Key Points
Question
What are the population-based distributions and pathologic characteristics of melanocytic proliferations, ranging from benign to malignant, as diagnosed via skin biopsies?
Findings
Using natural language processing applied to 80 368 pathology reports, we found that 23% of biopsies performed were of melanocytic lesions and 77% were of nonmelanocytic lesions. When the melanocytic lesions were subclassified by MPATH-Dx category, we found that about 83% were class I; 8% class II, 5% class III, 2% class IV, and 2% class V.
Meaning
These population-based estimates provide important new data on the frequency of melanocytic proliferations and the characteristics of their diagnostic spectrum.
Abstract
Importance
Population-based information on the distribution of histologic diagnoses associated with skin biopsies is unknown. Electronic medical records (EMRs) enable automated extraction of pathology report data to improve our epidemiologic understanding of skin biopsy outcomes, specifically those of melanocytic origin.
Objective
To determine population-based frequencies and distribution of histologically confirmed melanocytic lesions.
Design, Setting, and Participants
A natural language processing (NLP)-based analysis of EMR pathology reports of adult patients who underwent skin biopsies at a large integrated health care delivery system in the US Pacific Northwest from January 1, 2007, through December 31, 2012.
Exposures
Skin biopsy procedure.
Main Outcomes and Measures
The primary outcome was histopathologic diagnosis, obtained using an NLP-based system to process EMR pathology reports. We determined the percentage of diagnoses classified as melanocytic vs nonmelanocytic lesions. Diagnoses classified as melanocytic were further subclassified using the Melanocytic Pathology Assessment Tool and Hierarchy for Diagnosis (MPATH-Dx) reporting schema into the following categories: class I (nevi and other benign proliferations such as mildly dysplastic lesions typically requiring no further treatment), class II (moderately dysplastic and other low-risk lesions that may merit narrow reexcision with <5-mm margins), class III (eg, melanoma in situ and other higher-risk lesions warranting reexcision with 5-mm to 1-cm margins), and class IV/V (invasive melanoma requiring wide reexcision with ≥1-cm margins and potential adjunctive therapy). Health system cancer registry data were used to define the percentage of invasive melanoma cases within MPATH-Dx class IV (stage T1a) vs V (≥stage T1b).
Results
A total of 80 368 skin biopsies, performed on 47 529 patients, were examined. Nearly 1 in 4 skin biopsies were of melanocytic lesions (23%; n = 18 715), which were distributed according to MPATH-Dx categories as follows: class I, 83.1% (n = 15 558); class II, 8.3% (n = 1548); class III, 4.5% (n = 842); class IV, 2.2% (n = 405); and class V, 1.9% (n = 362).
Conclusions and Relevance
Approximately one-quarter of skin biopsies resulted in diagnoses of melanocytic proliferations. These data provide the first population-based estimates across the spectrum of melanocytic lesions ranging from benign through dysplastic to malignant. These results may serve as a foundation for future research seeking to understand the epidemiology of melanocytic proliferations and optimization of skin biopsy utilization.
This large-scale analysis of pathology reports in electronic medical records, using the natural language processing technique, evaluates population-based distributions and pathologic characteristics of melanocytic proliferations.
Introduction
The number of skin biopsies performed in the United States increases by approximately 6% annually, and nearly 1 in 10 older adults undergoes a skin biopsy procedure each year. This utilization amounts to millions of skin biopsies per year, yet little is known about the outcomes of these biopsies from a population perspective. The paucity of information exists in part because registry-based reporting is not mandated in the United States for dermatological diseases other than cutaneous melanoma. This knowledge gap is compounded by variable diagnostic classifications as well as variable integration of pathology reporting and claims-based billing data, such that final pathology skin biopsy diagnoses are not consistently recorded using typical billing or insurance claims.
Accordingly, the epidemiology of most diseases for which skin biopsies are performed remains largely unknown. In general, research in this area has been limited to case series and small cohort studies with designs unable to provide reliable population-based estimates. Alternatively, studies have used insurance claims–based analyses of specific conditions (such as nonmelanoma skin cancers) that are often restricted to specific patient subpopulations (eg, Medicare participants) using repurposed data of varying fidelity and narrow statistical approaches.
