenome of Do. paradoxus
| Gene | Direction | Nucleotide no. | Size (bp) | Anticodon | IGS | OL | Start codon | Stop codon |
|---|---|---|---|---|---|---|---|---|
| tRNA Ile | F | 1–64 | 64 | GAT | 3 | |||
| tRNA Gln | R | 68–136 | 69 | TTG | 1 | |||
| tRNA Met | F | 136–204 | 69 | CAT | ||||
| ND2 | F | 205–1,213 | 1009 | ATG | T | |||
| tRNA Trp | F | 1,214–1,280 | 67 | TCA | 1 | |||
| tRNA Cys | R | 1,282–1,344 | 63 | GCA | 9 | |||
| tRNA Tyr | R | 1,354–1,418 | 65 | GTA | 1 | |||
| COI | F | 1,420–2,953 | 1534 | AAT | T | |||
| tRNA Leu(UUR) | F | 2,954–3,018 | 65 | TAA | ||||
| COII | F | 3,019–3,699 | 681 | 3 | ATC | TAG | ||
| tRNA Lys | F | 3,703–3,772 | 70 | TTT | ||||
| tRNA Asp | F | 3,773–3,845 | 73 | GTC | ||||
| ATP8 | F | 3,846–4,001 | 156 | 7 | ATC | TAA | ||
| ATP6 | F | 3,995–4,668 | 674 | ATG | TA | |||
| COIII | F | 4,669–5,457 | 789 | 6 | ATG | TAA | ||
| tRNA Gly | F | 5,464–5,527 | 64 | TCC | ||||
| ND3 | F | 5,528–5,880 | 353 | ATT | TA | |||
| tRNA Ala | F | 5,881–5,946 | 66 | TGC | 1 | |||
| tRNA Arg | F | 5,946–6,011 | 66 | TCG | 1 | |||
| tRNA Asn | F | 6,011–6,075 | 65 | GTT | ||||
| tRNA Ser(AGN) | F | 6,076–6,142 | 67 | TCT | ||||
| tRNA Glu | F | 6,143–6,206 | 64 | TTC | 2 | |||
| tRNA Phe | R | 6,209–6,273 | 65 | GAA | ||||
| ND5 | R | 6,274–7,994 | 1,721 | ATT | TA | |||
| tRNA His | R | 7,995–8,063 | 69 | GTG | ||||
| ND4 | R | 8,064–9,396 | 1,333 | 1 | ATG | T | ||
| ND4L | R | 9,398–9,677 | 280 | ATG | T | |||
| tRNA Thr | F | 9,690–9,754 | 65 | TGT | ||||
| tRNA Pro | R | 9,755–9,820 | 66 | TGG | 2 | |||
| ND6 | F | 9,823–10,331 | 509 | ATT | TA | |||
| Cytb | F | 10,332–11,473 | 1,142 | ATG | TA | |||
| tRNA Ser(UCN) | F | 11,474–11,541 | 68 | TGA | 21 | |||
| ND1 | R | 11,563–12,513 | 951 | TTG | TAA | |||
| tRNA Leu(CUN) | R | 12,514–12,579 | 66 | TAG | ||||
| lrRNA | R | 12,580–13,868 | 1,289 | |||||
| tRNA Val | R | 13,869–13,938 | 70 | TAC | ||||
| srRNA | R | 13,939–14,742 | 804 | |||||
| A+T-rich region | 14,743–15,922 | 1,180 |
The direction of the genes is presented as F for forward and R for reverse direction. OL denotes inside genes overlap. IGS represents the intergenic spacer sequences.
The nucleotide composition of the mitogenome of Do. paradoxus is biased toward As and Ts, which account for 71.13% of the base pairs (A 39.67%, G 11.00%, T 31.45%, and C 17.87%). The bias value is the lowest among the completely sequenced Cerambycidae species (Table 1).
The AT skew of the forward strand of the mitogenome of Do. paradoxus is slightly positive (0.116), indicating the occurrence of more As than Ts. This case is the largest among previously sequenced Cerambycidae mitogenomes. The GC skew of Do. paradoxus mitogenome is -0.238, which is well within the range of all analyzed Cerambycidae mitogenomes.
