Abstract
Gut microbiota is key to the development and modulation of the mucosal immune system. It plays a central role in several physiological functions, in the modulation of inflammatory signaling and in the protection against infections. In healthy states, there is a perfect balance between commensal and pathogens, and microbiota and the immune system interact to maintain gut homeostasis. The alteration of such balance, called dysbiosis, determines an intestinal bacterial overgrowth which leads to the disruption of the intestinal barrier with systemic translocation of pathogens. The pancreas does not possess its own microbiota, and it is believed that inflammatory and neoplastic processes affecting the gland may be linked to intestinal dysbiosis. Increasing research evidence testifies a correlation between intestinal dysbiosis and various pancreatic disorders, but it remains unclear whether dysbiosis is the cause or an effect. The analysis of specific alterations in the microbiome profile may permit to develop novel tools for the early detection of several pancreatic disorders, utilizing samples, such as blood, saliva, and stools. Future studies will have to elucidate the mechanisms by which gut microbiota is modulated and how it tunes the immune system, in order to be able to develop innovative treatment strategies for pancreatic disorders.
1. Introduction
The human gastrointestinal tract hosts more than 1014 microorganisms, a number 10 to 20 times greater than the total number of cells of the human body, and includes at least 1000 different microbial species, including bacteria, fungi, yeast, viruses, and archaea [1–3]. The ensemble of these populations constitutes the so-called gut microbiota. Instead, the collection of the whole genome sequence of gut microbiota species is called microbiome and consists of more than 5,000,000 genes [4–7].
Gut microbiota is central to the development and modulation of the mucosal innate and adaptive immune system and exerts an important role in the protection against pathogenic microbes by maintaining gut integrity and regulating intestinal barrier permeability. It weighs about 900–1200 g and participates in several physiological functions. Indeed, gut microbiota is constantly involved in facilitating digestion, storing nutrients, secreting vitamins, activating metabolic functions, and shaping intestinal architecture [8]. It is composed of various microbial populations, the most prevalent being the Firmicutes and Bacteroidetes phyla which together represent about 80–90% of the whole gut microbiota [9]. These microbial populations are separated from intestinal epithelial cells by a physical-chemical barrier composed of mucus, mucin glycoproteins, and multiple antibacterial molecules, including alpha-defensins, C-type lectins, lysozyme, phospholipase A2, and secretory IgA [10]. In healthy conditions, all gut microbial species are in a mutualistic or commensal symbiotic state contributing to a perfect and constant homeostasis [11]. In such state, the interaction between gut microbiota, intestinal epithelial cells, and the mucosal immune system creates an environment which controls overgrowth of the host pathogenic flora [12] and limits the colonization of the intestinal tract by foreign pathogens [13–16].
The breakdown of this balance between gut microbiota, the immune system, and the intestinal epithelial barrier results in a pathological condition called dysbiosis [17]. In recent years, several diseases and dysfunctions have been linked to intestinal dysbiosis, including celiac disease, inflammatory bowel disease (IBD), and irritable bowel syndrome (IBS), as well as other conditions [18–24]. In a similar way, given that pancreas is known not to have its own microbial collection, gut microbiota may be involved in the pathogenesis of pancreatic disorders [25]. In this article, we will review the currently available data linking gut microbiota-immune system crosstalk and several pancreatic disorders, such as pancreatitis, diabetes, and pancreatic cancer.
2. Inflammatory Pancreatic Diseases
Acute pancreatitis is an inflammatory disease frequently associated with gallstones or alcohol consumption with a high risk of mortality.
Chronic pancreatitis, instead, is a long-standing, inflammatory disease leading to severe alterations in pancreatic structure and function. The typical clinical manifestations are recurrent episodes of acute pancreatitis in a previously compromised pancreatic gland or a pancreatic exocrine insufficiency due to persistent chronic damage [26].
In either acute or chronic pancreatitis, several alterations in gut microbiota composition have been reported [27].
2.1. Acute Pancreatitis
Hallmark of an acute pancreatitis is an inflammatory state [28, 29] due to an imbalance between pro- and anti-inflammatory cytokines. Recently, Chen et al., in a necrotizing pancreatitis mouse model, demonstrated an overexpression of several proinflammatory cytokines and chemokines, such as TNF-alpha, IL-1beta, IL-6, IL-17A, CXCL1, and IL-18, and a parallel decrease in the Paneth cell-related antimicrobial peptides, such as alpha-defensins and lysozyme [30, 31].
Indeed, pancreatic acinar and Paneth cell-related antimicrobial peptides are essential for gut homeostasis, intestinal immunity integrity, and even for the control of microbiome composition [32]. Recently, in a mouse model, Ahuja et al. have demonstrated that deletion of the Ca2+ channel Orai1 in pancreatic acinar cells (Orai1−/− mice) induces several signs of gut inflammation and bacterial overgrowth, leading to bacterial translocation, systemic infection, and death [33]. These experimental findings further confirm the critical role played by antimicrobial pancreatic secretion in modulating gut/pancreatic homeostasis and gut immune system integrity.
As response to inflammation-mediated tissue damage, acinar pancreatic cells produce several molecules that may have the function of damage-associated molecular patterns (DAMPs) [34], such as high-mobility group box protein 1 (HMGB1), heat shock protein 70 (Hsp70), cytosolic protease-caspase 1, nucleotide-binding domain (NLRP3), adenosine triphosphate (ATP), and DNA [35–37]. DAMPs promote activation of the Toll-like-receptors (TLRs) germline-encoded type I transmembrane receptors present on epithelial cells, immune cells, macrophages, and other cells. TLRs act as pathogen recognition receptors (PRRs) and are able to identify pathogen-associated molecular patterns (PAMPs) [38]. To date, in humans, a total of at least 10 different TLRs have been recognized [39]. The TLRs most frequently implicated in the interactions with intestinal bacteria are TLR2 and TLR4, but several other TLRs may be implicated in the pathogenesis of acute pancreatitis [38, 40]. Nishio et al. demonstrated that in mice genetically deficient in the anti-inflammatory cytokine IL-10, the repeated administration of TLR4 and TLR9 ligands was able to induce pancreatic injury [41]. Matas-Cobos et al. comparing 269 acute pancreatitis patients to 269 healthy controls demonstrated that polymorphisms in TLR3 and TLR6 genes were associated with increased severity of pancreatitis [42].
Each TLR responds to distinct DAMPs, leading to the activation of specific intracellular signaling pathways, and to the production of inflammatory cytokines and chemokines [43]. Notably, in the blood of severe acute pancreatitis patients, an increase of TNF-alpha, IL-1, IL-6, and IL-10 has been documented [28, 29]. However, TLR activation is also linked to the transcription of several genes related to some nuclear factors, such as nuclear factor kappa-B (NF-kB), MAP kinase p38, JNK, and IRF-3, crucial in the control of infection and inflammation [11]. Thus, TLRs may be initially responsible for the inflammatory state, but subsequently, they protect the host, repair damaged tissue, and promote a mucosal immune response [38].
Recently, Watanabe et al. proposed that pancreatitis should be thought as a unique form of immune-mediated inflammation [44]. In this model, a pivotal role is played by TLRs (activated by pathogens related DAMPs), in inducing NF-kB-related adaptive immune system cytokines. In this proinflammatory context, damaged acinar cells begin to produce the proinflammatory cytokine IL-33 that, in turn, determines the activation and recruitment of T-cell subpopulations which participate in pancreatic inflammation.
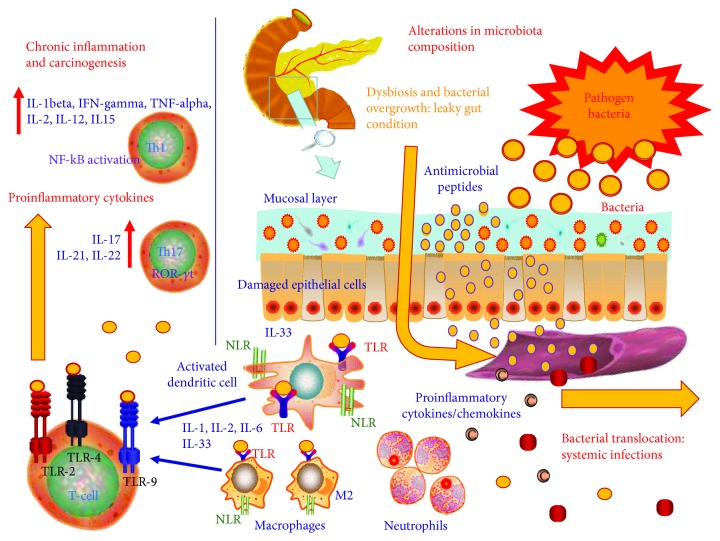
In the context of acute pancreatitis, the inflammation produces intestinal damage by several concomitant pathogenic mechanisms, such as alterations in microcirculation, vasoconstriction in the splanchnic district, and ischemia-reperfusion damage [45, 46]. This, in turn, alters intestinal permeability and leads to a condition known as leaky gut (Figure 1). When there is bacterial overgrowth, leaky gut facilitates the translocation of bacteria and toxins to the pancreas. This worsens pancreatic inflammation resulting in further damage leading to fibrosis or even, in severe cases, necrosis. The bacterial translocation may also be responsible for secondary infections that are associated with a high mortality risk [47].