In particular, our understanding of the epidemiology associated with skin biopsies revealing nonmalignant melanocytic neoplasms remains limited. The lack of information regarding the prevalence and distribution of benign nevi vs dysplastic nevi has, in turn, constrained our ability to evaluate potential overdiagnosis and underdiagnosis of these lesions, despite the emergence of promising standardized tools enabling consistent and meaningful histopathologic grading. While multiple prior studies report variability in the interpretation of melanocytic lesions, with some dermatopathologists recommending expert consensus review for challenging cases, it is not known what percentage of all skin biopsies result in diagnoses of melanocytic lesions across the spectrum from benign through intermediate to malignant lesions.
Rising adoption of electronic medical record (EMR) systems, in concert with advances in machine learning–based algorithms, may enable improved understanding of the diagnostic outcomes associated with skin biopsy procedures that could overcome previous challenges. These challenges include reliance on diagnostic and procedural codes that are primarily designed for insurance reimbursement and not epidemiologic research. Accuracy can sometimes be enhanced by manual medical record review, but this may be expensive, time-consuming, and subject to inherent variability across human abstractors, in addition to posing potential risks to patient privacy.
Natural language processing (NLP)—an array of computational methods for evaluating machine-readable, unstructured text—has recently emerged as an alternative or adjunctive approach for gathering rich clinical details embedded within EMR systems for large-scale analyses. For example, various medical specialties have successfully used NLP to perform granular analyses of radiographic imaging and pathology reports. These NLP-based approaches have been found to perform as well as, or better than, manual medical record review.
We used an NLP-based system to describe the distribution of diagnoses applied to skin biopsies, a basic question that has, to date, remained unanswered. Although a similar NLP-based approach has been used to assess lymph node status in patients with invasive melanoma, the overall population-based estimates of cutaneous melanocytic lesions is unknown, and more specifically, the distribution of melanocytic proliferations ranging from benign to malignant has not been previously characterized.
We report the results of an NLP-based approach to evaluate skin biopsy pathology reports from patients in a large, integrated health system. Our primary goals were (1) to determine the percentage of all skin biopsies diagnosed as melanocytic proliferations and (2) to categorize and characterize the distribution of these melanocytic proliferations using the Melanocytic Pathology Assessment Tool and Hierarchy for Diagnosis (MPATH-Dx) schema, a standardized classification system for melanocytic lesions ranging from class I (eg, benign melanocytic lesions) to class V (eg, ≥pT1b invasive melanomas).
Methods
Study Population and EMR-Documents
This study was conducted at Kaiser Permanente Washington (formerly Group Health Cooperative), an integrated health care delivery system in Washington State, from January 1, 2007, to December 31, 2012. Clinical documents for all patients were obtained from EMR systems and included all available machine-readable pathology reports. These were chosen as the primary source of skin biopsy–associated diagnoses because pathology reports provide the strongest evidence regarding the outcome of the skin biopsies and are often linguistically simpler than other clinical text contained within an EMR. This study was approved by the Kaiser Permanente Washington institutional review board, waiving written informed consent for deidentified data.
Inclusion/Exclusion Criteria
The study population included all patients ages 18 years or older enrolled in the health plan during the study period who underwent a skin biopsy. We defined skin biopsies using corresponding Healthcare Common Procedure Coding System (HCPCS)/Current Procedural Terminology 4 (CPT-4) and International Classification of Diseases, Ninth Revision (ICD-9) codes (eAppendix 1 in the Supplement). Twelve months of continuous patient enrollment, defined as enrollment gaps no longer than 92 days prior to skin biopsy, was required for study inclusion.
Study Design, Exposures, and Outcomes
The primary study design was cross-sectional, with the primary exposure constituting receipt of a skin biopsy and the primary outcomes defined as (1) frequency and percentage of nonmelanocytic vs melanocytic histologic diagnoses and (2) frequency and percentage of melanocytic proliferations classified according to the MPATH-Dx system.
