Abstract
Hepatocellular carcinoma (HCC) is one of the most prevalent cancers. It has been demonstrated that various cellular microRNAs (miRNAs) play an important role in HCC development. Here, we analyzed the miRNA profile in HCC tissues by Solexa sequencing, and we identified a novel microRNA, miR-HCC1, which is upregulated in HCC tissues. Further experiments showed that miR-HCC1 promoted HCC cell proliferation in vivo and in vitro, and migration and invasion resulting from the epithelial-mesenchymal transition (EMT) process. Nuclear factor I/X (NFIX), which inhibited cell proliferation, migration and invasion in HCC cells, was identified as a direct and functional target of miR-HCC1. Furthermore, lymphoid enhancer binding factor 1 (LEF1), a transcription factor, was shown to bind the promoter of miR-HCC1 and activate its expression. Collectively, these results indicate that LEF1-upregulated miR-HCC1 functions as an oncogene through the negative regulation of NFIX expression, which links the LEF1/miR-HCC1/NFIX axis to contribute to cell proliferation, migration and invasion of HCC cells and could provide novel insights into miRNA function and hepatocarcinogenesis and potential biomarkers for HCC.
Subject terms: Cell growth, Metastasis
Introduction
Hepatocellular carcinoma (HCC) is one of the most common neoplasms and most frequent cause of cancer death. In China, Hepatitis B virus (HBV) is a major etiologic agent of HCC; almost 80% of HCC patients are associated with hepatitis B virus (HBV) infection. The early metastasis and rapid proliferation of cancer cells play critical roles in the development of HCC. Although locoregional or surgical treatments and chemotherapy are used in clinical settings, the 5-year survival rate of HCC patients remains poor, which is largely due to late stage diagnosis and metastasis.1–3 Therefore, it is essential to explore the mechanisms underlying pathogenesis to facilitate the development of new therapeutic strategies to improve the prognosis of HCC patients.
MicroRNAs (miRNAs) are a class of short endogenous non-coding RNAs approximately 22 nucleotides in length that suppress gene expression by targeting mRNA 3′untranslated regions (3′UTR) or enhance gene expression by binding mRNA 3′UTR in a G-rich RNA sequence binding factor 1 (GRSF1)-dependent manner.4–7 miRNAs participate in a variety of biological progresses, including proliferation, differentiation, and apoptosis.8 Over the last decade, researchers have established an association between aberrant miRNA expression and tumorigenesis of HCC. For example, miR-122 is required for hepatitis C virus (HCV) replication, and robust Argonaute 2 (Ago2) binding to HCV 5′UTR during HCV infection can result in the de-repression of miR-122 targets providing an oncogenic potential between cancer and miRNAs.9 Our lab recently demonstrated that miR-1269b promotes proliferation, cell cycle and migration in HCC cells by binding CDC40 3′UTR to enhance CDC40 expression and function as an oncogene in HCC cells.10 miR-1236 inhibits proliferation, migration and invasion by targeting AFP in HCC cells and it is downregulated in HCC tissues compared with adjacent tissues, which suggests that miR-1236 could act as a tumor suppressor in HCC.11 miRNAs could be oncogenes or tumor suppressors causing the upregulation of oncogenes or the inhibition of tumor suppressor genes or genes with functions related to cell differentiation or apoptosis in HCC.12 Moreover, tumor cells can escape immune surveillance within the tumor microenvironment. miRNAs and multiple proteins participate in this process. For example, TGF-β regulated miR-34a promotes venous metastases of HBV-positive HCC by CCL22 signaling-recruited immunosuppressive cells and promotes the escape of HCC cells from immune surveillance.13 Previous reports also showed that potential novel miRNAs were expressed neither in normal hepatic stem cells nor in differentiated HCC cells but rather in hepatic cancer stem cells (CSCs).14
The transition of epithelial cells to a mesenchymal phenotype (EMT) plays an important role in the migration and invasion of various cancer cells, including HCC.15 EMT is characterized by the decreased expression of epithelial markers, such as E-cadherin, β-catenin, and cytokeratin, as well as the increased expression of mesenchymal markers including ICAM-1, vimentin and fibronectin.16 Therefore, EMT is one of the main molecular mechanisms involved during oncogenesis to promote cancer metastasis. For example, miR-10a promotes HCC cell migration and invasion by targeting EphA4 which be regulated by EMT.17 Previous studies also demonstrated that some factors participate in EMT, lymphoid enhancer binding factor 1 (LEF1), a member of the LEF1/T-cell-specific factor (TCF) family is involved in the development of human cancers, such as colorectal cancer, pancreatic cancer and rhabdomyosarcoma.18–21 Another factor, nuclear factor I/X (NFIX), belongs to the nuclear factor I (NFI) family, which contains four members, NFIa, NFIb, NFIc, and NFIx and encodes proteins with a conserved N-terminal DNA-binding domain, dimerization domain and a C-terminal transactivation/repression domain.22 One of the NFI family numbers, NFIX, is downregulated in esophageal cancer and can inhibit cancer progression in esophageal squamous cell carcinoma (ESCC).23
In this study, Solexa sequencing was applied to profile the miRNA transcriptome of HCC tissues. Surprisingly, beyond the known miRNAs, some new small RNAs were obtained. Among them, a novel miRNA named miR-HCC1 was chosen for further study. miR-HCC1 was found to be upregulated in HCC tissues compared to adjacent non-tumor tissue, and it functions as an oncogene by downregulating the nuclear factor I/X (NFIX) to facilitate cell proliferation, migration and invasion by accelerating EMT process in HCC cells and promoting tumor growth in a xenograft mouse model. Furthermore, LEF1 was shown to activate the transcription of miR-HCC1 via directly binding to its promoter. Thus, these findings demonstrate that a new activated pathway, LEF1-miR-HCC1-NFIX, contributes to oncogenic activities in HCC, which could enhance the understanding between miRNAs and HCC development and could have potential value for clinical application in treating HCC.
Results
MiR-HCC1 is identified and upregulated in HCC tissues and sera
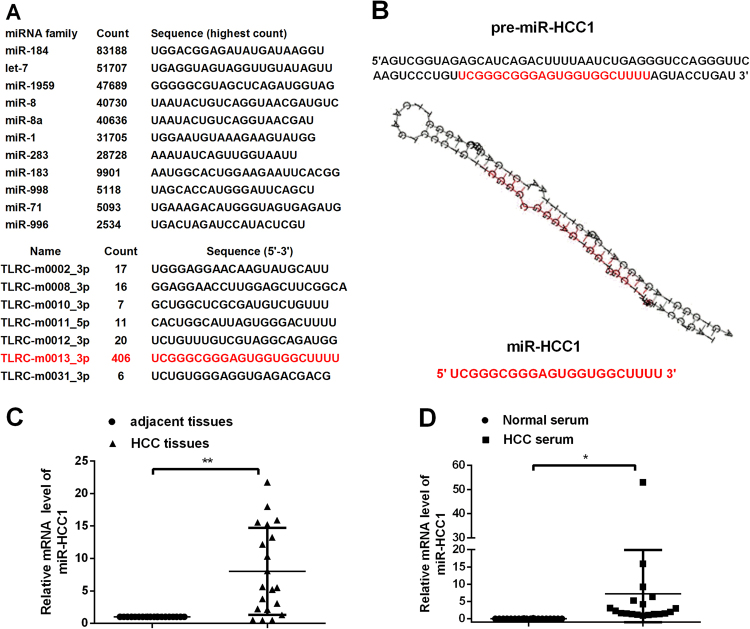
To globally identify novel miRNAs specifically altered in HCC tissues, we performed a Solexa sequencing approach using HCC tissue samples. Sequencing analysis showed approximately 3 million reads and their size distribution was enriched in 21–24 nT. The abundance of almost miRNAs was relatively high; we listed the ten most highly expression known miRNAs (Fig. 1a up), and seven novel miRNAs were discovered, but only one of them was more than 100 count. The other count was relatively low (Fig. 1a down). The novel miRNA with the highest count (406 count) in HCC tissues named miR-HCC1 has the sequence “UUCGGGCGGGAGUGGUGGCUUUU” and it was located in Homo sapiens chromosome 6 genomic scaffold, GRCh38.p7 (NCBI Reference Sequence: NT_113891.3). The pre-miR-HCC1 and its secondary structure were predicted (Fig. 1b). To confirm the Solexa sequencing result, RT-qPCR was applied to detect the level of miR-HCC1 in HCC tissues compared with adjacent noncancerous tissues. As shown in Fig. 1c, miR-HCC1 expression levels were increased ~8-fold in 20 pairs of HCC tissues compared to those in the corresponding paired adjacent noncancerous tissues. We also detected the level of miR-HCC1 in normal sera from 20 people and HCC sera from 20 HCC patients by RT-qPCR. The expression level of miR-HCC1 in HCC sera was ~7.5-fold higher than in normal sera (Fig. 1d). Thus, miR-HCC1 expression levels in HCC tissues and sera were higher than those in the adjacent noncancerous tissues and control sera, which suggest that miR-HCC1 could be involved in carcinogenesis in HCC.