Figure 1.
Role of leaky gut in pancreatic inflammation and carcinogenesis. The breakdown of the relationship among physiologic and pathogenic bacteria, the immune system, and intestinal epithelial barrier leads to dysbiosis. The pancreas does not possess its own microbiota, and thus, inflammatory and neoplastic processes affecting the gland may be linked to intestinal dysbiosis. In this way, during bacterial overgrowth, leaky gut is responsible for the translocation of bacteria and toxins to the pancreas. Bacterial translocation is involved in pancreatic inflammation due to toxin diffusion and complications like fibrosis, digestive and absorption disorders, diabetes, or cancer. TLR: Toll-like receptors; NLRs: NOD-like receptors; IL: interleukin; IFN: interferon; TNF: tumor necrosis factor; ROR-γt: RAR-related orphan receptor-gamma t; NF-kB: nuclear factor kappa-B.
Moreover, several studies have investigated the relation between inflammatory patterns and microbiota composition during acute pancreatitis. In general, during acute pancreatitis, there is an increase of pathogenic bacteria of the Enterobacteriaceae and Firmicutes families and a decrease of beneficial Bacteroidetes and Lactobacillales [28]. Gerritsen et al. in a mouse model documented that the normal intestinal flora is replaced by an “acute pancreatitis-associated microbiota” [30]. In 2015, Tan et al. published the results of a multicentre prospective clinical study involving 108 acute pancreatitis patients compared to healthy controls [28]. The authors analyzed the 10 predominant bacteria and measured several serum markers of systemic inflammation, such as plasma endotoxin, TNF-alpha, IL-1, IL-6, and IL-10. The findings have shown that the pathogenetic Enterococcus, of the phylum Firmicutes (order Lactobacillales), is increased while Bifidobacterium, of the phylum Actinobacteria (order Bifidobacteriales), is decreased. Additionally, IL-6 serum levels correlated directly with Enterobacteriaceae and Enterococcus number and inversely with the Bifidobacterium and Clostridium cluster XI number. The study by Tan et al. was also able to demonstrate that the extent of gut microbiota modifications predicts pancreatitis severity and the occurrence of systemic complications.
It is notable that in the context of acute pancreatitis several commensal bacteria populations have also been identified. These are associated with reduced levels of inflammatory cytokines, such as IL-1beta, TNF-alpha, CXCL1, and IL-18, and are inversely correlated with pancreatitis severity and systemic infectious complications. Thus, it can be hypothesized that the restoration of a physiological gut microbiota composition may be a useful strategy to treat acute pancreatitis [48]. Indeed, the use of probiotics in this clinical setting has been tested, but results are controversial [49]. Qin et al. in 76 acute pancreatitis patients demonstrated that the restoration of a physiological commensal/pathogens ratio is able to limit the systemic infectious complications [50]. On the other hand, in several other studies, oral administration of probiotics showed no significant impact on disease outcome or on the prevention of complications [48, 51, 52].
2.2. Chronic Pancreatitis
Chronic pancreatitis results from a long-standing inflammation leading to a chronic damage and severe functional impairment of the gland [53, 54].
It has been reported that about one-third of chronic pancreatitis patients are affected by intestinal bacterial overgrowth but the specific alterations in microbiota composition are not yet fully known [55–59]. Some authors have observed an increase in Firmicutes and a relative decrease in Bacteroidetes [27]. Recently, Jandhyala et al. published a study analyzing three groups of patients: chronic pancreatitis with and without diabetes and healthy controls. Regardless of diabetes, in pancreatitis patients, it was documented a progressive, duration-dependent reduction of the commensal bacteria Faecalibacterium prausnitzii [27]. Notably, Faecalibacterium prausnitzii promotes the homeostasis of intestinal epithelium favoring mucin production and tight-junction protein synthesis [60], induces the anti-inflammatory cytokine IL-10 [61], and regulates gut T-cell responses. Thus, the progressive reduction in Faecalibacterium prausnitzii observed in chronic pancreatitis patients testifies to a duration-dependent disruption of gut mucosal integrity [27]. Furthermore, Faecalibacterium prausnitzii levels negatively correlated with plasma endotoxin ones and an increase of endotoxin levels was associated with an impairment of glucose metabolism. Thus, the reduction in Faecalibacterium prausnitzii observed in chronic pancreatitis patients is an additional factor favoring the onset of diabetes or worsening its course. Then, Jandhyala et al. reported a reduction of Ruminococcus bromii in chronic pancreatitis patients [27]. Ruminococcus bromii has an important physiologic role in the degradation of starch in human colon [62]. Its reduction is related to the disruption of the gut mucosal barrier and is responsible of an alteration of the glucose metabolism.
In other studies, a reduction of Bacteroidetes, a Gram-negative bacteria source of lipopolysaccharide (LPS), has consistently been reported. LPS is considered a potent mediator of inflammation. In fact, in binding TLR4, LPS may activate NF-kB-related proinflammatory cytokine production [63]. Chronic pancreatitis patients have higher LPS and endotoxin levels than healthy controls, and these correlate with disease duration. LPS may induce an impairment of pancreatic beta-cells further worsening glucose metabolism [64]. The inflammatory process targets pancreatic islets, and also, T-cell recruitment occurs. In this way, literature data testifies that during chronic pancreatitis there is an increase in both Th1 and Th17 cells [65] and their related proinflammatory cytokines, such as IFN-gamma in pancreatic islets [66].
2.3. Autoimmune Pancreatitis
Pancreatic inflammation may elicit an immune response in the exocrine tissue, leading to either acute or chronic damage. Autoimmune pancreatitis (AIP) accounts for about 5% of all pancreatitis, and it is usually associated with other autoimmune diseases [67]. An increase in serum immunoglobulin G4 (IgG4) is a diagnostic criterion [68]. While genetic factors have been hypothesized [69], the pathogenesis of AIP remains unknown [70].
Interestingly, the gastric Helicobacter pylori infection has been shown to be associated with AIP [71, 72]. This bacterium is known to trigger immune responses against host tissues via several molecular mimicry pathways [73]. Guarneri et al. reported a homology between the human carbonic anhydrase II (CA-II) and alpha-carbonic anhydrase of Helicobacter pylori (HpCA). CA-II is an enzyme of the pancreatic epithelium whose specific serum antibodies are characteristics of AIP, and the bacterial homolog segments contain the binding motif of the high-risk HLA-DR alleles. These data demonstrated that Helicobacter pylori may trigger AIP in genetically predisposed subjects [74].
Other suggestions link bacterial infections with the development of AIP. In a mouse model, Escherichia coli induces a severe pancreatic inflammation and fibrosis similar to the human AIP [75]. Numerous studies have reported that specific microbial antigens may trigger the development of AIP activating immune responses. Gram-negative bacteria-associated LPS is able to activate immune response via-TLRs [41]. Several TLRs (TLR2, TLR3, TLR4, TLR5, and TLR7) have been linked with the development of AIP [76–78]. Among these, TLR3 typically recognizes microbial dsRNA activating the Fas/FasL-mediated cytotoxicity, responsible for chronic inflammation [79]. Finally, TLR7 is able to recognize viral ssRNA, thus activating proinflammatory signaling cascades [80].
3. Diabetes
3.1. Type 1 Diabetes
Type 1 diabetes (T1D) is characterized by a loss of insulin secretion due to damage to pancreatic beta-cells caused by an autoimmune process triggered by microbial infections.
Several alterations in gut microbiota composition have been related to the development of T1D. In a recent study on 76 children at high genetic risk, it has been demonstrated that early changes in gut microbiome composition predict T1D onset [81]. In particular, in the microbiome of these T1D predisposed children, Bacteroides dorei and Bacteroides vulgatus are increased. Instead, in people with late-onset T1D, there is not only a similar increase in Bacteroides species but also a reduction of Clostridium leptum [38, 82].
Furthermore, several bacterial or viral antigens recognized in children and teenagers have been associated later to the development of T1D [83], including antigens from Coxsackievirus A and B, Echovirus, Enterovirus, and so forth.
During the course of T1D, profound alterations in gut microbiota composition and related metabolites take place [84, 85]. Of importance, changes in the ratio of butyrate-producing Bacteroidetes and Firmicutes bacteria occur [86–88]. Other butyrate-producing and mucin-degrading bacteria, such as Prevotella and Akkermansia muciniphila, are decreased [89] while short-chain fatty acid- (SCFAs-) producing bacteria such as Klebsiella are increased.