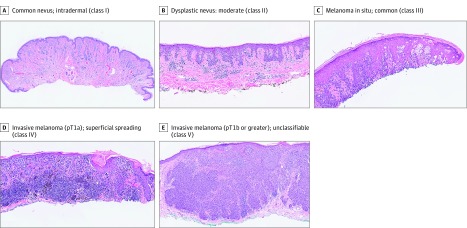
The MPATH-Dx classification system was developed as a tool to standardize and improve communication about melanocytic lesions. The development and evaluation of this tool has been previously reported. Briefly, the histologic diagnosis of these lesions can be subject to discordance and errors, potentially leading to inappropriate treatment and harm. The lack of standardization in diagnostic terminology can lead to confusion for clinical care and challenges for investigators. The diverse terminologies are stratified by commonalities of treatments into a 5-class MPATH-Dx system moving from benign lesions to the highest grade of invasive melanoma (Figure).
Figure. Example Histopathologic Images for Each of the 5 MPATH-Dx Classes.
MPATH-Dx indicates Melanocytic Pathology Assessment Tool and Hierarchy for Diagnosis.
The study was performed within a Surveillance, Epidemiology, and End Results Registry (SEER) location so that we could take advantage of SEER estimates for delineating the invasive melanoma cases. The NLP-based system used to extract information from pathology reports was not designed to distinguish between MPATH-Dx class IV (invasive melanoma stage T1a) and class V (invasive melanoma ≥stage T1b); thus, estimates for these categories were initially combined into a single class IV/class V category. Thereafter, corresponding population-level estimates of invasive melanoma by stage were obtained from the local integrated health system cancer registry data used in SEER registry reporting (for all enrollees 18 years or older from 2007 through 2012) (eAppendix 2 in the Supplement). The relative population-level percentages of stage T1a and stage T1b or higher invasive melanomas were then applied to the combined MPATH-Dx class IV/class V category to derive estimates of class IV vs class V frequencies and percentages.
We conducted a secondary analysis to describe the diagnoses from skin biopsies of melanocytic proliferations over time at the level of the patient. A retrospective cohort design was used to account for additional diagnoses that may arise over time as subsequent skin biopsies are performed. For this secondary analysis, each patient undergoing skin biopsy was included only once, identified according to the date of the index or first skin biopsy. Each patient was followed over 1 year from the date of index skin biopsy to determine if additional skin biopsies were performed and if the patient received a higher-level diagnosis after subsequent biopsies. Subsequent biopsies (if any) performed following the index biopsy were identified and stratified by prespecified time intervals (index biopsy, 90 days, and 365 days). We report person-level pathology diagnosis distributions for men and women by age group.
NLP classification
Original pathology reports (n = 289) were identified with a goal of capturing a stratified distribution of pathology reports across each of the MPATH-Dx classes. The original reports were independently reviewed and classified into the MPATH-Dx system by 2 experienced dermatologists (J.P.L. and E.K.), and any cases with disagreements were reviewed in conjunction to reach consensus. A string search method was initially used to extract information from the text with phrases used to create a simple context-free grammar that generated 6455 different phrases, all linked to their associated MPATH-Dx class. As each phrase is created, it is turned into a regular expression that allows for flexible spelling and spacing between words.
The second step was to incorporate linking and negation rules using a modified version of the NegEx algorithm. These linking rules describe conjunctions such as “and,” “or,” and commas to ensure that linked phrases are appropriately negated according to their intended linguistic meaning. These rules ensure that such phrases as “no melanocytes or nevus detected” are interpreted correctly by the NLP algorithm as meaning “no melanocytes” as well as “no nevus.” Positive predictive value (PPV) (also called precision [P]), sensitivity (also called recall [R]) and the summary F1 score were computed according to the following equation to determine the performance of the search method and query classification used in this project: Fβ = ([1 + β2] PR)/β2P + R, where β is the balance between precision and recall. This is a harmonic weighted mean of precision and recall. Commonly, the F score is used with a β = 1 (β times as much importance to recall as precision) and then called the F1 score.
Given that individual pathology reports may contain multiple diagnoses associated with multiple biopsies performed during a single dermatology visit, coding for separation of multiple diagnoses per pathology report were implemented. A detailed description of the NLP-system and classification is included in eAppendix 2 in the Supplement.