Coding Regions of the Do. paradoxus Mitogenome
Among 13 PCGs, 11 have a typical ATN start codon: 6 have an ATG (ND2, ATP6, COIII, ND4, ND4L, Cytb), 2 have an ATC (COII, ATP8), and 3 have an ATT (ND3, ND5, ND6). The remaining two PCGs begin with atypical codons: COI with AAT and ND1 with TTG, as found in Aeolesthes oenochrous (Fairmaire) (Coleoptera: Cerambycidae) (Chiu et al. 2016). AAT (Asparagine) is highly conserved in coleopteran Polyphaga (Sheffield et al. 2008), and TTG (Leucine) has been proposed as the start codon of ND1 in Anopheles quadrimaculatus Say (Diptera: Culicidae) and Tricholepidion gertschi Wygodzinsky (Insecta, Zygentoma) (Mitchell et al. 1993, Nardi et al. 2003). Only 4 of the 13 PCGs harbor a complete termination codon (three TAA and one TAG), the remaining 9 possess incomplete termination codons (4 T and 5 TA).
The Do. paradoxus mitogenome bears a typical set of 22 tRNA genes (1 specific for each amino acid and 2 each for Leucine and Serine). These tRNAs are scattered throughout the mitogenome, between rRNAs and PCGs, with sizes ranging from 63 bp for tRNACys to 73 bp for tRNAAsp. The anticodons are identical to those observed in other Cerambycidae insects. With the exception of tRNASer(AGN), which lacks a stable stem-loop structure in the DHU arm, the other 21 tRNAs can fold into a typical clover-leaf secondary structures. This abnormal structure of tRNASer(AGN) has been observed in many metazoan species, including insects (Wolstenholme 1992).
Two rRNA genes (lrRNA and srRNA), located between tRNALeu (CUN), and tRNAVal, and between tRNAVal and the A+T-rich region, respectively, were identified in the mitogenome of Do. paradoxus. The length of lrRNA and srRNA were 1,289 and 804 bp, respectively, which were well within the range observed in the completely sequenced Cerambycidae insects.
Noncoding Regions of the Do. paradoxus Mitogenome
The Do. paradoxus mitochondrial genes harbor a total of 49-bp intergenic spacer sequences, which are spread over 10 regions and range in size from 1 to 21 bp. The longest one is located between tRNASer (UCN) and ND1 contains the 5-bp-long motif (TACTA), which is conserved across Cerambycidae species (Fig. 2A). This region is present in most insect mitochondrial DNA, even though the nucleotide sequence can be quite divergent (Cameron and Whiting 2008, Kim et al. 2009). This motif has been suggested to be a possible recognition site for mtTERM protein, the transcription termination peptide (Taanman 1999).
Fig. 2.
Sequence alignment results. (A) The intergenic spacer region between tRNASer(UCN) and ND1 of Cerambycidae species. The bold nucleotides indicate the conserved 5 bp-long motif (TACTA) found in all sequenced Cerambycidae insects. (B) Tandem repeat units detected in the Do. paradoxus A+T-rich region. The nucleotide position is provided at each end of the sequence with regard to the mitogenome of Do. paradoxus.
The Do. paradoxus mitogenome has a total of 10-bp overlapping sequences that vary in length from 1 to 7 bp in four regions. The longest overlap of 7-bp overlap was found with the reading frames of ATP8/ATP6, which is common in insect mitogenomes (Boore 1999). The remaining three overlaps are involved in tRNAs.
The 1,180-bp A+T-rich region, located between srRNA and tRNAIle, exhibits the highest A+T content of 82.71% (A 44.07%, G 6.95%, T 38.64%, C 10.34%) in the mitogenome. In coleopteran insects, the presence of tandem repeats in the mitochondrial A+T-rich region has been observed, including 21 copies of a 58-bp repeat element in Trachypachus holmbergi Mannerheim (Coleoptera: Trachypachidae), 6 tandem repeats of a 312-bp sequence in Priasilpha obscura (Herbst) (Coleoptera: Phloeostichidae), 3 ~96-bp tandem repeat copies in Chaetosoma scaritides Westwood (Coleoptera: Melyridae), and 7 copies of 57-bp tandem repeats in Psacothea hilaris Pascoe (Coleoptera: Cerambycidae) (Sheffield et al. 2008, Kim et al. 2009). In the Do. paradoxus A+T-rich region, a tandem repeat comprising two 47-bp-long copies was identified, beginning from nucleotide number 15,315 with regard to the Do. paradoxus mitogenome. The nucleotide sequence of repeat 1 is almost identical to repeat 2 (Fig. 2B). The repeat sequence comprises >91.67% A+T, which is higher than that of the whole A+T-rich region. Moreover, the Do. paradoxus A+T-rich region includes two poly-T stretches: one is 12 bp long (position: 15,132–15,143) in the majority strand, and the other is 21-bp long (position: 15,841–15,861) in the minority strand near the tRNAIle. It has been verified that the latter poly-T stretch is a structural signal for the recognition of proteins that are involved in replication initiation, at least among holometabolous insects (Zuker et al. 1999).