Recently, Semenkovich et al. demonstrated bidirectional relationships between gut microbiota alterations and T1D-related inflammation. In fact, in a NOD mouse model, gut microbiota was able to instruct hormonal changes in the testosterone axis (in males) which led to T1D susceptibility, and the hormonal levels, in turn, were able to alter the microbial niches in the gut. This phenomenon may be a possible explanation for the different susceptibility between sexes [84, 90].
In a murine T1D model associated with a reduction in Lactobacillus and Bifidobacterium species [91], a coexisting high-grade lymphopenia [92] and an upregulation of Th17 cells have been shown [93]. These findings lend support to the hypothesis that alterations in gut microbiota composition are associated with abnormalities of the mucosal immune system and that both mechanisms participate in T1D pathogenesis [94]. In addition, a leaky gut exacerbates T1D either indirectly via beta-cell damage, due to bacterial translocation and related antigen presentation [95], or directly via beta-cell function impairment mediated by microbial toxins, such as streptozotocin [94].
Diet modification and pharmacological treatment have been similarly studied. Recently, a nonobese diabetic mouse study found that exposure to acidified water is able to increase the presence of mucosal and spleen T-regulatory cells (Tregs) and to decrease Th17 cells, thus decreasing the onset of T1D [96]. A mouse model revealed that insulin treatment is able to somewhat restore microbial populations, positively modulating the microbiota composition towards the normal, healthy state [97]. Xenobiotics have also been implicated in the pathogenesis of T1D. In a recently published study, the neonatal oral administration of vancomycin in a nonobese diabetic mouse reduced the presence of several major genera of Gram-positive and Gram-negative bacteria, with one single species (Akkermansia muciniphila) becoming dominant [98].
Furthermore, in T1D pathogenesis, a special role is played by mucosal innate and adaptive immunity. To elucidate the role of innate immunity in the susceptibility to T1D, the nucleotide-binding oligomerization domain-containing protein 2 (Nod2) has been identified as a key factor [99]. Nod2, mainly expressed in neutrophils and monocytes/macrophages, recognizes bacterial molecules which possess the muramyl dipeptide (MDP) moiety and stimulates an immune reaction, inducing CD4+ Th1 and CD4+ Th17 cells in pancreatic tissue, contributing to autoantibody production and tissue damage [100, 101].
Recently, Li et al. have generated Nod2−/− nonobese diabetic (NOD) mice with a different gut microbiota composition compared to Nod2+/+ NOD mice. Nod2−/− NOD mice appear to be significantly protected from diabetes and present a significant reduction in the proinflammatory cytokine-secreting immune cells and an increase in Tregs [99]. Interestingly, when Nod2−/− NOD mice were housed with Nod2+/+ NOD mice, they lost the protection from diabetes, and this evidence confirmed that T1D susceptibility in Nod2−/− NOD mice is dependent on the alteration of gut microbiota, which modulated the frequency and function of IgA-secreting beta-cells and IL-10 promoting T-regulatory cells. Thus, this study has confirmed the close relationship between gut microbiota and T1D susceptibility and the strong interaction between gut microbiota and the immune system.
Several studies have specifically investigated the role of adaptive immune cells in the pathogenesis of T1D. There is evidence that pancreatic islets infiltrating lymphocytes induce beta-cell damage via CD8+ cytotoxic T-cells. This abnormal activation is believed to be the consequence of mechanisms of molecular mimicry and of microbial infections triggering an immune response. Recent studies have focused on the possible role of TLRs. Pancreatic beta-cells express TLR4 which make them sensitive to LPS, promoting and activating transcription of NF-kB-related proinflammatory genes that mediate an immune response against microbial invasion. Thus, the upregulation of TLR4 is a further mechanism to understand the pathogenesis of T1D [71].
3.2. Metabolic Syndrome and Type 2 Diabetes
Metabolic syndrome is defined by a complex cluster of various elements, including visceral obesity, abnormal glucose metabolism, dyslipidaemia, and arterial hypertension. Metabolic syndrome is associated with an increased risk of type 2 diabetes (T2D) and cardiovascular diseases [102]. The disease is characterized by an increased cytokine production (mainly TNF-alpha and IL-1beta) [103], with a persistent low-grade inflammation [104]. This, in turn, generates a continuous recruitment of immune cells in metabolically active tissues, such as adipose tissue, the pancreatic gland, thyroid, liver, and muscle [105, 106]. T2D is a multifactorial disease, and several factors are involved in its pathogenesis, including diet, obesity, and gut dysbiosis [107].
Gut microbiota has conclusively been linked to the pathogenesis of both metabolic syndrome and T2D. Recently, Guo et al. developed a mouse model with high-fat feeding and demonstrated that the diet was able to alter gut microbial communities, the Paneth cell-related antimicrobial peptide production, and even to increase circulating proinflammatory cytokines, such as TNF-alpha, IL-6, and IL-1beta [108]. Thus, it is the intestinal dysbiosis related to diet, rather than adipose tissue per se, that has a pivotal role in developing intestinal inflammation.
Hence, gut microbiota by affecting the production and storage of energy could influence body weight and obesity [8], tissue proinflammatory activity, peripheral insulin resistance, pancreatic intestinal hormone production, and finally bile acid metabolism [109]. Consequently, in metabolic syndrome, the increase in the Firmicutes/Bacteroidetes ratio corresponds to body weight and promotes the hydrolysis of nondigestible polysaccharides in the gut, which in turn favors an increase in calories extracted from food [110, 111]. Several metagenomic studies performed on metabolic syndrome and T2D patient stools compared to healthy subjects revealed an increase in the order Lactobacillales with a decrease in Roseburia intestinalis, Faecalibacterium prausnitzii, Bacteroides, Prevotella genera, Bifidobacterium animalis, and Methanobrevibacter smithii. On the other hand, Staphylococcus aureus, Escherichia coli, and Lactobacillus reuteri have been found to be elevated and to predict the development of obesity [107].
Certain types of bacteria, such as Tannerella spp., are associated with oral infections and periodontal disease. These are typically characterized by an increase of several proinflammatory cytokines like TNF-alpha, IL-1beta, and IL-6 [112]. Gram-negative bacteria-induced LPS is able to trigger an immune response via LPS-binding protein (LBP), which in turn binds the macrophage receptor CD14. The complex formed by LPS-LBP and CD14 may activate NF-kB and AP-1 proinflammatory genes via TLR4 [113]. LPS may also activate the macrophage and dendritic cell NOD-like receptors (NLRs) that induce NF-kB in association with TLR4 [114]. In this way, a mouse model demonstrated that the lack of TLR4 protects against insulin resistance [115].
Finally, recent evidences demonstrated that intestinal dysbiosis may also mediate alterations in the Th17 cells/Tregs balance. So, the breakdown in the physiological equilibrium between pro- and anti-inflammatory T-cell subpopulations may be responsible for the development and progression of several inflammatory diseases, both in the gastrointestinal tract and in the systemic ones, including obesity-associated metabolic syndrome and T2D [104]. Thus, intestinal dysbiosis is intimately linked to significant alterations in Th17/Tregs balance contributing to obesity, metabolic syndrome, and T2D. Understanding the complex mechanisms responsible for this alteration will allow to develop novel translational therapeutic strategies to potentially treat these widespread diseases.
4. Pancreatic Cancer
Pancreatic cancer is extremely aggressive, with a very poor prognosis. Only 25% of pancreatic cancer can be surgically removed at the time of diagnosis. About 95% of them are adenocarcinomas that originate from gland, ductal, or acinar cells of the exocrine pancreas [116].
A link among dysbiosis, chronic inflammation, and pancreatic cancer has been well established [117–120]. Importantly, dysbiosis is considered not to have a direct mutagenic action disrupting cell cycle control, activating oncogenic signaling pathways, and producing tumor-promoting metabolites [121–124]. However, intestinal dysbiosis can activate the immune system through several pathways involving tumor-infiltrating lymphocytes (TILs) and their related cytokines, innate immune cells, TLRs, and others. In this way, TILs produce proinflammatory mediators inducing STAT3 and NF-kB pathways that act as tumorigenic factors increasing cellular proliferation and suppressing apoptosis [125–127].
Several germ-free mouse models have allowed to understand the significant impact of gut microbiome in carcinogenesis. In fact, germ-free animals have a significant reduction in cancer development, probably due to decreased gut dysbiosis and related chronic inflammation [1, 128]. In the same way, a reduction in cancer development has been observed in mice after antibiotic treatment that may be responsible for the reduction of the pathogen load in the gut mucosa [117]. Other experimental evidence has highlighted the close relationship among diet, xenobiotics, gut microbiota, and cancer [129]. In one study, mice genetically predisposed to colorectal cancer displayed increased tumor progression in a context characterized by a specific microbiota composition. This tumor-predisposing phenotype could be transferred to healthy mice after microbiota transplant using fecal samples. Interestingly, in these mice, antibiotics were able to limit tumor development, probably blocking the tumor-inducing gut microbiota [129]. However, antibiotics could also have a detrimental role. In a recent case-control study conducted on a very large cancer population, Boursi et al. proved that repeated antibiotic exposure is able to promote cancer formation, probably due to a change in microbiota [130]. This study revealed that especially the use of penicillin was associated with an elevated risk of developing colorectal, esophageal, gastric, and pancreatic cancers [130].