Results
In early 2007, before digitization of all pathology report text was standardized within the integrated health care system EMR, 11 987 reports were not readable by the NLP algorithm. Thus, this study sample included patients undergoing skin biopsies from mid-2007 through the end of 2012. During this period, a total of 80 368 skin biopsies were performed on 47 529 adult patients. Most patients underwent only 1 skin biopsy (n = 32 262 patients) or 2 biopsies (n = 9015 patients). The mean number of skin biopsies per patient was 1.9 (range, 1-34). Compared with consensus diagnoses obtained from independent manual medical record review, the NLP system yielded the following performance characteristics: PPV, 82.4%; sensitivity, 81.7%; and F1 measure, 0.82.
Of the 80 368 skin biopsies, 61 653 (77%) were of nonmelanocytic lesions, and 18 715 were of melanocytic lesions (23%). The distribution of the 18 715 melanocytic lesions using the MPATH-Dx classification system is detailed in the Table. The overall distribution by MPATH-Dx class was as follows: class I, 83.1% (n = 15 558); class II, 8.3% (n = 1548); class III, 4.5% (n = 842); class IV, 2.2% (n = 405); and class V, 1.9% (n = 362).
Table. Distribution of Melanocytic Skin Biopsy Lesions Noted Over a 6-Year Study Period Among Adult Patients in a Large Integrated Health System.
| MPATH-Dx Class | Example Clinical Diagnosis | Total Melanocytic Skin Biopsies, No. (%) (n = 18 715) |
|
|---|---|---|---|
| NLP Results | Results Including Health Plan Data on Distribution of Invasive Melanoma Classes | ||
| I | Mild dysplastic nevi | 15 558 (83.1) | 15 558 (83.1) |
| II | Moderately dysplastic nevi | 1548 (8.3) | 1548 (8.3) |
| III | Melanoma in situ | 842 (4.5) | 842 (4.5) |
| IV | Invasive melanoma stage T1a | 767 (4.1)a | 405 (2.2)a |
| V | Invasive melanoma stage ≥T1b | 362 (1.9)a | |
Abbreviations: MPATH-Dx, Melanocytic Pathology Assessment Tool and Hierarchy for Diagnosis; NLP, natural language processing.
This is a combination total because the NLP processing technique was not developed to distinguish MPATH-Dx class IV from MPATH-Dx class V; thus SEER data from the health plan were used to approximate the percentage distribution of the invasive melanomas (See eAppendix 2 in the Supplement).
While these results describe outcomes at a skin biopsy level, we performed secondary analyses at the level of the individual patient, since patients often undergo multiple skin biopsies in clinical practice. We present data for the index biopsy and showing the classifications at 90 days and 365 days after the index biopsy at the patient level (eAppendix 3 in the Supplement). The results suggest that over the course of 1 year, an upgrading of MPATH-Dx diagnosis classification occurs for a small number of patients after follow-up. We display stratified results for men and women and by age groups in eAppendix 3 in the Supplement.
Discussion
In this study, we successfully used NLP techniques to review more than 80 000 pathology reports of skin biopsies performed over a 6-year period. We found that about 1 out of every 4 skin biopsies (23%) were of melanocytic lesions, highlighting the importance of a classification system that pathologists can use for these diagnostically challenging lesions. We were also able to quantify the breakdown of these melanocytic lesions by MPATH-Dx class as follows: class I, 83.1%; class II, 8.3%; class III, 4.5%; class IV, 2.2%; and class V, 1.9%.
This preliminary study of NLP-based analysis yielded an F1 score of 0.82. Previous research has found that a human annotator will achieve an F1 score around 0.88 on a similar task. Given that NLP algorithms may continue to be iteratively refined and improved given ongoing implementation across data sets, our initial approach may be considered to have yielded excellent performance characteristics, particularly given the complexity of the task.
While this study was performed at a single site, and the results of skin biopsies may be different in other geographic and clinical settings, the underlying health system patient population is large and representative of adults living in the region. Additionally, the MPATH-Dx tool is not currently used in all clinical practices, nor do all pathologists grade melanocytic lesions.
A striking array of terms are currently used by pathologists when interpreting the same melanocytic lesion. Thus, collapsing the plethora of terms used by practicing pathologists into a smaller number of classes using the MPATH-Dx tool may improve communication and the ease of abstracting information from EMRs. Moving forward, as more pathologists use the MPATH-Dx tool to classify melanocytic lesions internationally, these diagnostic classes will enable more rapid and accurate NLP assessment of large bodies of EMR data. National guidelines on phraseology in pathology reporting have long been suggested, and adopting such guidelines would improve our ability to extract helpful information from EMRs using NLP.