Phylogenetic Relationships
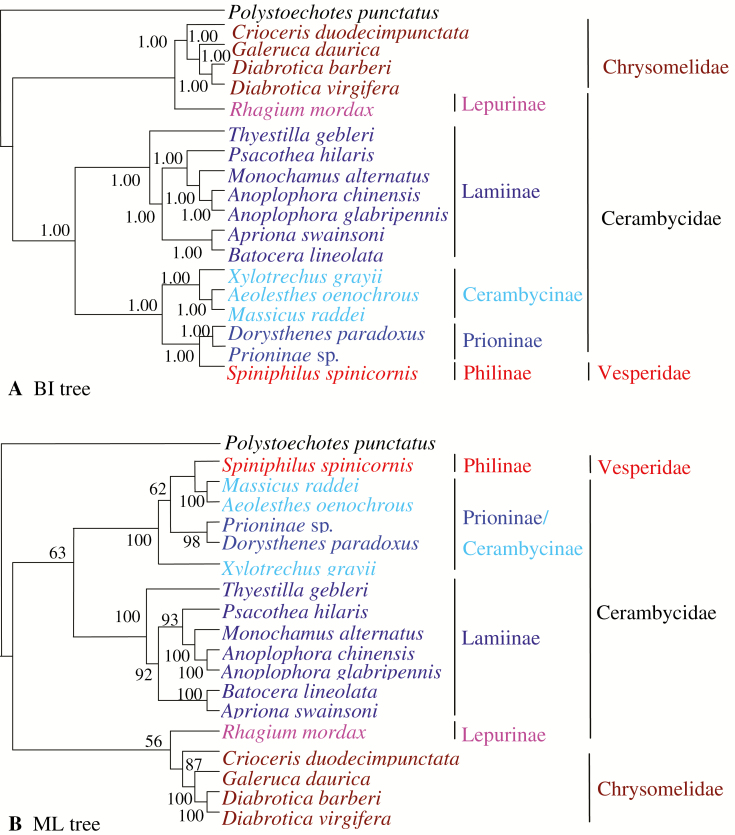
In the present study, we intended to use the 18 complete or near complete mitogenomes available to date, including 13 Cerambycidae, four Chrysomelidae and one Vesperidae, to evaluate the phylogenetic relationship within the superfamily Chrysomeloidea. According to the morphological characters, the 13 Cerambycidae mitogenomes used in this study are derived from four subfamilies, i.e., Prioninae (Do. paradoxus, Prioninae sp.), Cerambycinae (Ae. oenochrous, Ma. raddei, Xy. grayii), Lamiinae (An. chinensis, An. glabripennis, Ap. swainsoni, Ba. lineolata, Mo. alternatus, Ps. hilaris, Th. gebleri), and Lepturinae (Rh. mordax). The mitogenome of Sp. spinicornis, belonging to Philinae of Vesperidae was also included due to its problematic status (Nie et al. 2017). Their phylogenetic relationships were reconstructed based on the nucleotide sequences of 13 PCGs with BI and ML methods (Fig. 3A and B). The phylogenetic analyses yielded almost identical topological relationships, indicating the presence of two distinct groups: Cerambycidae and Chrysomelidae, which is consistent with the morphological analysis and the previous findings (Nie et al. 2017). In our phylogenetic analyses, Lepturinae (Rh. mordax) is closely related to Chrysomelidae, rather than Cerambycidae, indicating that the taxonomy status of Rh. mordax needs to be reconsidered.
Fig. 3.
Phylogeny of Chrysomeloidea species. Phylogenetic tree inferred from the nucleotide sequences of 13 PCGs by Bayesian inference (A) and maximum likelihood (B). The numbers close to the nodes specify posterior probability values and bootstrap percentages, and values higher than 50% are shown.
Both BI and ML analyses confirmed the subfamily rank of Cerambycidae ([Prioninae + Cerambycinae] + Lamiinae) with high support values (100% for BI and 63% for ML), the monophyly of Lamiinae, with the nearest relationship being between Philinae (Sp. spinicornis) and Prioninae/Cerambycinae. BI analysis supported the monophyly of Prioninae and Cerambycinae, and the nearest relationship between Prioninae and Philinae with 100% posterior probabilities. However, the monophyly of Prioninae and Cerambycinae, and the nearest relationship between Prioninae and Philinae were not recovered in ML analysis. These results suggested that the monophyly of Prioninae and Cerambycinae and their relationship with Prioninae should be further studied in the future.