In chronic pancreatitis people who harbor a KRAS mutation, there is an increased risk of cancer [131, 132]. In these individuals, gut dysbiosis is able to accelerate pancreatic carcinogenesis due to the mutated KRAS hyperstimulation by the LPS-driven inflammation and by the TLR-mediated NF-kB proinflammatory gene transcription [133, 134]. The role of Gram-negative LPS-TLR4 interaction in inducing chronic inflammation and cancer has been well recognized [135]. In a recent study, Ochi et al. specifically demonstrated their impact in the pathogenesis of pancreatic cancer [136]. In a mouse model, the administration of LPS was able to significantly accelerate carcinogenic progression. On the other hand, the inhibition of TLR4 limited cancer progression, while the inhibition of the TLR adapter protein myeloid differentiation primary response gene 88 (MyD88) unpredictably worsened pancreatic inflammation and cancer development. The procancerogenetic and inflammatory actions of MyD88 inhibition are mediated by dendritic cells (DCs), which were able to induce pancreatic antigen-restricted Th2 cells and promote the transition from pancreatitis to pancreatic cancer [136].
Pathogens are able to act as carcinogenetic agents after infecting the pancreatic gland through intestinal translocation. Among these, a special role is played by Helicobacter pylori [72]. In fact, it has been well established that it may promote the carcinogenesis of the stomach, liver, and pancreas, by inducing the activation of the nuclear factor NF-kB and its proinflammatory cytokines, such as IL-1beta [137]. Fusobacterium species have also been linked to the development of pancreatic cancer, and they are associated with worse prognosis [138].
Recently, Ren et al. studied the microbiota profile of 85 pancreatic cancer patients compared to 57 healthy people [139]. This study revealed that gut microbial diversity is significantly reduced in pancreatic cancer and this tumor is characterized by a unique microbial profile. In particular, the microbial alterations in pancreatic cancer regarded an increase in several pathogens, such as Veillonella, Klebsiella, and Selenomonas, and LPS-producing bacteria including Prevotella, Hallella, and Enterobacter, and a related decrease in several commensals, such as Bifidobacterium, and some butyrate-producing bacteria, such as Coprococcus, Clostridium IV, Blautia, Flavonifractor, and Anaerostipes [139]. The evidence of the increase in the LPS-producing bacteria confirms the role of dysbiosis in mediating chronic inflammation and oxidative damage activating the NF-kB pathway and its related proinflammatory cytokine production. In this way, long-standing chronic inflammation and oxidative damage participate in the development of cancer.
Likewise, it has been shown that pancreatic cancer is associated with an alteration of the physiological oral microbiota composition [140]. Oral microbiota is composed of more than 700 bacteria species which contribute to health and physiology of the mouth, teeth, and oral cavity [117]. Alterations in the taxa dominance and diversity among oral microbial communities, particularly regarding those related to the periodontal disease, may be associated with an increased pancreatic cancer risk [140]. Farrell et al. performed a study analyzing salivary microbiota of several pancreatic cancer and chronic pancreatitis patients compared to healthy subjects [141]. These authors demonstrated that pancreatic cancer is related to a specific alteration in salivary microbiota composition. In particular, it was shown that Neisseria elongata, Corynebacterium spp., and Streptococcus mitis decreased, while Granulicatella adiacens and Porphyromonas gingivalis increased [140, 141]. Recently, Torres et al. conducted a cross-sectional study showing an increase in Leptotrichia spp. and a reduction in Porphyromonas spp. in pancreatic cancer patient saliva; thus, a higher Leptotrichia : Porphyromonas (L : P) ratio may become an important pancreatic cancer diagnostic biomarker [142]. Otherwise, Michaud et al. demonstrated that high antibody titer against gut commensal bacteria was associated with a reduction of 45% in the risk of pancreatic cancer compared to those with a lower antibody titer [143]. In the same way, these authors revealed that the highest concentration of serum antibodies to the pathogenetic bacteria Porphyromonas gingivalis (associated with periodontal disease) was linked to a 2-fold increased risk of pancreatic cancer [143].
Altogether, these evidences highlight the potential to develop future novel diagnostic tools to detect early pancreatic cancer, utilizing samples easy to collect, such as blood, saliva, and stools. However, at the present time, it is not possible to discriminate whether these gut microbial alterations exert a causal role in the developing of pancreatic cancer or, instead, are a result of cancer formation.
Importantly, it should be noted that chronic inflammation-related pancreatic cancer development may occur even without the presence of bacteria. This type of sterile inflammation may be triggered by distant intestinal dysbiosis or translocation of bacteria components, such as LPS, and it is guided by the activation of the immune system through TLRs. In this way, TLR2, TLR4, and TLR9 have been recently shown to be associated with pancreatic cancer development [144, 145].
Finally, recent evidences have shown that gut microbiota and antibiotics may alter tumor response to chemotherapy by modulating tumor microenvironment [146, 147]. Hence, gut microbiota may modify the efficacy of traditional cancer chemotherapies, the novel immune-target drugs, such as anti-CTLA4 and anti-CD274 therapies, but also the tumor recurrence after pancreatic surgery [121].
In conclusion, pancreatic cancer is considered a very insidious and aggressive disease characterized by late diagnosis and no effective screening methods. In this way, in the one hand, it may be too early to hope in the routine use of gut microbiome modulation for therapeutic purposes, and on the other hand, gut microbiome profiling may have important diagnostic tools in the prediction of pancreatic cancer development, thus improving the survival rates associated with this disease.
5. Conclusions
Gut microbiota is central to the development and modulation of the intestinal homeostasis and mucosal immune system integrity and exerts an important role in the protection against pathogenic microbes by maintaining gut integrity and regulating intestinal barrier permeability.
The pancreas does not possess its own microbiota, and the available evidence demonstrates that alteration of gut microbiota determining dysbiosis and bacterial translocation (Table 1) is correlated with the duration and prognosis of several pancreatic disorders, including pancreatitis, diabetes, and cancer. However, whether gut dysbiosis is the cause or an effect of such pathological conditions remains unclear.
Table 1.
Gut microbiota alterations in pancreatic pathologies.
|
Bacterium
(phylum) |
Acute pancreatitis [28] | Chronic pancreatitis [27] | Autoimmune pancreatitis (AIP) | Type 1 diabetes (T1D) | Metabolic syndrome and type 2 diabetes (T2D) | Pancreatic cancer |
|---|---|---|---|---|---|---|
| Increase |
Enterococcus spp. (Firmicutes) Enterobacteriaceae |
Firmicutes |
Helicobacter pylori
(Molecular mimicry mechanism) [72–75] Escherichia coli (Trigger mechanism) [75] |
Bacteroides dorei and vulgates (Bacteroidetes) [38, 82] Klebsiella spp. (Enterobacteriaceae) [89] Coxsackievirus A and B, Echovirus, Enterovirus [83] |
Lactobacillales, Staphylococcus aureus (Firmicutes) [107] Escherichia coli (Proteobacteria) [107] Tannerella spp. (Bacteroidetes) [112] |
Helicobacter pylori
(Proteobacteria) [72] Fusobacterium [138] Leptotrichia [142] (Fusobacteria) Veillonella spp. Selenomonas spp. (Firmicutes) [139] Klebsiella spp. Enterobacter spp. (Enterobacteriaceae) [139] Prevotella spp. Hallella spp. (Bacteroidetes) [139] Salivary microbiota: [140, 141] Granulicatella adiacens (Firmicutes) Porphyromonas gingivalis (Bacteroidetes) |
|
| ||||||
| Decrease | Bacteroidetes Bifidobacterium spp. (Actinobacteria) Lactobacillus spp. Clostridium cluster XI (Firmicutes) |
Bacteroidetes Faecalibacterium prausnitzii (Firmicutes) Ruminococcus bromii (Firmicutes) |
— |
Lactobacillus spp. Clostridium leptum (Firmicutes) [38, 82] Bifidobacterium spp. (Actinobacteria) [91] Prevotella spp. (Bacteroidetes) [89] Akkermansia muciniphila (Verrucomicrobia) [89] |
Bacteroides
Prevotella (Bacteroidetes) [107] Roseburia intestinalis Faecalibacterium prausnitzii (Firmicutes) [107] Bifidobacterium animalis (Actinobacteria) [107] Methanobrevibacter smithii (Methanobacteria) [107] |
Bifidobacterium spp. (Actinobacteria) [139] Coprococcus spp. Clostridium cluster IV Blautia spp. Flavinofractor spp. (Firmicutes) [139] Salivary microbiota: [140, 141] Neisseria elongata (Proteobacteria) Streptococcus mitis (Firmicutes) Corynebacterium spp. (Actinobacteria) |
In principle, the pharmacological modulation of gut microbiota may be beneficial in the treatment of pancreatic conditions and related complications. However, the use of prebiotics, probiotics, antibiotics, and anti-inflammatory drugs or the fecal microbiota transplantation either as a preventative or as a therapeutic strategy remains controversial. These procedures have not yet been a subject to the rigorous efficacy and safety testing necessary to recommend their routine use.