Limitations
Our study has limitations. We did not include skin biopsies from children, and biopsy outcomes might be different in other populations. Additionally, although our study classifies melanocytic skin lesions according to the MPATH-Dx tool, this classification system is not currently universally adopted nor accepted. However, in a national study of pathologists using the MPATH-Dx classification system for diagnostic interpretations, the majority of pathologists (96%) thought it somewhat to very likely that patient care would be improved by the use of a standardized taxonomy such as the MPATH-Dx tool in the diagnosis of melanocytic skin lesions. Nearly all participants in that study (98%) also stated that they would likely adopt a standardized taxonomy in their own clinical practice if available. Additionally, we recognize that there may be errors in data fidelity associated with skin biopsy identification and associated pathology outcomes arising from potential inaccuracies in EMR data as well as incompletely optimized NLP identification and classification. However, we believe that these anticipated limitations have minimal effect on our main results and acknowledge that performance of NLP applied to this novel area of research will likely continue to improve.
Future work should include validating the NLP-based system’s performance in other institutional settings and incorporating machine learning to enhance the accuracy of status annotations (eg, negation, uncertainty). Additional areas of focus should also explore combining NLP-based methods with structured data algorithms based on diagnostic and procedural codes to improve the accuracy of melanoma classification. If clinical documents describing some diagnostic outcomes are ambiguous or incomplete, structured data may help to clarify diagnoses, thereby improving opportunities to conduct population-based research within this important area of dermatology. Such research may be necessary for ongoing efforts to optimize delivery of dermatologic care, for which sound and sufficiently large population-based research is critical.
Conclusions
In summary, we successfully used an NLP technique to quantify and characterize the outcomes of skin biopsies. Given the prevalence of melanocytic proliferations noted in this population-based study, estimated at 1 of every 4 skin biopsies performed, the importance of reliable and accurate diagnoses on these challenging diagnostic cases is emphasized.
eAppendix 1. Skin Biopsy Identification and Melanoma Identification Using Corresponding HCPCS/CPT 4, ICD-9, and ICD-O-3 Codes
eAppendix 2. Additional Details on NLP Methods and Health Plan SEER Data
eAppendix 3. Diagnosis at patient level at the index biopsy and when the patient record was followed forward to 90 days and 365 days to evaluate follow-up biopsy results. Results are shown overall and stratified by sex.
References
- 1.Weinstock MA, Lott JP, Wang Q, et al. Skin biopsy utilization and melanoma incidence among Medicare beneficiaries. Br J Dermatol. 2017;176(4):949-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Welch HG, Woloshin S, Schwartz LM. Skin biopsy rates and incidence of melanoma: population based ecological study. BMJ. 2005;331(7515):481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langan S, Schmitt J, Coenraads PJ, Svensson A, von Elm E, Williams H; European Dermato-Epidemiology Network (EDEN) . The reporting of observational research studies in dermatology journals: a literature-based study. Arch Dermatol. 2010;146(5):534-541. [DOI] [PubMed] [Google Scholar]
- 4.Eide MJ, Krajenta R, Johnson D, et al. Identification of patients with nonmelanoma skin cancer using health maintenance organization claims data. Am J Epidemiol. 2010;171(1):123-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogers HW, Weinstock MA, Feldman SR, Coldiron BM. Incidence estimate of nonmelanoma skin cancer (keratinocyte carcinomas) in the US population, 2012. JAMA Dermatol. 2015;151(10):1081-1086. [DOI] [PubMed] [Google Scholar]
- 6.Rogers HW, Weinstock MA, Harris AR, et al. Incidence estimate of nonmelanoma skin cancer in the United States, 2006. Arch Dermatol. 2010;146(3):283-287. [DOI] [PubMed] [Google Scholar]