Acknowledgments
We are thankful to Prof. Hong Fang (Department of Entomology, College of Plant Protection, Shenyang Agricultural University, China) for species identification. This work was partially supported by grants from the National Natural Science Foundation of China (No. 31672493; 31372372), the China Agriculture Research System (No. CARS-18), the scientific research program for agricultural commonweal from Ministry of Agriculture (201403031), and the Natural Science Foundation of Liaoning Province (2014027002).
References Cited
- Beckenbach A. T., and Stewart J. B.. 2009. Insect mitochondrial genomics 3: the complete mitochondrial genome sequences of representatives from two neuropteroid orders: a dobsonfly (order Megaloptera) and a giant lacewing and an owlfly (order Neuroptera). Genome. 52: 31–38. [DOI] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M., Donath A., Jühling F., Externbrink F., Florentz C., Fritzsch G., Pütz J., Middendorf M., and Stadler P. F.. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69: 313–319. [DOI] [PubMed] [Google Scholar]
- Boore J. L. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27: 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron S. L., and Whiting M. F.. 2008. The complete mitochondrial genome of the tobacco hornworm, Manduca sexta, (Insecta: Lepidoptera: Sphingidae), and an examination of mitochondrial gene variability within butterflies and moths. Gene. 408: 112–123. [DOI] [PubMed] [Google Scholar]
- Chiu W. C., Yeh W. B., Chen M. E., and Yang M. M.. 2016. Complete mitochondrial genome of Aeolesthes oenochrous (Fairmaire) (Coleoptera: Cerambycidae): an endangered and colorful longhorn beetle. Mitochondrial DNA A. DNA Mapp. Seq. Anal. 27: 686–687. [DOI] [PubMed] [Google Scholar]
- Coates B. S. 2014. Assembly and annotation of full mitochondrial genomes for the corn rootworm species, Diabrotica virgifera virgifera and Diabrotica barberi (Insecta: Coleoptera: Chrysomelidae), using next generation sequence data. Gene. 542: 190–197. [DOI] [PubMed] [Google Scholar]
- Fang J., Qian L., Xu M., Yang X., Wang B., and An Y.. 2016. The complete nucleotide sequence of the mitochondrial genome of the Asian longhorn beetle, Anoplophora glabripennis (Coleoptera: Cerambycidae). Mitochondrial DNA A. DNA Mapp. Seq. Anal. 27: 3299–3300. [DOI] [PubMed] [Google Scholar]
- Guindon S. and Gascuel O.. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Hao C., Yan C.-X., Zhang G.-H., and Yu G.-F.. 1999. Morphological characteristics of the digestive tract of Dorysthenes paradoxus. J. Shanxi. Agri. Univ. 19: 7–8. [Google Scholar]
- Hassanin A. 2006. Phylogeny of Arthropoda inferred from mitochondrial sequences: strategies for limiting the misleading effects of multiple changes in pattern and rates of substitution. Mol. Phylogenet. Evol. 38: 100–116. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J. P., and Ronquist F.. 2001. MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics. 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Hunt T., Bergsten J., Levkanicova Z., Papadopoulou A., John O. S., Wild R., Hammond P. M., Ahrens D., Balke M., Caterino M. S.,. et al. 2007. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science. 318: 1913–1916. [DOI] [PubMed] [Google Scholar]
- Kim K. G., Hong M. Y., Kim M. J., Im H. H., Kim M. I., Bae C. H., Seo S. J., Lee S. H., and Kim I.. 2009. Complete mitochondrial genome sequence of the yellow-spotted long-horned beetle Psacothea hilaris (Coleoptera: Cerambycidae) and phylogenetic analysis among coleopteran insects. Mol. Cells. 27: 429–441. [DOI] [PubMed] [Google Scholar]
- Li F., Zhang H., Wang W., Weng H., and Meng Z.. 2016a. Complete mitochondrial genome of the Japanese pine sawyer, Monochamus alternatus (Coleoptera: Cerambycidae). Mitochondrial DNA A. DNA Mapp. Seq. Anal. 27: 1144–1145. [DOI] [PubMed] [Google Scholar]