In the foreseeable future, the analysis of specific alterations in the microbiome profile may permit to develop novel tools for the early detection of several pancreatic disorders, utilizing samples easy to collect, such as blood, saliva, and stools.
In conclusion, the ways in which gut microbiota is modulated and interacts with the immune system need to be further elucidated to enter a new era of treatment modalities.
Acknowledgments
The authors thank Dr. C. A. Piccirillo for his valuable expert assistance and suggestions.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
- 1.Leal-Lopes C., Velloso F. J., Campopiano J. C., Sogayar M. C., Correa R. G. Roles of commensal microbiota in pancreas homeostasis and pancreatic pathologies. Journal of Diabetes Research. 2015;2015:20. doi: 10.1155/2015/284680.284680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Savage D. C. Microbial ecology of the gastrointestinal tract. Annual Review of Microbiology. 1977;31(1):107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 3.Whitman W. B., Coleman D. C., Wiebe W. J. Prokaryotes: the unseen majority. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(12):6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen B., Sun L., Zhang X. Integration of microbiome and epigenome to decipher the pathogenesis of autoimmune diseases. Journal of Autoimmunity. 2017;83:31–42. doi: 10.1016/j.jaut.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Ehrlich S. D. The human gut microbiome impacts health and disease. Comptes Rendus Biologies. 2016;339(7-8):319–323. doi: 10.1016/j.crvi.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Forbes J. D., Van Domselaar G., Bernstein C. N. The gut microbiota in immune-mediated inflammatory diseases. Frontiers in Microbiology. 2016;7:p. 1081. doi: 10.3389/fmicb.2016.01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lloyd-Price J., Abu-Ali G., Huttenhower C. The healthy human microbiome. Genome Medicine. 2016;8(1):p. 51. doi: 10.1186/s13073-016-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang C., Li J. Pathogenic microorganisms and pancreatic cancer. Gastrointestinal Tumors. 2015;2(1):41–47. doi: 10.1159/000380896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucas López R., Grande Burgos M. J., Gálvez A., Pérez Pulido R. The human gastrointestinal tract and oral microbiota in inflammatory bowel disease: a state of the science review. APMIS. 2017;125(1):3–10. doi: 10.1111/apm.12609. [DOI] [PubMed] [Google Scholar]
- 10.Gallo R. L., Hooper L. V. Epithelial antimicrobial defence of the skin and intestine. Nature Reviews Immunology. 2012;12(7):503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frosali S., Pagliari D., Gambassi G., Landolfi R., Pandolfi F., Cianci R. How the intricate interaction among toll-like receptors, microbiota, and intestinal immunity can influence gastrointestinal pathology. Journal of Immunology Research. 2015;2015:12. doi: 10.1155/2015/489821.489821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pagliari D., Piccirillo C. A., Larbi A., Cianci R. The interactions between innate immunity and microbiota in gastrointestinal diseases. Journal of Immunology Research. 2015;2015:3. doi: 10.1155/2015/898297.898297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooper L. V., Macpherson A. J. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nature Reviews Immunology. 2010;10(3):159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 14.Longman R. S., Yang Y., Diehl G. E., Kim S. V., Littman D. R. Microbiota: host interactions in mucosal homeostasis and systemic autoimmunity. Cold Spring Harbor Symposia on Quantitative Biology. 2013;78:193–201. doi: 10.1101/sqb.2013.78.020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macpherson A. J., Uhr T. Compartmentalization of the mucosal immune responses to commensal intestinal bacteria. Annals of the New York Academy of Sciences. 2004;1029(1):36–43. doi: 10.1196/annals.1309.005. [DOI] [PubMed] [Google Scholar]
- 16.Macpherson A. J., Geuking M. B., McCoy K. D. Innate and adaptive immunity in host-microbiota mutualism. Frontiers in Bioscience. 2012;4:685–698. doi: 10.2741/293. [DOI] [PubMed] [Google Scholar]
- 17.Lin L., Zhang J. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunology. 2017;18(1):p. 2. doi: 10.1186/s12865-016-0187-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen J. J., Sartor R. B. Therapeutic manipulation of the microbiome in IBD: current results and future approaches. Current Treatment Options in Gastroenterology. 2015;13(1):105–120. doi: 10.1007/s11938-014-0042-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sartor R. B. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134(2):577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 20.Uronis J. M., Mühlbauer M., Herfarth H. H., Rubinas T. C., Jones G. S., Jobin C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS One. 2009;4(6, article e6026) doi: 10.1371/journal.pone.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marietta E. V., Gomez A. M., Yeoman C., et al. Low incidence of spontaneous type 1 diabetes in non-obese diabetic mice raised on gluten-free diets is associated with changes in the intestinal microbiome. PLoS One. 2013;8(11, article e78687) doi: 10.1371/journal.pone.0078687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo J., Kim S. Probiotics and prebiotics: present status and future perspectives on metabolic disorders. Nutrients. 2016;8(3):p. 173. doi: 10.3390/nu8030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amar J., Chabo C., Waget A., et al. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Molecular Medicine. 2011;3(9):559–572. doi: 10.1002/emmm.201100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burcelin R., Serino M., Chabo C., Blasco-Baque V., Amar J. Gut microbiota and diabetes: from pathogenesis to therapeutic perspective. Acta Diabetologica. 2011;48(4):257–273. doi: 10.1007/s00592-011-0333-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Signoretti M., Roggiolani R., Stornello C., Delle Fave G., Capurso G. Gut microbiota and pancreatic diseases. Minerva Gastroenterologica e Dietologica. 2017;63(4):399–410. doi: 10.23736/S1121-421X.17.02387-X. [DOI] [PubMed] [Google Scholar]
- 26.Conwell D. L., Banks P. A., Sandhu B. S., et al. Validation of demographics, etiology, and risk factors for chronic pancreatitis in the USA: a report of the North American Pancreas Study (NAPS) group. Digestive Diseases and Sciences. 2017;62(8):2133–2140. doi: 10.1007/s10620-017-4621-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jandhyala S. M., Madhulika A., Deepika G., et al. Altered intestinal microbiota in patients with chronic pancreatitis: implications in diabetes and metabolic abnormalities. Scientific Reports. 2017;7, article 43640 doi: 10.1038/srep43640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan C., Ling Z., Huang Y., et al. Dysbiosis of intestinal microbiota associated with inflammation involved in the progression of acute pancreatitis. Pancreas. 2015;44(6):868–875. doi: 10.1097/MPA.0000000000000355. [DOI] [PubMed] [Google Scholar]
- 29.Kylanpaa M. L., Repo H., Puolakkainen P. A. Inflammation and immunosuppression in severe acute pancreatitis. World Journal of Gastroenterology. 2010;16(23):2867–2872. doi: 10.3748/wjg.v16.i23.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerritsen J., Timmerman H. M., Fuentes S., et al. Correlation between protection against sepsis by probiotic therapy and stimulation of a novel bacterial phylotype. Applied and Environmental Microbiology. 2011;77(21):7749–7756. doi: 10.1128/AEM.05428-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J., Huang C., Wang J., et al. Dysbiosis of intestinal microbiota and decrease in paneth cell antimicrobial peptide level during acute necrotizing pancreatitis in rats. PLoS One. 2017;12(4, article e0176583) doi: 10.1371/journal.pone.0176583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong W. Shaping the gut microbiome from the pancreas. Science Signaling. 2017;10(472, article eaan3016) doi: 10.1126/scisignal.aan3016. [DOI] [PubMed] [Google Scholar]