Fig. 1. MiR-HCC1 is identified and deregulated in HCC tissues and sera.
a novel-miRNAs in HCC tissues detected by Solexa sequencing. b The sequence and predicted secondary structure of pre-miR-HCC1 and the mature sequences of miR-HCC1. c The relative levels of miR-HCC1 in HCC tissues and adjacent tissues (n = 20). U6 RNA was used for normalization. d The relative levels of miR-HCC1 in HCC sera and control sera (n = 20) using miR-16 as a normalization control. All data were compared with corresponding control groups. The data are shown as the mean ± SD (*P < 0.05; **P < 0.01)
MiR-HCC1 facilitates cell proliferation of HCC cells in vitro and in vivo and promotes migration and invasion of HCC cells
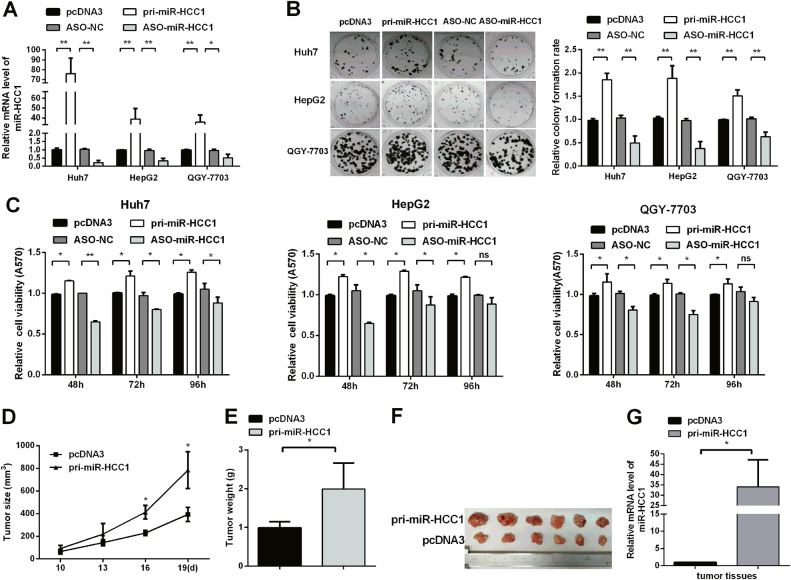
To investigate the role of miR-HCC1 in HCC cells, we constructed the miR-HCC1 overexpression plasmid (pri-miR-HCC1) and commercially synthesized 2′-O-methyl-modified antisense oligonucleotides (ASO-miR-HCC1) to alter the expression levels of miR-HCC1 in HCC cells. As shown in Fig. 2a, miR-HCC1 levels were upregulated 76-fold, 39-fold, and 36-fold in Huh7, HepG2, and QGY-7703 cells transfected with pri-miR-HCC1, respectively, while miR-HCC1 expression level was decreased almost 80%, 70%, and 50% in Huh7, HepG2, and QGY-7703 cells transfected with ASO-miR-HCC1, respectively. Next, MTT and colony formation assays were performed to evaluate the effect of miR-HCC1 on cell proliferation in Huh7, HepG2, and QGY-7703 cells. Pri-miR-HCC1 increased colony formation rate approximately 1.5-fold, but ASO-miR-HCC1 suppressed the colony formation rate ~50% compared with the respective empty vector (Fig. 2b). MTT assays showed that pri-miR-HCC1 increased the cell viability from 1.15-fold to 1.25-fold while ASO-miR-HCC1 significantly inhibited the cell viability from 80 to 65% (Fig. 2c). To further explore the facilitative effect of miR-HCC1 on tumor growth in vivo, we performed animal experiments using a nude mouse tumor xenograft model. The average growth curve (Fig. 2d), weight and volume (Fig. 2e, f) of tumors derived from QGY-7703 cells were significant greater in the miR-HCC1 group than in the control group. The mRNA level of miR-HCC1 in tumor tissues with pri-miR-HCC1 was higher than the control groups (Fig. 2g). The results indicate that miR-HCC1 promotes the growth of HCC cells in vivo and in vivo.
Fig. 2. MiR-HCC1 facilitates cell proliferation of HCC cells in vitro and in vivo.
a RT-qPCR was performed to examine the alteration of miR-HCC1 expression in Huh7, HepG2, and QGY-7703 cells after transfection with pri-miR-HCC1 and ASO-miR-HCC1. b The cell clonogenicity in vitro was assessed by colony formation assays. Huh7, HepG2, and QGY-7703 cells were harvested at 7, 15, and 10 days after transfection, respectively. c MTT assay was applied to determine the cell viability of Huh7, HepG2, and QGY-7703 cell lines. d The nude mouse tumor xenograft model was applied to detect the effect of miR-HCC1 on tumor growth in vivo. The tumor size was measured every 3 days, beginning 10 days after the injection of the QGY-7703 cells. e The tumor weight was measured after the tumors were isolated from the mice. f The mice were euthanized, and the tumors were isolated 19 days after implantation. g miR-HCC1 expression level of nodules in mice. All data were compared with corresponding control groups. The data are shown as the mean ± SD (*P < 0.05; **P < 0.01; ns not significant)
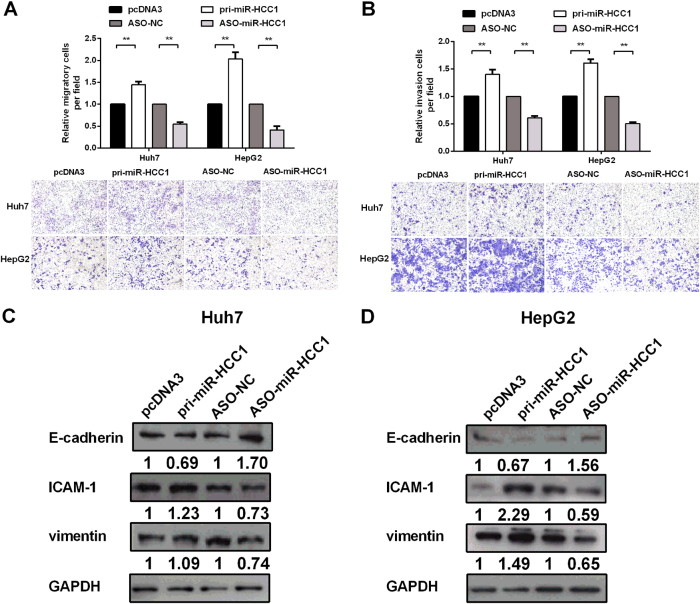
Migration and invasion assays were used to evaluate the migration and invasion capability of HCC cells affected by miR-HCC1. As shown in Fig. 3a, b, miR-HCC1 increased cell migration capacity and invasion capacity in Huh7 and HepG2 cells. Conversely, blocking of miR-HCC1 inhibited cell migration and invasion capacity in Huh7 and HepG2 cells. We also investigated the effect of miR-HCC1 on EMT. EMT-related protein markers were analyzed by Western blot. Pri-miR-HCC1 decreased the protein level of E-cadherin but increased ICAM-1 and vimentin protein levels, whereas miR-HCC1-blocked cells exhibited the opposite results (Fig. 3c, d). Collectively, these results reveal that miR-HCC1 facilitates cell migration and invasion in HCC cells by promoting EMT.