- Li W., Yang X., Qian L., An Y., and Fang J.. 2016b. The complete mitochondrial genome of the citrus long-horned beetle, Anoplophora chinensis (Coleoptera: Cerambycidae). Mitochondrial DNA A. DNA Mapp. Seq. Anal. 27: 4665–4667. [DOI] [PubMed] [Google Scholar]
- Lohse M., Drechsel O., Kahlau S., and Bock R.. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41: W575–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S. E., Cockburn A. F., and Seawright J. A.. 1993. The mitochondrial genome of Anopheles quadrimaculatus species A: complete nucleotide sequence and gene organization. Genome. 36: 1058–1073. [DOI] [PubMed] [Google Scholar]
- Nardi F., Spinsanti G., Boore J. L., Carapelli A., Dallai R., and Frati F.. 2003. Hexapod origins: monophyletic or paraphyletic?Science. 299: 1887–1889. [DOI] [PubMed] [Google Scholar]
- Nie R., Lin M., Xue H., Bai M., and Yang X.. 2017. Complete mitochondrial genome of Spiniphilus spinicornis (Coleoptera: Vesperidae: Philinae) and phylogenetic analysis among Cerambycoidea. Mitochondrial DNA A. DNA Mapp. Seq. Anal. 28: 145–146. [DOI] [PubMed] [Google Scholar]
- Page R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12: 357–358. [DOI] [PubMed] [Google Scholar]
- Perna N. T. and Kocher T. D.. 1995. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. j. Mol. Evol. 41: 353–358. [DOI] [PubMed] [Google Scholar]
- Sama G., Buse J., Orbach E., Friedman A. L. L., Rittner O., and Chikatunov V.. 2010. A new Catalogue of the Cerambycidae (Coleoptera) of Israel with notes on their distribution and host plants. Munis. Entomol. Zool. 5: 1–51. [Google Scholar]
- Sheffield N. C., Song H., Cameron S. L., and Whiting M. F.. 2008. A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol. Biol. Evol. 25: 2499–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C., Buckley T. R., Frati F., Stewart J. B., and Beckenbach A. T.. 2006. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 37: 545–579. [Google Scholar]
- Stewart J. B. and Beckenbach A. T.. 2003. Phylogenetic and genomic analysis of the complete mitochondrial DNA sequence of the spotted asparagus beetle Crioceris duodecimpunctata. Mol. Phylogenet. Evol. 26: 513–526. [DOI] [PubMed] [Google Scholar]
- Taanman J. W. 1999. The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta. 1410: 103–123. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., and Higgins D. G.. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhou Z.-Y., Lu C., Xiang Z.-H., Yang B., and Zeng H.-M.. 1997. DNA extraction from single egg of silkworm. J. Southwest. Agri. Univ. 19: 434–437. [Google Scholar]
- Wang C., Feng Y., and Chen X.. 2012. Complete sequence and gene organization of the mitochondrial genome of Batocera lineolata Chevrolat (Coleoptera: Cerambycidae). Chinese. Sci. Bull. 57: 3578–3585. [Google Scholar]
- Wang Y. T., Liu Y. X., Tong X. L., Ren Q. P., and Jiang G. F.. 2016. The complete mitochondrial genome of the longhorn beetle, Massicus raddei. Mitochondrial DNA A. DNA Mapp. Seq. Anal. 27: 209–211. [DOI] [PubMed] [Google Scholar]
- Wolstenholme D. R. 1992. Animal mitochondrial DNA: structure and evolution. Int. Rev. Cytol. 141: 173–216. [DOI] [PubMed] [Google Scholar]
- Yan C.-X., Hao C., and Wang M.-Q.. 1997. A preliminary study on Dorysthenes paradoxus. J. Shanxi. Agri. Univ. 17: 342–345. [Google Scholar]
- Zhang H., Liu N., Han Z., and Liu J.. 2016. Phylogenetic analyses and evolutionary timescale of Coleoptera based on mitochondrial sequence. Biochem. Syst. Ecol. 66: 229–238. [Google Scholar]
- Zhou X., Han H., Pang B., and Zhang P.. 2016. The complete mitochondrial genome of Galeruca daurica (Joannis) (Coleoptera: Chrysomelidae). Mitochondrial DNA A. DNA Mapp. Seq. Anal. 27: 2891–2892. [DOI] [PubMed] [Google Scholar]
- Zuker M., Mathews D. H., and Turner D. H.. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: A practical guide, pp. 11–43. In: J. Barciszewski and B. F. C. Clark (eds.), RNA biochemistry and biotechnology. Springer, Dordrecht, the Netherlands. [Google Scholar]