- 33.Ahuja M., Schwartz D. M., Tandon M., et al. Orai1-mediated antimicrobial secretion from pancreatic acini shapes the gut microbiome and regulates gut innate immunity. Cell Metabolism. 2017;25(3):635–646. doi: 10.1016/j.cmet.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kono H., Rock K. L. How dying cells alert the immune system to danger. Nature Reviews Immunology. 2008;8(4):279–289. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bui F. Q., Johnson L., Roberts J. A., et al. Fusobacterium nucleatum infection of gingival epithelial cells leads to NLRP3 inflammasome-dependent secretion of IL-1β and the danger signals ASC and HMGB1. Cellular Microbiology. 2016;18(7):970–981. doi: 10.1111/cmi.12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee B. C., Lee J. Cellular and molecular players in adipose tissue inflammation in the development of obesity-induced insulin resistance. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2014;1842(3):446–462. doi: 10.1016/j.bbadis.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keyel P. A. How is inflammation initiated? Individual influences of IL-1, IL-18 and HMGB1. Cytokine. 2014;69(1):136–145. doi: 10.1016/j.cyto.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Kramer C. D., Genco C. A. Microbiota, immune subversion, and chronic inflammation. Frontiers in Immunology. 2017;8:p. 255. doi: 10.3389/fimmu.2017.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Achek A., Yesudhas D., Choi S. Toll-like receptors: promising therapeutic targets for inflammatory diseases. Archives of Pharmacal Research. 2016;39(8):1032–1049. doi: 10.1007/s12272-016-0806-9. [DOI] [PubMed] [Google Scholar]
- 40.Hoque R., Malik A. F., Gorelick F., Mehal W. Z. Sterile inflammatory response in acute pancreatitis. Pancreas. 2012;41(3):353–357. doi: 10.1097/MPA.0b013e3182321500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishio A., Asada M., Uchida K., Fukui T., Chiba T., Okazaki K. The role of innate immunity in the pathogenesis of experimental autoimmune pancreatitis in mice. Pancreas. 2011;40(1):95–102. doi: 10.1097/MPA.0b013e3181f3a5d4. [DOI] [PubMed] [Google Scholar]
- 42.Matas-Cobos A. M., Redondo-Cerezo E., Alegría-Motte C., et al. The role of Toll-like receptor polymorphisms in acute pancreatitis occurrence and severity. Pancreas. 2015;44(3):1–33. doi: 10.1097/MPA.0000000000000272. [DOI] [PubMed] [Google Scholar]
- 43.Evavold C. L., Kagan J. C. How inflammasomes inform adaptive immunity. Journal of Molecular Biology. 2018;430(2):217–237. doi: 10.1016/j.jmb.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe T., Kudo M., Strober W. Immunopathogenesis of pancreatitis. Mucosal Immunology. 2017;10(2):283–298. doi: 10.1038/mi.2016.101. [DOI] [PubMed] [Google Scholar]
- 45.Lutgendorff F., Nijmeijer R. M., Sandström P. A., et al. Probiotics prevent intestinal barrier dysfunction in acute pancreatitis in rats via induction of ileal mucosal glutathione biosynthesis. PLoS One. 2009;4(2, article e4512) doi: 10.1371/journal.pone.0004512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Capurso G., Zerboni G., Signoretti M., et al. Role of the gut barrier in acute pancreatitis. Journal of Clinical Gastroenterology. 2012;46:S46–S51. doi: 10.1097/MCG.0b013e3182652096. [DOI] [PubMed] [Google Scholar]
- 47.Petrov M. S., Shanbhag S., Chakraborty M., Phillips A. R. J., Windsor J. A. Organ failure and infection of pancreatic necrosis as determinants of mortality in patients with acute pancreatitis. Gastroenterology. 2010;139(3):813–820. doi: 10.1053/j.gastro.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 48.Rychter J. W., van Minnen L. P., Verheem A., et al. Pretreatment but not treatment with probiotics abolishes mouse intestinal barrier dysfunction in acute pancreatitis. Surgery. 2009;145(2):157–167. doi: 10.1016/j.surg.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 49.van Baal M. C., Kohout P., Besselink M. G., et al. Probiotic treatment with Probioflora in patients with predicted severe acute pancreatitis without organ failure. Pancreatology. 2012;12(5):458–462. doi: 10.1016/j.pan.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 50.Qin H. L., Zheng J. J., Tong D. N., et al. Effect of Lactobacillus plantarum enteral feeding on the gut permeability and septic complications in the patients with acute pancreatitis. European Journal of Clinical Nutrition. 2008;62(7):923–930. doi: 10.1038/sj.ejcn.1602792. [DOI] [PubMed] [Google Scholar]
- 51.Sharma B., Srivastava S., Singh N., Sachdev V., Kapur S., Saraya A. Role of probiotics on gut permeability and endotoxemia in patients with acute pancreatitis: a double-blind randomized controlled trial. Journal of Clinical Gastroenterology. 2011;45(5):442–448. doi: 10.1097/MCG.0b013e318201f9e2. [DOI] [PubMed] [Google Scholar]
- 52.Besselink M. G. H., van Santvoort H. C., Buskens E., et al. Probiotic prophylaxis in predicted severe acute pancreatitis: a randomised, double-blind, placebo-controlled trial. Lancet. 2008;371(9613):651–659. doi: 10.1016/S0140-6736(08)60207-X. [DOI] [PubMed] [Google Scholar]
- 53.Frulloni L., Falconi M., Gabbrielli A., et al. Italian consensus guidelines for chronic pancreatitis. Digestive and Liver Disease. 2010;42(Supplement 6):S381–S406. doi: 10.1016/S1590-8658(10)60682-2. [DOI] [PubMed] [Google Scholar]
- 54.Etemad B., Whitcomb D. C. Chronic pancreatitis: diagnosis, classification, and new genetic developments. Gastroenterology. 2001;120(3):682–707. doi: 10.1053/gast.2001.22586. [DOI] [PubMed] [Google Scholar]
- 55.Capurso G., Signoretti M., Archibugi L., Stigliano S., Delle Fave G. Systematic review and meta-analysis: small intestinal bacterial overgrowth in chronic pancreatitis. United European Gastroenterology Journal. 2016;4(5):697–705. doi: 10.1177/2050640616630117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Signoretti M., Stigliano S., Valente R., Piciucchi M., Fave G. D., Capurso G. Small intestinal bacterial overgrowth in patients with chronic pancreatitis. Journal of Clinical Gastroenterology. 2014;48:S52–S55. doi: 10.1097/MCG.0000000000000238. [DOI] [PubMed] [Google Scholar]
- 57.Mancilla A. C., Madrid S. A., Hurtado H. C., et al. Small intestine bacterial overgrowth in patients with chronic pancreatitis. Revista Médica de Chile. 2008;136(8):976–980. doi: 10.4067/S0034-98872008000800003. [DOI] [PubMed] [Google Scholar]
- 58.Grace E., Shaw C., Whelan K., Andreyev H. J. N. Review article: small intestinal bacterial overgrowth – prevalence, clinical features, current and developing diagnostic tests, and treatment. Alimentary Pharmacology and Therapeutics. 2013;38(7):674–688. doi: 10.1111/apt.12456. [DOI] [PubMed] [Google Scholar]
- 59.Kumar K., Ghoshal U. C., Srivastava D., Misra A., Mohindra S. Small intestinal bacterial overgrowth is common both among patients with alcoholic and idiopathic chronic pancreatitis. Pancreatology. 2014;14(4):280–283. doi: 10.1016/j.pan.2014.05.792. [DOI] [PubMed] [Google Scholar]
- 60.Wrzosek L., Miquel S., Noordine M. L., et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biology. 2013;11(1):p. 61. doi: 10.1186/1741-7007-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rossi O., van Berkel L. A., Chain F., et al. Faecalibacterium prausnitzii A2-165 has a high capacity to induce IL-10 in human and murine dendritic cells and modulates T cell responses. Scientific Reports. 2016;6(1, article 18507) doi: 10.1038/srep18507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ze X., Duncan S. H., Louis P., Flint H. J. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. The ISME Journal. 2012;6(8):1535–1543. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Newton K., Dixit V. M. Signaling in innate immunity and inflammation. Cold Spring Harbor Perspectives in Biology. 2012;4(3) doi: 10.1101/cshperspect.a006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amyot J., Semache M., Ferdaoussi M., Fontés G., Poitout V. Lipopolysaccharides impair insulin gene expression in isolated islets of Langerhans via Toll-like receptor-4 and NF-κB signalling. PLoS One. 2012;7(4, article e36200) doi: 10.1371/journal.pone.0036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pavan Kumar P., Radhika G., Rao G. V., et al. Interferon γ and glycemic status in diabetes associated with chronic pancreatitis. Pancreatology. 2012;12(1):65–70. doi: 10.1016/j.pan.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 66.Talukdar R., Sasikala M., Pavan Kumar P., Rao G. V., Pradeep R., Reddy D. N. T-helper cell–mediated islet inflammation contributes to β-cell dysfunction in chronic pancreatitis. Pancreas. 2016;45(3):434–442. doi: 10.1097/MPA.0000000000000479. [DOI] [PubMed] [Google Scholar]
- 67.Majumder S., Takahashi N., Chari S. T. Autoimmune pancreatitis. Digestive Diseases and Sciences. 2017;62(7):1762–1769. doi: 10.1007/s10620-017-4541-y. [DOI] [PubMed] [Google Scholar]
- 68.Hamano H., Kawa S., Horiuchi A., et al. High serum IgG4 concentrations in patients with sclerosing pancreatitis. The New England Journal of Medicine. 2001;344(10):732–738. doi: 10.1056/NEJM200103083441005. [DOI] [PubMed] [Google Scholar]
- 69.Klöppel G., Lüttges J., Löhr M., Zamboni G., Longnecker D. Autoimmune pancreatitis: pathological, clinical, and immunological features. Pancreas. 2003;27(1):14–19. doi: 10.1097/00006676-200307000-00002. [DOI] [PubMed] [Google Scholar]
- 70.Watanabe T., Yamashita K., Arai Y., et al. Chronic fibro-inflammatory responses in autoimmune pancreatitis depend on IFN-α and IL-33 produced by plasmacytoid dendritic cells. The Journal of Immunology. 2017;198(10):3886–3896. doi: 10.4049/jimmunol.1700060. [DOI] [PubMed] [Google Scholar]