Fig. 3. MiR-HCC1 promotes migration and invasion of HCC cells.
a Migration and invasion assays were used to evaluate the migration and invasion ability of Huh7 cells. b Migration and invasion assays were used to evaluate the migration and invasion ability of HepG2 cells. c The protein levels of EMT-associated markers including E-cadherin, ICAM-1 and vimentin in Huh7 cells assessed by Western blot assays. d The protein levels of EMT-associated markers in HepG2 cells assessed by Western blot assays. All data were compared with corresponding control groups. The data are shown as the mean ± SD (*P < 0.05; **P < 0.01)
MiR-HCC1 directly targets NFIX
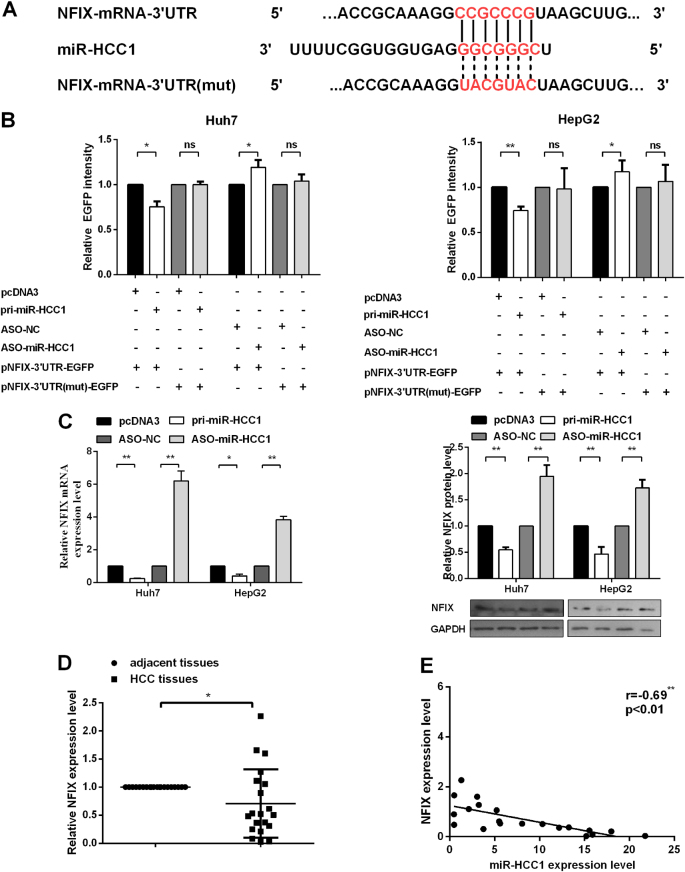
miRNAs can potentially target multiple genes to exert various functions. To determine the target gene that mediates the effects of miR-HCC1 in HCC, bioinformatics analysis was used to predict hundreds of potential targets of miR-HCC1, in which NFIX 3′UTR contains a putative miR-HCC1 binding site (Fig. 4a). We chose NFIX for further study as a miR-HCC1 target gene. To determine a direct interaction between miR-HCC1 and NFIX, EGFP reporter assays were employed. The 3′UTR of NFIX containing the miR-HCC1 binding sites or mutant sites (Fig. 4a) was cloned into the EGFP reporter vectors (pNFIX-3′UTR-EGFP and pNFIX-3′UTR-mut-EGFP). The EGFP fluorescence intensity was reduced by pri-miR-HCC1 but enhanced by ASO-miR-HCC1 when co-transfected with pNFIX-3′UTR-EGFP in Huh7 and HepG2 cells. However, pri-miR-HCC1 or ASO-miR-HCC1 did not affect the EGFP intensity when co-transfected with pNFIX-3′UTR-mut-EGFP (Fig. 4b). In addition, RT-qPCR and Western blot showed that miR-HCC1 overexpression was reduced, but blocking miR-HCC1 increased endogenous mRNA and the NFIX protein level (Fig. 4c). We next examined the expression levels of NFIX by RT-qPCR in 20 pairs of HCC tissues, and we found that NFIX was downregulated in HCC tissues compared with adjacent noncancerous tissues (Fig. 4d). The correlation analysis between NFIX and miR-HCC1 is consitent with the miR-HCC1 negatively regulating NFIX (Fig. 4e). All these results indicate that miR-HCC1 directly targets and downregulates NFIX expression by targeting its 3′UTR.
Fig. 4. MiR-HCC1 targets and downregulates NFIX.
a Putative and mutant binding sites of miR-HCC1 on 3’UTR of NFIX. b Fluorescent reporter assay was conducted 48 h after transfection with pri-miR-HCC1 or ASO-miR-HCC1 in Huh7 and HepG2 cells. c mRNA and protein levels of NFIX at 48 h after transfection with miR-HCC1 or empty vector. d mRNA levels of NFIX in HCC tissues and adjacent noncancerous tissues were evaluated by RT-qPCR (n = 20). e Pearson’s correlation analysis between miR-HCC1 and NFIX expression in HCC tissues. All data were compared with the corresponding control groups. The data are shown as the mean ± SD (*P < 0.05; **P < 0.01; ns not significant)
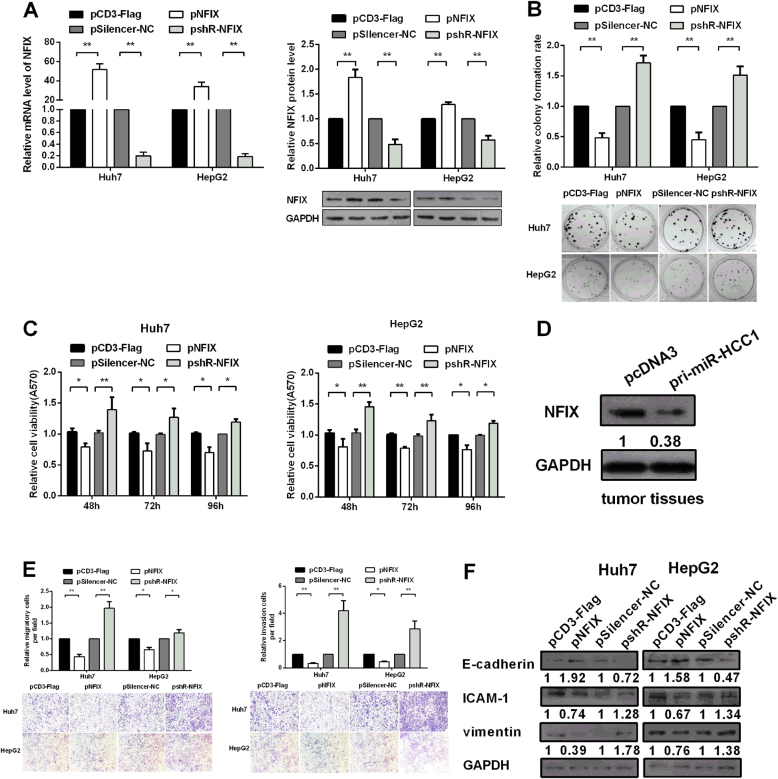
NFIX suppresses HCC cells proliferation, migration and invasion in vitro
NFIX suppresses cancer progression in esophageal squamous cell carcinoma (ESCC) and Murine subventricular zone (SVZ) neurogenic niche.23, 24 To investigate the effects of NFIX in HCC cells, we constructed a pNFIX plasmid to express NFIX fused with Flag tag and a pshR-NFIX plasmid to silence NFIX expression. RT-qPCR and Western blot assays showed that transfection of pNFIX noticeably increased the expression of NFIX, whereas pshR-NFIX significantly decreased NFIX expression in Huh7 and HepG2 cells (Fig. 5a). Colony formation and MTT assays demonstrated that NFIX decreased cell viability, while knockdown of NFIX increased cell viability in Huh7 and HepG2 cells (Fig. 5b, c). We also detected NFIX protein level in tumor tissues from our mouse xenograft study, and the results showed that NFIX protein level in tumor tissues with pri-miR-HCC1 was decreased ~60% compared with the control groups (Fig. 5d). These results showed that NFIX inhibited HCC cell proliferation in vitro.