- 71.Frulloni L., Lunardi C., Simone R., et al. Identification of a novel antibody associated with autoimmune pancreatitis. The New England Journal of Medicine. 2009;361(22):2135–2142. doi: 10.1056/NEJMoa0903068. [DOI] [PubMed] [Google Scholar]
- 72.Rabelo-Goncalves E. M., Roesler B. M., Zeitune J. M. Extragastric manifestations of Helicobacter pylori infection: possible role of bacterium in liver and pancreas diseases. World Journal of Hepatology. 2015;7(30):2968–2979. doi: 10.4254/wjh.v7.i30.2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kountouras J., Zavos C., Gavalas E., Tzilves D. Challenge in the pathogenesis of autoimmune pancreatitis: potential role of Helicobacter pylori infection via molecular mimicry. Gastroenterology. 2007;133(1):368–369. doi: 10.1053/j.gastro.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 74.Guarneri F., Guarneri C., Benvenga S. Helicobacter pylori and autoimmune pancreatitis: role of carbonic anhydrase via molecular mimicry? Journal of Cellular and Molecular Medicine. 2005;9(3):741–744. doi: 10.1111/j.1582-4934.2005.tb00506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yanagisawa N., Haruta I., Shimizu K., et al. Identification of commensal flora-associated antigen as a pathogenetic factor of autoimmune pancreatitis. Pancreatology. 2014;14(2):100–106. doi: 10.1016/j.pan.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 76.Umemura T., Katsuyama Y., Hamano H., et al. Association analysis of Toll-like receptor 4 polymorphisms with autoimmune pancreatitis. Human Immunology. 2009;70(9):742–746. doi: 10.1016/j.humimm.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 77.Fukui Y., Uchida K., Sakaguchi Y., et al. Possible involvement of Toll-like receptor 7 in the development of type 1 autoimmune pancreatitis. Journal of Gastroenterology. 2015;50(4):435–444. doi: 10.1007/s00535-014-0977-4. [DOI] [PubMed] [Google Scholar]
- 78.Watanabe T., Yamashita K., Fujikawa S., et al. Involvement of activation of toll-like receptors and nucleotide-binding oligomerization domain–like receptors in enhanced IgG4 responses in autoimmune pancreatitis. Arthritis & Rheumatism. 2012;64(3):914–924. doi: 10.1002/art.33386. [DOI] [PubMed] [Google Scholar]
- 79.Soga Y., Komori H., Miyazaki T., et al. Toll-like receptor 3 signaling induces chronic pancreatitis through the Fas/Fas ligand-mediated cytotoxicity. The Tohoku Journal of Experimental Medicine. 2009;217(3):175–184. doi: 10.1620/tjem.217.175. [DOI] [PubMed] [Google Scholar]
- 80.Ochi A., Graffeo C. S., Zambirinis C. P., et al. Toll-like receptor 7 regulates pancreatic carcinogenesis in mice and humans. The Journal of Clinical Investigation. 2012;122(11):4118–4129. doi: 10.1172/JCI63606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davis-Richardson A. G., Ardissone A. N., Dias R., et al. Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Frontiers in Microbiology. 2014;5:p. 678. doi: 10.3389/fmicb.2014.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sysi-Aho M., Ermolov A., Gopalacharyulu P. V., et al. Metabolic regulation in progression to autoimmune diabetes. PLoS Computational Biology. 2011;7(10, article e1002257) doi: 10.1371/journal.pcbi.1002257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Frisk G., Nilsson E., Tuvemo T., Friman G., Diderholm H. The possible role of Coxsackie A and echo viruses in the pathogenesis of type I diabetes mellitus studied by IgM analysis. The Journal of Infection. 1992;24(1):13–22. doi: 10.1016/0163-4453(92)90814-M. [DOI] [PubMed] [Google Scholar]
- 84.Semenkovich C. F., Danska J., Darsow T., et al. American Diabetes Association and JDRF research symposium: diabetes and the microbiome. Diabetes. 2015;64(12):3967–3977. doi: 10.2337/db15-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Paun A., Yau C., Danska J. S. The influence of the microbiome on type 1 diabetes. The Journal of Immunology. 2017;198(2):590–595. doi: 10.4049/jimmunol.1601519. [DOI] [PubMed] [Google Scholar]
- 86.Giongo A., Gano K. A., Crabb D. B., et al. Toward defining the autoimmune microbiome for type 1 diabetes. The ISME Journal. 2011;5(1):82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Knip M., Siljander H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nature Reviews Endocrinology. 2016;12(3):154–167. doi: 10.1038/nrendo.2015.218. [DOI] [PubMed] [Google Scholar]
- 88.Lernmark B., Johnson S. B., Vehik K., et al. Enrollment experiences in a pediatric longitudinal observational study: The Environmental Determinants of Diabetes in the Young (TEDDY) study. Contemporary Clinical Trials. 2011;32(4):517–523. doi: 10.1016/j.cct.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brown C. T., Davis-Richardson A. G., Giongo A., et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6(10, article e25792) doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Markle J. G. M., Frank D. N., Mortin-Toth S., et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339(6123):1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 91.Roesch L. F. W., Lorca G. L., Casella G., et al. Culture-independent identification of gut bacteria correlated with the onset of diabetes in a rat model. The ISME Journal. 2009;3(5):536–548. doi: 10.1038/ismej.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brugman S., Klatter F. A., Visser J. T. J., et al. Antibiotic treatment partially protects against type 1 diabetes in the bio-breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia. 2006;49(9):2105–2108. doi: 10.1007/s00125-006-0334-0. [DOI] [PubMed] [Google Scholar]
- 93.Lau K., Benitez P., Ardissone A., et al. Inhibition of type 1 diabetes correlated to a Lactobacillus johnsonii N6.2-mediated Th17 bias. The Journal of Immunology. 2011;186(6):3538–3546. doi: 10.4049/jimmunol.1001864. [DOI] [PubMed] [Google Scholar]
- 94.Aw W., Fukuda S. Understanding the role of the gut ecosystem in diabetes mellitus. Journal of Diabetes Investigation. 2018;9(1):5–12. doi: 10.1111/jdi.12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Concannon P., Rich S. S., Nepom G. T. Genetics of type 1A diabetes. The New England Journal of Medicine. 2009;360(16):1646–1654. doi: 10.1056/NEJMra0808284. [DOI] [PubMed] [Google Scholar]
- 96.Wolf K. J., Daft J. G., Tanner S. M., Hartmann R., Khafipour E., Lorenz R. G. Consumption of acidic water alters the gut microbiome and decreases the risk of diabetes in NOD mice. The Journal of Histochemistry & Cytochemistry. 2014;62(4):237–250. doi: 10.1369/0022155413519650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wirth R., Bódi N., Maróti G., et al. Regionally distinct alterations in the composition of the gut microbiota in rats with streptozotocin-induced diabetes. PLoS One. 2014;9(12, article e110440) doi: 10.1371/journal.pone.0110440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hansen C. H. F., Krych L., Nielsen D. S., et al. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia. 2012;55(8):2285–2294. doi: 10.1007/s00125-012-2564-7. [DOI] [PubMed] [Google Scholar]
- 99.Li Y. Y., Pearson J. A., Chao C., et al. Nucleotide-binding oligomerization domain-containing protein 2 (Nod2) modulates T1DM susceptibility by gut microbiota. Journal of Autoimmunity. 2017;82:85–95. doi: 10.1016/j.jaut.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Costa F. R. C., Françozo M. C. S., de Oliveira G. G., et al. Gut microbiota translocation to the pancreatic lymph nodes triggers NOD2 activation and contributes to T1D onset. The Journal of Experimental Medicine. 2016;213(7):1223–1239. doi: 10.1084/jem.20150744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wen L., Ley R. E., Volchkov P. Y., et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature. 2008;455(7216):1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alberti K. G., Eckel R. H., Grundy S. M., et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120(16):1640–1645. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- 103.Nordmann T. M., Dror E., Schulze F., et al. The role of inflammation in β-cell dedifferentiation. Scientific Reports. 2017;7(1):p. 6285. doi: 10.1038/s41598-017-06731-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Luo A., Leach S. T., Barres R., Hesson L. B., Grimm M. C., Simar D. The microbiota and epigenetic regulation of T helper 17/regulatory T cells: in search of a balanced immune system. Frontiers in Immunology. 2017;8:p. 417. doi: 10.3389/fimmu.2017.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sell H., Habich C., Eckel J. Adaptive immunity in obesity and insulin resistance. Nature Reviews Endocrinology. 2012;8(12):709–716. doi: 10.1038/nrendo.2012.114. [DOI] [PubMed] [Google Scholar]