Fig. 5. NFIX suppresses HCC cell proliferation, migration and invasion in vitro.
a RT-qPCR and Western blot assays were performed to verify the efficiency of pNFIX or pshR-NFIX vector. b Colony formation assays were used to detect cell proliferation in Huh7 and HepG2 cells. c The cell viability of Huh7 and HepG2 cells were analyzed by MTT assay. d Western blot was used to detect the NFIX protein level in tumor tissues. e Alteration of the migration and invasion capability of Huh7 and HepG2 cells transfected with empty vector or NFIX. f Western blot was applied to detect the protein levels of EMT-associated markers-affected by NFIX in Huh7 and HepG2 cells. All data were compared with the corresponding control groups. The data are shown as the mean ± SD (*P < 0.05; **P < 0.01)
Migration and invasion assays showed that NFIX overexpression decreased, but knockdown of NFIX by pshR-NFIX increased the migration and invasion abilities in Huh7 and HepG2 cells (Fig. 5e). We further examined EMT markers including E-cadherin, ICAM-1 and vimentin protein level. Western blot indicated that NFIX promoted the protein level of E-cadherin but decreased ICAM-1 and vimentin in Huh7 and HepG2 cells. Conversely, knockdown of NFIX decreased the protein level of E-cadherin but enhanced the protein levels of ICAM-1 and vimentin (Fig. 5f). These results indicate that NFIX suppresses HCC cell proliferation and enhances EMT to cause oncogenic activities in HCC cells.
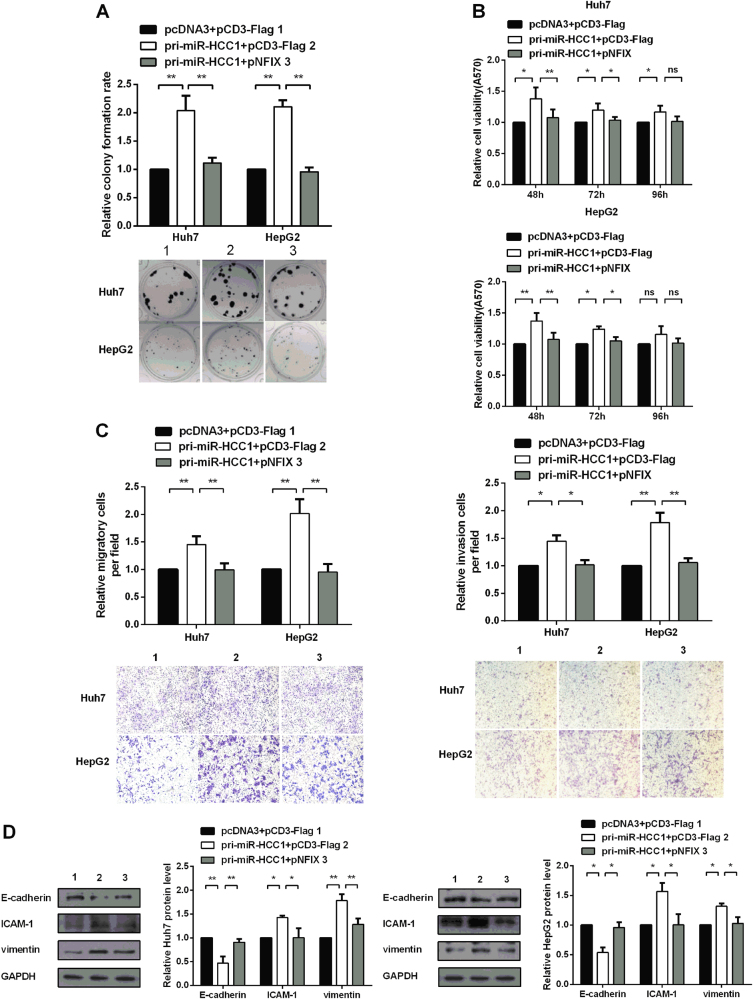
Restoration of NFIX expression counteracts the effects of miR-HCC1 on HCC
To determine whether miR-HCC1 regulated cell proliferation, migration and invasion through targeting NFIX, we performed a rescue experiment in which pri-miR-HCC1 and pNFIX were co-transfected into Huh7 and HepG2 cell lines. As expected, the effects of miR-HCC1 on cell proliferation, migration and invasion were impaired when restoring the NFIX expression level (Fig. 6a–c). Western blot showed that the expression levels of the epithelial marker (E-cadherin) and mesenchymal markers (ICAM-1, vimentin) were rescued by restoring NFIX expression in Huh7 and HepG2 cells (Fig. 6d). Taken together, our results demonstrate that NFIX overexpression counteracted the effect of miR-HCC1 on cell proliferation, migration and invasion. All these results suggest that miR-HCC1 could function as an oncogene at least partially by downregulating NFIX expression.
Fig. 6. Restoration of NFIX expression counteracts the effect of miR-HCC1.
a Cell proliferation was detected by colony formation assay in a rescue experiment. b The cell viability of Huh7 and HepG2 cells were determined by MTT assay. c Migration and invasion assays were used to investigate the migration and invasion capability of Huh7 and HepG2 cells. d EMT-associated markers were detected by Western blot in a rescue experiment. All data were compared with the corresponding control groups. The data are shown as the mean ± SD (*P < 0.05; **P < 0.01; ns not significant)
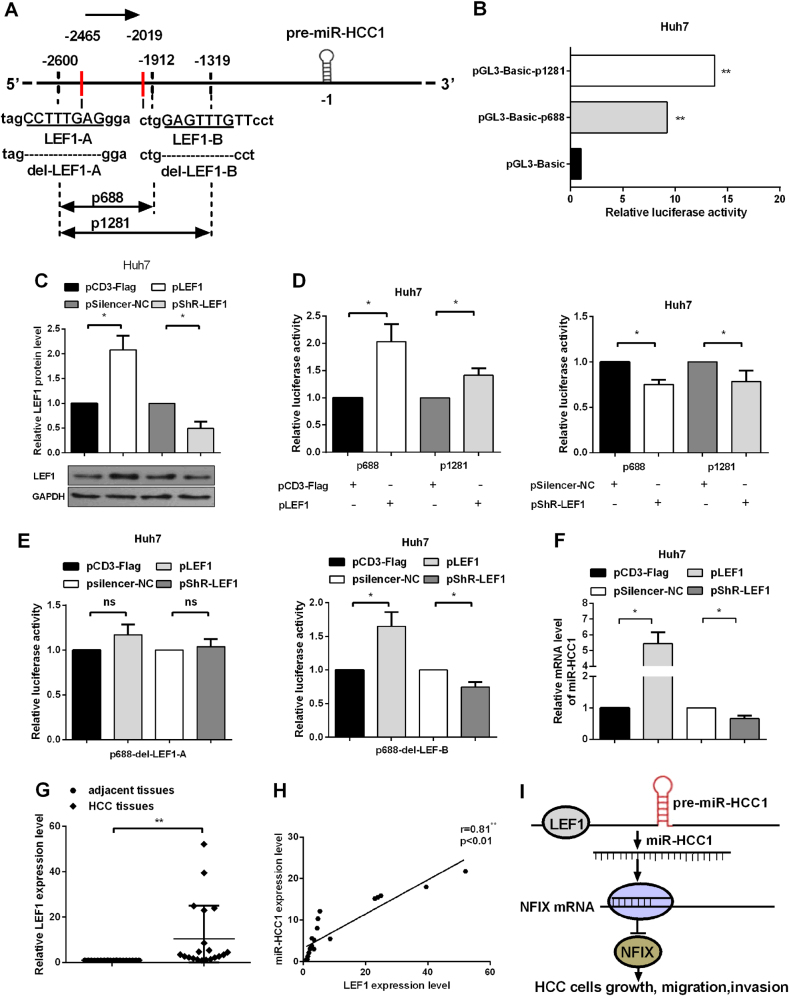
LEF1 binds promoter and enhances the transcription of miR-HCC1
To explore the mechanism of miR-HCC1 upregulation in HCC cells, we applied bioinformatics methods (TRANSPLORER and TFSEARCH) to predict promoter and potential transcription factor binding sites. The predicted promoter region of miR-HCC1 contains two LEF1 binding sites, which are located in the region approximately 2.5 kb upstream from the pre-miR-HCC1 (Fig. 7a). A fragment containing the putative promoter and two LEF1 fragments (−2600 to −1912 kb, −2600 to −1319 kb) were cloned into pGL3-Basic vectors, named pGL3-Basic-p688 and pGL3-Basic-p1281, respectively. The reporter gene activities driven by the fragments were analyzed by luciferase reporter assays. As shown in Fig. 7b, the luciferase activity of pGL3-Basic-p688 and pGL3-Basic-p1281 were greater than 10-fold higher compared with the pGL3-Basic control, suggesting the fragments −2600 to 1319 kb contained a functional promoter.
Fig. 7. LEF1 binds the promoter region and enhances the transcription of miR-HCC1.
a A schematic predicting the regulatory region of miR-HCC1 in the genome. Two transcription initiation sites are indicated. Sequences of two putative LEFs binding sites are shown. b Dual luciferase assays indicate the activity of the miR-HCC1 promoter fragments reporter in Huh7 cells. c Western blot assays were performed to examine the efficiency of pLEF1 or pshR-LEF1 vector in Huh7 cells. d Dual-luciferase assays indicated the activity of the miR-HCC1 promoter fragments by co-transfecting with pLEF1 or pshR-LEF1. e Dual-luciferase assays were conducted to determine the activity of p688-del-LEF1-A and p688-del-LEF-B when transfected with pLEF1 or pshR-LEF1. f miR-HCC1 primary transcript was detected by RT-qPCR when transfected with pLEF1 or pshR-LEF1. g LEF1 expression level detected by RT-qPCR in 20 pairs of HCC tissues. h Pearson’s correlation analysis between LEF1 and miR-HCC1 expression level in HCC tissues. All data were compared with the corresponding control groups. i The proposed model of the transcription factor LEF1 directly binding to promoter of miR-HCC1 to enhance its expression which targets NFIX to contribute to the growth, migration and invasion in HCC. The data are shown as the mean ± SD (*P < 0.05; **P < 0.01)
We next constructed overexpression and knockdown of LEF1 vectors to verify the regulation of LEF1 on the miR-HCC1 promoter, pGL3-Basic-p688 and pGL3-Basic-p1281. Western blot assay was performed to validate the efficiency of pLEF1 and pShR-LEF1 (Fig. 7c). Subsequently, we co-transfected pLEF1 or pShR-LEF1 vector and miR-HCC1 promoter fragments to determine whether LEF1 regulates miR-HCC1 promoter activity. The results indicated that ectopic expression or knockdown of LEF1 enhanced or attenuated the promoter activity of miR-HCC1 in Huh7 cells, thus demonstrating that LEF1 can promote the activity of the miR-HCC1 promoter (Fig. 7d). Next, to demonstrate which LEF1 binding site can regulate the expression level of miR-HCC1, we constructed the plasmids which deleted the predicted LEF1 binding sites in the promoter of pGL3-Basic-p688, named p688-del-LEF1-A (LEF1 binding site A, −2465) and p688del-LEF1-B (LEF1 binding site B, −1912). Luciferase reporter assays showed that alterations of LEF1 did not affect p688-del-LEF1-A activity (Fig. 7e, left panel), but still influenced p688-del-LEF1-B (Fig. 7e, right panel), which suggests that the predicted LEF1 binding site A of miR-HCC1 promoter could be response for binding LEF1. We also tested whether LEF1 affected the endogenous miR-HCC1 primary transcript levels. LEF1 overexpression plasmid or knockdown plasmid was transfected into Huh7 cells and the primary miR-HCC1 was detected by RT-qPCR. As shown in Fig. 7f, the pri-miR-HCC1 level increased when LEF1 was overexpressed in Huh7 cells. By contrast, the pri-miR-HCC1 level decreased upon the knockdown of LEF1. To analyze the correlations of miR-HCC1 and LEF1 in HCC tissues, we examined the expression level of LEF1 in 20 pairs of HCC tissues. The average level of LEF1 mRNA was approximately 10-fold higher compared with the adjacent noncancerous tissues (Fig. 7g). The correlation analysis between LEF1 and miR-HCC1 (Fig. 1c) showed that LEF1 positively regulated miR-HCC1 (Fig. 7h). These results demonstrate that LEF1 activates the transcription of miR-HCC1 by binding to the promoter of miR-HCC1.
Discussion
miRNAs play an important role in the initiation and progression of HCC, and aberrant miRNA expression levels are associated with cancer development and metastasis.25 To explore the underlying mechanisms of the effect of miRNAs on HCC development, we utilized Solexa sequencing to identify the profile of miRNAs in HCC tissues. Notably, some novel miRNAs were discovered. Among them, the highest expression level of a novel miRNA, miR-HCC1, was chosen for investigation in this study. Compared to the miRNAs highly expressed in HCC, such as miR-184, let-7, the miR-HCC1 expression level was relatively low. RT-qPCR showed that miR-HCC1 was upregulated in HCC tissues and sera than in control groups. So we reasonably hypothesized that miR-HCC1 could play roles in the process of HCC. To verify our hypothesis, a series of cell functional experiments were performed, the results showed that miR-HCC1 can promote HCC cell proliferation, migration and invasion capacity, and can accelerate tumor growth in vivo in the nude mouse tumor xenograft model.