- 106.Ray I., Mahata S. K., De R. K. Obesity: an immunometabolic perspective. Frontiers in Endocrinology. 2016;7:p. 157. doi: 10.3389/fendo.2016.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Million M., Maraninchi M., Henry M., et al. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. International Journal of Obesity. 2012;36(6):817–825. doi: 10.1038/ijo.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 108.Guo X., Li J., Tang R., et al. High fat diet alters gut microbiota and the expression of Paneth cell-antimicrobial peptides preceding changes of circulating inflammatory cytokines. Mediators of Inflammation. 2017;2017:9. doi: 10.1155/2017/9474896.9474896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Han J. L., Lin H. L. Intestinal microbiota and type 2 diabetes: from mechanism insights to therapeutic perspective. World Journal of Gastroenterology. 2014;20(47):17737–17745. doi: 10.3748/wjg.v20.i47.17737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., Gordon J. I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 111.Zmora N., Bashiardes S., Levy M., Elinav E. The role of the immune system in metabolic health and disease. Cell Metabolism. 2017;25(3):506–521. doi: 10.1016/j.cmet.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 112.Stafford G., Roy S., Honma K., Sharma A. Sialic acid, periodontal pathogens and Tannerella forsythia: stick around and enjoy the feast! Molecular Oral Microbiology. 2012;27(1):11–22. doi: 10.1111/j.2041-1014.2011.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Neal M. D., Leaphart C., Levy R., et al. Enterocyte TLR4 mediates phagocytosis and translocation of bacteria across the intestinal barrier. The Journal of Immunology. 2006;176(5):3070–3079. doi: 10.4049/jimmunol.176.5.3070. [DOI] [PubMed] [Google Scholar]
- 114.Boulangé C. L., Neves A. L., Chilloux J., Nicholson J. K., Dumas M. E. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Medicine. 2016;8(1):p. 42. doi: 10.1186/s13073-016-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shi H., Kokoeva M. V., Inouye K., Tzameli I., Yin H., Flier J. S. TLR4 links innate immunity and fatty acid–induced insulin resistance. The Journal of Clinical Investigation. 2006;116(11):3015–3025. doi: 10.1172/JCI28898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bailey P., Chang D. K., Nones K., et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 117.Schwabe R. F., Jobin C. The microbiome and cancer. Nature Reviews Cancer. 2013;13(11):800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zambirinis C. P., Pushalkar S., Saxena D., Miller G. Pancreatic cancer, inflammation, and microbiome. The Cancer Journal. 2014;20(3):195–202. doi: 10.1097/PPO.0000000000000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Balkwill F., Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357(9255):539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 120.Malka D., Hammel P., Maire F., et al. Risk of pancreatic adenocarcinoma in chronic pancreatitis. Gut. 2002;51(6):849–852. doi: 10.1136/gut.51.6.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mima K., Nakagawa S., Sawayama H., et al. The microbiome and hepatobiliary-pancreatic cancers. Cancer Letters. 2017;402:9–15. doi: 10.1016/j.canlet.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 122.Garrett W. S. Cancer and the microbiota. Science. 2015;348(6230):80–86. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.García-Castillo V., Sanhueza E., McNerney E., Onate S. A., García A. Microbiota dysbiosis: a new piece in the understanding of the carcinogenesis puzzle. Journal of Medical Microbiology. 2016;65(12):1347–1362. doi: 10.1099/jmm.0.000371. [DOI] [PubMed] [Google Scholar]
- 124.Grivennikov S. I., Greten F. R., Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cianci R., Pagliari D., Pietroni V., Landolfi R., Pandolfi F. Tissue infiltrating lymphocytes: the role of cytokines in their growth and differentiation. Journal of Biological Regulators & Homeostatic Agents. 2010;24(3):239–249. [PubMed] [Google Scholar]
- 126.Francescone R., Hou V., Grivennikov S. I. Microbiome, inflammation, and cancer. The Cancer Journal. 2014;20(3):181–189. doi: 10.1097/PPO.0000000000000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pandolfi F., Cianci R., Pagliari D., et al. The immune response to tumors as a tool toward immunotherapy. Clinical and Developmental Immunology. 2011;2011:12. doi: 10.1155/2011/894704.894704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li Y., Kundu P., Seow S. W., et al. Gut microbiota accelerate tumor growth via c-jun and STAT3 phosphorylation in APC Min/+ mice. Carcinogenesis. 2012;33(6):1231–1238. doi: 10.1093/carcin/bgs137. [DOI] [PubMed] [Google Scholar]
- 129.Schulz M. D., Atay C., Heringer J., et al. High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature. 2014;514(7523):508–512. doi: 10.1038/nature13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Boursi B., Mamtani R., Haynes K., Yang Y. X. Recurrent antibiotic exposure may promote cancer formation – another step in understanding the role of the human microbiota? European Journal of Cancer. 2015;51(17):2655–2664. doi: 10.1016/j.ejca.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Guerra C., Schuhmacher A. J., Cañamero M., et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11(3):291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 132.di Magliano M. P., Logsdon C. D. Roles for KRAS in pancreatic tumor development and progression. Gastroenterology. 2013;144(6):1220–1229. doi: 10.1053/j.gastro.2013.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Daniluk J., Liu Y., Deng D., et al. An NF-κB pathway–mediated positive feedback loop amplifies Ras activity to pathological levels in mice. The Journal of Clinical Investigation. 2012;122(4):1519–1528. doi: 10.1172/JCI59743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huang H., Daniluk J., Liu Y., et al. Oncogenic K-Ras requires activation for enhanced activity. Oncogene. 2014;33(4):532–535. doi: 10.1038/onc.2012.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kojima M., Morisaki T., Izuhara K., et al. Lipopolysaccharide increases cyclo-oxygenase-2 expression in a colon carcinoma cell line through nuclear factor-κB activation. Oncogene. 2000;19(9):1225–1231. doi: 10.1038/sj.onc.1203427. [DOI] [PubMed] [Google Scholar]
- 136.Ochi A., Nguyen A. H., Bedrosian A. S., et al. MyD88 inhibition amplifies dendritic cell capacity to promote pancreatic carcinogenesis via Th2 cells. The Journal of Experimental Medicine. 2012;209(9):1671–1687. doi: 10.1084/jem.20111706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang W., Abbruzzese J. L., Evans D. B., Larry L., Cleary K. R., Chiao P. J. The nuclear factor-κB RelA transcription factor is constitutively activated in human pancreatic adenocarcinoma cells. Clinical Cancer Research. 1999;5(1):119–127. [PubMed] [Google Scholar]
- 138.Mitsuhashi K., Nosho K., Sukawa Y., et al. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget. 2015;6(9):7209–7220. doi: 10.18632/oncotarget.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ren Z., Jiang J., Xie H., et al. Gut microbial profile analysis by MiSeq sequencing of pancreatic carcinoma patients in China. Oncotarget. 2017;8(56):95176–95191. doi: 10.18632/oncotarget.18820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ertz-Archambault N., Keim P., Von Hoff D. Microbiome and pancreatic cancer: a comprehensive topic review of literature. World Journal of Gastroenterology. 2017;23(10):1899–1908. doi: 10.3748/wjg.v23.i10.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Farrell J. J., Zhang L., Zhou H., et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2012;61(4):582–588. doi: 10.1136/gutjnl-2011-300784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Torres P. J., Fletcher E. M., Gibbons S. M., Bouvet M., Doran K. S., Kelley S. T. Characterization of the salivary microbiome in patients with pancreatic cancer. PeerJ. 2015;3, article e1373 doi: 10.7717/peerj.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Michaud D. S., Izard J., Wilhelm-Benartzi C. S., et al. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut. 2013;62(12):1764–1770. doi: 10.1136/gutjnl-2012-303006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Grimmig T., Moench R., Kreckel J., et al. Toll like receptor 2, 4, and 9 signaling promotes autoregulative tumor cell growth and VEGF/PDGF expression in human pancreatic cancer. International Journal of Molecular Sciences. 2016;17(12) doi: 10.3390/ijms17122060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Michaud D. S., Izard J. Microbiota, oral microbiome, and pancreatic cancer. The Cancer Journal. 2014;20(3):203–206. doi: 10.1097/PPO.0000000000000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vetizou M., Pitt J. M., Daillere R., et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Iida N., Dzutsev A., Stewart C. A., et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342(6161):967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]