Numerous studies have shown that miRNAs can regulate cancer invasion and metastasis by epithelial to mesenchymal transition (EMT)-related mechanisms.16 The EMT process that converts epithelial cells into mesenchymal cells is activated during cancer invasion and metastasis, and cells lose their epithelial features and acquire mesenchymal properties.26 A characteristic of EMT is the loss of E-cadherin and cytokeratin-1 and the acquisition of vimentin and ICAM-1. As we discovered miR-HCC1 can promote HCC cell migration and invasion capacity, we assume that miR-HCC1 may affect EMT process. Then detecting the protein level of E-cadherin, ICAM-1 and vimentin, the results showed that miR-HCC1 decreases the E-cadherin and increases the ICAM-1 and vimentin protein levels. These findings demonstrated that miR-HCC1 affects the migration and invasion via facilitating the EMT process in HCC cells, which suggests that miR-HCC1 could function as an oncogene in hepatocarcinogenesis.
miRNA regulates target gene expression by the RISC complex or in a GRSF1-dependent manner.6,7, 27 miRNAs post-transcriptionally regulate gene expression by binding the 3’UTR of target mRNA, consequently inducing its degradation.8 Bioinformatics combined with gene function analysis were utilized to predict hundreds of candidate target genes of miR-HCC1. One of the candidate genes, NFIX, was found to regulate proliferation and differentiation in radial glial and as a transcription factor to represses the myostatin promoter to regulate satellite cell differentiation and muscle regeneration.28, 29 NFIX also plays crucial roles in regulating the transcriptional switch from embryonic to fetal myogenesis.30 Recently, NFIX was found to be downregulated in esophageal cancer and inhibited cancer progression in esophageal squamous cell carcinoma (ESCC).23 Thus, although the function of NFIX in HCC cells was unclear, we hypothesized that upregulated novel miR-13 could target and suppress the expression of NF1X to promote HCC progression; thus, we selected it as a target of miR-HCC1 for further study. We applied EGFP reporter assays to elucidate that miR-HCC1 can bind the 3′UTR of NF1X, and miR-HCC1 negatively regulated the mRNA and protein levels of NFIX. In addition, we found that NFIX had a lower expression level in HCC tissues compared to adjacent non-tumor tissues, which is inversely correlated with upregulated miR-HCC1 in HCC tissues. Next, we addressed the role of NF1X in HCC and a functional target of miR-HCC1. A series assays showed that NFIX decreased HCC cell growth, proliferation, migration and invasion capacity. Consistently, NFIX inhibited EMT process in HCC cell lines. The rescue experiments also demonstrated that restoration of NFIX expression abrogates the effect of miR-HCC1 on HCC. All the results reveal that miR-HCC1 promotes oncogenic activities of HCC at least partly by downregulating NFIX.
To explore the mechanism of miR-HCC1 upregulation in HCC cells, we cloned the promoter of miR-HCC1 and the upstream transcription regulatory factors. Bioinformatics analysis predicted that miR-HCC1 promoter contains two LEF1 binding sites and that the highly conserved binding element is composed of a consensus sequence (5′-CTTTG[A/T][A/T]-3′) in the promoter region. Researchers have shown that LEF1 can function as a transcription factor to transactivate the expression of the tyrosinase (TYR) gene by physically interacting with its promoter.31 LEF1 facilitated the heparanase (HPSE) transcription via directly binding to its promoter in neuroblastoma (NB) cell lines.32 Therefore, we hypothesize that LEF1 could bind to the miR-HCC1 promoter region to affect the expression level of miR-HCC1. Additional luciferase reporter assays indicated that overexpression of LEF1 increased the promoter fragment activity of miR-HCC1, and the knockdown of LEF1 obtained the opposite results. Mutation assay showed that LEF1 binds to the conserved region (CCTTTGAG, LEF1 binding site A) −2465 kb from the pri-miR-HCC1. We also demonstrated that LEF1 can activate miR-HCC1 expression in Huh7 cells. The LEF1 expression level in HCC tissues was unclear, so we examined the LEF1 expression level in 20 pairs of HCC tissues. As expected, the LEF1 expression level in HCC tissues was higher than the adjacent noncancerous tissues, and a positive correlation was observed with miR-HCC1. Thus, we conclude that LEF1 binds to the promoter to activate miR-HCC1 expression in HCC. Furthermore, LEF1 participates in Wnt signaling pathway which is associated with differentiation, cell polarity and migration.33 So, miR-HCC1 may play roles in Wnt signaling pathways to regulate cell viability, migration, etc.
In summary, we characterized a novel miRNA, miR-HCC1, which is upregulated in HCC tissues and functions as an oncogene that promotes cell proliferation in vitro and in vivo, migration and invasion in vitro and accelerated EMT in HCC. The functions of miR-HCC1 were at least partially caused by the downregulation of NFIX expression. The transcription factor LEF1 directly binds promoter fragments of miR-HCC1 to enhance its transcription in HCC cells and tissues (Fig. 7i). In addition, most of HCC patients are associated with hepatitis B virus (HBV) infection, so we speculate that HBV may regulate the expression level of miR-HCC1, in reverse, the expression level of miR-HCC1 may effect the further infection of HBV in HCC, that’s what we are going to research. Understanding the LEF1/miR-HCC1/NFIX axis will provide insights into the molecular mechanism of tumorigenesis and could be valuable for the development of HCC diagnostics and therapeutics.
Materials and methods
Human HCC cancer tissue, serum specimens and cell lines
The HCC tissue samples used for Solexa sequencing approach and twenty HCC tissues and adjacent non-tumor tissues were all obtained from Tianjin Cancer Hospital and Cancer Institute, Tianjin Medical University. These diagnoses were verified by pathological analysis. Sera from twenty HCC patients and twenty normal people were all obtained from Tangshan People Hospital. Written informed consent was obtained from every patient, and ethics approval for this work was granted by the Ethics Committee of Tianjin Medical University. All the methods were carried out in accordance with the relevant guidelines, including any relevant details. All cell lines used in the experiment were validated and had no bacterial infection.
Vector constructions
The fragment containing pri-miR-HCC1 was inserted into the pcDNA3 vector between its BamHI and EcoRI sites (Promega, Madison, WI), and the antisense oligonucleotide of miR-HCC1 (ASO-miR-HCC1) was synthesized as the inhibitor of miR-HCC1, ASO-NC was used as a control. We constructed the enhanced green fluorescent protein (EGFP) reporter plasmid with the wild type or mutant forms of NFIX mRNA 3′UTR using BamHI and EcoRI sites. Similarly, the fragment containing the NFIX coding sequence was cloned into a pcDNA3-Flag vector using KpnI and XhoI sites. Two signal strands were annealed and cloned into the vector pSilencer 2.1-U6 neo (Ambion, Austin, TX, USA) at BamHI and HindIII sites to construct the knockdown plasmid of NFIX. The promoters of pri-miR-HCC1, which contained the predicted LEF1 binding sites (pGL3-Basic-p688 and pGL3-Basic-p1281) were cloned into the KpnI and EcoRI sites of the pGL3-Basic vector. The promoter of LEF1 deletion-A and deletion-B fragments were cloned into the pGL3-Basic vector using DpnI site and KpnI-EcoRI sites, respectively. LEF1 fragments were amplified from a cDNA derived from Saier Biotechnology Inc. (Tianjin, China) and cloned into a pcDNA3-Flag vector using BamHI and EcoRI sites. The knockdown plasmid of LEF1 used two signal strands that were annealed and cloned into the vector pSilencer 2.1-U6 neo using BamHI and HindIII sites. All plasmids were confirmed by DNA sequencing. The primers used are listed in Table 1.
Table 1.
The primers and oligonucleotides used in this work
| Name | Sequence (5′-3′) |
|---|---|
| pri-miR-HCC1-forward | CGGGATCCCTCACTAGAATGTGCAGGAAG |
| pri-miR-HCC1-reverse | CGGAATTCGTTCTACAGGGTTTGGTG |
| ASO-miR-HCC1 | UUUUGCCUCCUCTCCCGCCCGU |
| ASO-NC | CAGUACUUUUGUGUAGUACAA |
| NFIX-3′UTR-S | GATCCGCGGACCGCAAAGGCCGCCCGTAAGCTTG |
| NFIX-3′UTR-AS | AATTCAAGCTTACGGGCGGCCTTTGCGGTCCGCG |
| NFIX-3′UTR-mut-S | GATCCGCGGACCGCAAAGGGGCCCGCTAAGCTTG |
| NFIX-3′UTR-mut-AS | AATTCAAGCTTAGCGGGCCCCTTTGCGGTCCGCG |
| NFIX-forward | GCGCGGTACCATGTACTCCCCGTACTGCC |
| NFIX-reverse | TGTGGCTCGAGGCGAAAGTTGCCGTCCCGGG |
| NFIX-shR-S | GATCCGGCTCTACAAGTCGCCTCAGTCTCGAGACTGAGGCGACTTGTAGAGCCTTTTTGA |
| NFIX-shR-AS | AGCTTCAAAAAGGCTCTACAAGTCGCCTCAGTCTCGAGACTGAGGCGACTTGTAGAGCCG |
| miR-HCC1-P1281-S | CGCGGGGTACCGCTCTTGAAGATAGAGGG |
| miR-HCC1-P1281-AS | CCGGAATTCGATCCACTGGGTGAAGCC |
| miR-HCC1-P688-S | CGCGGGGTACCGCTCTTGAAGATAGAGGG |
| miR-HCC1-P688-AS | CTAAGAATTCGGAACATCAGAAGGAAC |
| miR-HCC1-P688-LEF1-del-A-S | GTGCTAGACCTTTCTGCTGTGAGATTAAAG |
| miR-HCC1-P688-LEF1-del-A-AS | ACAGCAGAAAGGTCTAGCACAATGGTTGTC |
| miR-HCC1-P688-LEF1-del-B-S | CGCGGGGTACCGCTCTTGAAGATAGAGGG |
| miR-HCC1-P688-LEF1-del-B-AS | CTAAGAATTCGGAACATCAGAAGACACGTCACGT |
| LEF1-forward | CGCGGATCCATGCCCCAACTCTCCGGAG3 |
| LEF1-reverse | CCGGAATTCTCAGATGTAGGCAGCTGTCATTC |
| LEF1-shR-S | GATCCGCAAGAGACAATTATGGTAAGCTCGAGCTTACCATAATTGTCTCTTGCTTTTTGA |
| LEF1-shR-AS | AGCTTCAAAAAGCAAGAGACAATTATGGTAAGCTCGAGCTTACCATAATTGTCTCTTGCG |
| qPCR-β-actin-forward | CGTGACATTAAGGAGAAGCTG |
| qPCR-β-actin-reverse | CTAGAAGCATTTGCGGTGGAC |
| qPCR-NFIX-forward | GGGCAATGAGGGCAACAGG |
| qPCR-NFIX-reverse | ACCGAAACAGAACGAACGACG |
| qPCR-LEF1-forward | AGAACACCCCGATGACGGA |
| qPCR-LEF1-reverse | GAGGGTCCCTTGTTGTAGAGG |
| miR-HCC1 RT primer | GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAAAAGCCA |
| miR-HCC1 forward primer | TGCGGTCGGGCGGGAGTGGTGG |
| U6 RT primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATATGGAAC |
| U6 forward primer | TGCGGGTGCTCGCTTCGGCAGC |
| Reverse primer | CCAGTGCAGGGTCCGAGGT |
Cell transfection and RNA extraction
Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) was used for transient transfection following the manufacturer’s protocol. Total RNA from cell lines, tissue samples and serum samples was isolated using a mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) according to the manufacturer’s instructions.
Solexa sequencing approach
The HCC tissue samples were extracted total RNA, then sent to Beijing Genomics Institute (BGI) for sequencing.
Quantitative reverse transcription-polymerase chain reaction (RT-qPCR)
Two microgram total RNA was used in the reverse transcription reactions (Promega, Madison, WI, USA), qPCR was performed with SYBR Mixture (CWBIO, Beijing, China) and the primers are indicated in Table 1. β-Actin, U6 snRNA or miR-16 was applied for the endogenous control. The transcript levels were analyzed by 2−△△Ct method.
Fluorescent reporter assays and luciferase reporter assays
For fluorescent reporter assays, Huh7 and HepG2 cells were co-transfected with pcDNA3 or miR-HCC1 and ASO-NC or ASO-miR-HCC1 in 48-well plates along with pNFIX-3′UTR-EGFP or mutated NFIX 3′UTR. pDsRed2N11 (Clontech, Mountain View, CA, USA) was used for normalization. The fluorescence intensities of EGFP and RFP were detected with an F-4500 Fluorescence Spectrophotometer (Hitachi, Tokyo, Japan).
For luciferase reporter assays, cells were seeded into 48-well plates and transfected with miR-HCC1 promoter fragment or co-transfected with miR-HCC1 promoter fragment and LEF1. Renilla luciferase pRL-TK was used for normalization. Luciferase was detected 24 h after transfection.
MTT and colony formation assays
Cells were transfected with the indicated plasmids for 24 h and seeded into 96-well plates at a density of 3 × 103 cells/well. At 48 h, 72 h, 96 h after transfection, 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was measured to determine cell viability. The absorbance at 570 nm was measured using a μQuant Universal Microplate Spectrophotometer (Bio-Tek Instruments, Winooski, VT, USA). For the colony formation assays, the cells were counted and seeded in 12-well plates at a density of either 3 × 102 cells/well (Huh7, QGY-7703) or 1 × 103 cells/well (HepG2). The culture medium was changed every 3 days. When the number of colonies contained more than 50 cells, the cells were washed with 1 × PBS and they were stained by crystal violet.
Migration and invasion assays
Transwell chamber inserts with a pore size of 8 μm were used for migration and invasion assays without or with Matrigel. For transwell migration assays, 8 × 104 Huh7 cells or 6 × 104 HepG2 cells in 200 μl MEM-α medium were seeded into the transwell chamber. For invasion assays, 1 × 105 Huh7 cells or 1.2 × 105 HepG2 cells were seeded into the chamber of each insert coated with 40 μl Matrigel (2 mg/ml). The bottom of the insert was incubated in 650 μl MEM-α added to 30% FBS. After incubating for several hours (48 h for migration assays and 72 h for invasion assays), the cells were fixed and then stained.
Western blot
Cellular protein and tissue protein was extracted with RIPA lysis buffer. Protein lysates were separated by SDS-PAGE, transferred into PVDF membranes and then incubated with antibodies. The following antibodies were purchased from Saier Biotechnology Inc. (Tianjin, China): 1:500 rabbit polyclonal anti-E-cadherin antibodies (Catalog No: SRP05266), 1:1000 rabbit polyclonal anti-ICAM-1 antibodies (Catalog No: SRP01174), 1:3000 rabbit polyclonal anti-vimentin antibodies (Catalog No: SRP01327), 1:500 rabbit polyclonal anti-NFIX antibodies (Catalog No: SRP07969) and anti-LEF1 antibodies (Catalog No: SRP00646) and 1:2000 rabbit polyclonal anti-GAPDH antibodies (Catalog No: SRP00849). GAPDH served as an endogenous control.
Tumor xenograft model in nude mice
The QGY-7703 cells were transfected with pri-miR-HCC1 or control vector. In total, 1 × 107 transfected cells were suspended in 100 μl serum-free RPMI 1640 culture medium and were subcutaneously injected into the flank of six 5-week-old female BALB/c nude mice. Tumor size was measured every three days after 10 days of injection. The tumor volume was calculated as follows: length × width2 × 1/2. The tumor weight was measured after the tumors were isolated from the mice. All mice were sacrificed 19 days after implantation. The tumors were isolated from the mice and stored at −80 °C. All studies were performed under the Ethics Committee of Tianjin Medical University.
Statistical analysis
All data were shown as the mean ± SD using GraphPad Prism Software. Pearson’s coefficient correlation was used to analyze gene expression relationships. Other data were analyzed by t-test or analysis of variance (ANOVA). P < 0.05 was considered as statistically significant. Each experiment was independently repeated at least three times.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Nos: 91629302; 81572790; 31270818; 81602410), the Natural Science Foundation of Tianjin (12JCZDJC25100) and Excellent Talent Funding of TMU.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
This article has been retracted. Please see the retraction notice for more detail:https://doi.org/10.1038/s41389-021-00336-91
Yaqi Hu, Xu Guo, and Jinxia Wang contributed equally to this work.
Change history
7/15/2021
A Correction to this paper has been published: 10.1038/s41389-021-00336-9
References
- 1.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 2.Khalili K, et al. Optimization of imaging diagnosis of 1-2 cm hepatocellular carcinoma: an analysis of diagnostic performance and resource utilization. J. Hepatol. 2011;54:723–728. doi: 10.1016/j.jhep.2010.07.025. [DOI] [PubMed] [Google Scholar]
- 3.Papatheodoridis GV, Lampertico P, Manolakopoulos S, Lok A. Incidence of hepatocellular carcinoma in chronic hepatitis B patients receiving nucleos(t)ide therapy: a systematic review. J. Hepatol. 2010;53:348–356. doi: 10.1016/j.jhep.2010.02.035. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 5.Lombardi G, Perego S, Sansoni V, Banfi G. Circulating miRNA as fine regulators of the physiological responses to physical activity: Pre-analytical warnings for a novel class of biomarkers. Clin. Biochem. 2016;49:1331–1339. doi: 10.1016/j.clinbiochem.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 6.van Rooij E, Kauppinen S. Development of microRNA therapeutics is coming of age. EMBO Mol. Med. 2014;6:851–864. doi: 10.15252/emmm.201100899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song G, et al. miR-346 and miR-138 competitively regulate hTERT in GRSF1- and AGO2-dependent manners, respectively. Sci. Rep. 2015;5:15793. doi: 10.1038/srep15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luna JM, et al. Hepatitis C virus RNA functionally sequesters miR-122. Cell. 2015;160:1099–1110. doi: 10.1016/j.cell.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong XX, et al. HBx-induced MiR-1269b in NF-kappaB dependent manner upregulates cell division cycle 40 homolog (CDC40) to promote proliferation and migration in hepatoma cells. J. Transl. Med. 2016;14:189. doi: 10.1186/s12967-016-0949-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao R, et al. miR-1236 down-regulates alpha-fetoprotein, thus causing PTEN accumulation, which inhibits the PI3K/Akt pathway and malignant phenotype in hepatoma cells. Oncotarget. 2015;6:6014–6028. doi: 10.18632/oncotarget.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, Li J, Zheng TH, Bai L, Liu ZJ. MicroRNAs in the Occurrence and Development of Primary Hepatocellular Carcinoma. Adv. Clin. Exp. Med. 2016;25:971–975. doi: 10.17219/acem/36460. [DOI] [PubMed] [Google Scholar]
- 13.Yang P, et al. TGF-beta-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. 2012;22:291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji J, et al. Identification of microRNAs specific for epithelial cell adhesion molecule-positive tumor cells in hepatocellular carcinoma. Hepatology. 2015;62:829–840. doi: 10.1002/hep.27886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol. Oncol. 2017;11:28–39. doi: 10.1002/1878-0261.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 17.Yan Y, et al. MicroRNA-10a is involved in the metastatic process by regulating Eph tyrosine kinase receptor A4-mediated epithelial-mesenchymal transition and adhesion in hepatoma cells. Hepatology. 2013;57:667–677. doi: 10.1002/hep.26071. [DOI] [PubMed] [Google Scholar]
- 18.Eastman Q, Grosschedl R. Regulation of LEF-1/TCF transcription factors by Wnt and other signals. Curr. Opin. Cell. Biol. 1999;11:233–240. doi: 10.1016/S0955-0674(99)80031-3. [DOI] [PubMed] [Google Scholar]
- 19.Wang WJ, et al. Increased LEF1 expression and decreased Notch2 expression are strong predictors of poor outcomes in colorectal cancer patients. Dis. Markers. 2013;35:395–405. doi: 10.1155/2013/983981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singhi AD, et al. Overexpression of lymphoid enhancer-binding factor 1 (LEF1) in solid-pseudopapillary neoplasms of the pancreas. Mod. Pathol. 2014;27:1355–1363. doi: 10.1038/modpathol.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drager J, et al. LEF1 reduces tumor progression and induces myodifferentiation in a subset of rhabdomyosarcoma. Oncotarget. 2017;8:3259–3273. doi: 10.18632/oncotarget.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gronostajski RM. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249:31–45. doi: 10.1016/S0378-1119(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 23.Mao Y, Liu J, Zhang D, Li B. MiR-1290 promotes cancer progression by targeting nuclear factor I/X(NFIX) in esophageal squamous cell carcinoma (ESCC) Biomed. Pharmacother. 2015;76:82–93. doi: 10.1016/j.biopha.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Heng YH, et al. NFIX regulates proliferation and migration within the murine SVZ neurogenic niche. Cereb. Cortex. 2015;25:3758–3778. doi: 10.1093/cercor/bhu253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang XW, Heegaard NH, Orum H. MicroRNAs in liver disease. Gastroenterology. 2012;142:1431–1443. doi: 10.1053/j.gastro.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diaz-Martin J, et al. A core microRNA signature associated with inducers of the epithelial-to-mesenchymal transition. J. Pathol. 2014;232:319–329. doi: 10.1002/path.4289. [DOI] [PubMed] [Google Scholar]
- 27.Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piper M, et al. Nuclear factor one X regulates the development of multiple cellular populations in the postnatal cerebellum. J. Comp. Neurol. 2011;519:3532–3548. doi: 10.1002/cne.22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossi G, et al. Nfix regulates temporal progression of muscle regeneration through modulation of myostatin expression. Cell. Rep. 2016;14:2238–2249. doi: 10.1016/j.celrep.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messina G, et al. Nfix regulates fetal-specific transcription in developing skeletal muscle. Cell. 2010;140:554–566. doi: 10.1016/j.cell.2010.01.027. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, et al. LEF-1 regulates tyrosinase gene transcription in vitro. PLoS. One. 2015;10:e0143142. doi: 10.1371/journal.pone.0143142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qu H, et al. Smad4 suppresses the tumorigenesis and aggressiveness of neuroblastoma through repressing the expression of heparanase. Sci. Rep. 2016;6:32628. doi: 10.1038/srep32628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohammed MK, et al. Wnt/beta-catenin signaling plays an ever-expanding role in stem cell self-renewal, tumorigenesis and cancer chemoresistance. Genes. Dis. 2016;3:11–40. doi: 10.1016/j.gendis.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]