- 7.Piepkorn MW, Barnhill RL, Elder DE, et al. The MPATH-Dx reporting schema for melanocytic proliferations and melanoma. J Am Acad Dermatol. 2014;70(1):131-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lott JP, Elmore JG, Zhao GA, et al. ; International Melanoma Pathology Study Group . Evaluation of the Melanocytic Pathology Assessment Tool and Hierarchy for Diagnosis (MPATH-Dx) classification scheme for diagnosis of cutaneous melanocytic neoplasms: results from the International Melanoma Pathology Study Group. J Am Acad Dermatol. 2016;75(2):356-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerami P, Busam K, Cochran A, et al. Histomorphologic assessment and interobserver diagnostic reproducibility of atypical spitzoid melanocytic neoplasms with long-term follow-up. Am J Surg Pathol. 2014;38(7):934-940. [DOI] [PubMed] [Google Scholar]
- 10.Duncan LM, Berwick M, Bruijn JA, Byers HR, Mihm MC, Barnhill RL. Histopathologic recognition and grading of dysplastic melanocytic nevi: an interobserver agreement study. J Invest Dermatol. 1993;100(3):318S-321S. [DOI] [PubMed] [Google Scholar]
- 11.Duray PH, DerSimonian R, Barnhill R, et al. An analysis of interobserver recognition of the histopathologic features of dysplastic nevi from a mixed group of nevomelanocytic lesions. J Am Acad Dermatol. 1992;27(5 Pt 1):741-749. [DOI] [PubMed] [Google Scholar]
- 12.Corona R, Mele A, Amini M, et al. Interobserver variability on the histopathologic diagnosis of cutaneous melanoma and other pigmented skin lesions. J Clin Oncol. 1996;14(4):1218-1223. [DOI] [PubMed] [Google Scholar]
- 13.Swerlick RA, Chen S. The melanoma epidemic: is increased surveillance the solution or the problem? Arch Dermatol. 1996;132(8):881-884. [DOI] [PubMed] [Google Scholar]
- 14.Elmore JG, Barnhill RL, Elder DE, et al. Pathologists' diagnosis of invasive melanoma and melanocytic proliferations: observer accuracy and reproducibility study. BMJ. 2017;357:j2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Dijk MC, Aben KK, van Hees F, et al. Expert review remains important in the histopathological diagnosis of cutaneous melanocytic lesions. Histopathology. 2008;52(2):139-146. [DOI] [PubMed] [Google Scholar]
- 16.Shain AH, Yeh I, Kovalyshyn I, et al. The genetic evolution of melanoma from precursor lesions. N Engl J Med. 2015;373(20):1926-1936. [DOI] [PubMed] [Google Scholar]
- 17.Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc. 2011;18(5):544-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman C, Hripcsak G. Natural language processing and its future in medicine. Acad Med. 1999;74(8):890-895. [DOI] [PubMed] [Google Scholar]
- 19.Pons E, Braun LMM, Hunink MG, Kors JA. Natural language processing in radiology: a systematic review. Radiology. 2016;279(2):329-343. [DOI] [PubMed] [Google Scholar]
- 20.Hripcsak G, Austin JH, Alderson PO, Friedman C. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology. 2002;224(1):157-163. [DOI] [PubMed] [Google Scholar]
- 21.Chapman WW, Fizman M, Chapman BE, Haug PJ. A comparison of classification algorithms to automatically identify chest X-ray reports that support pneumonia. J Biomed Inform. 2001;34(1):4-14. [DOI] [PubMed] [Google Scholar]
- 22.Carrell DS, Halgrim S, Tran DT, et al. Using natural language processing to improve efficiency of manual chart abstraction in research: the case of breast cancer recurrence. Am J Epidemiol. 2014;179(6):749-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coden A, Savova G, Sominsky I, et al. Automatically extracting cancer disease characteristics from pathology reports into a Disease Knowledge Representation Model. J Biomed Inform. 2009;42(5):937-949. [DOI] [PubMed] [Google Scholar]
- 24.Crowley RS, Castine M, Mitchell K, Chavan G, McSherry T, Feldman M. caTIES: a grid based system for coding and retrieval of surgical pathology reports and tissue specimens in support of translational research. J Am Med Inform Assoc. 2010;17(3):253-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strauss JA, Chao CR, Kwan ML, Ahmed SA, Schottinger JE, Quinn VP. Identifying primary and recurrent cancers using a SAS-based natural language processing algorithm. J Am Med Inform Assoc. 2013;20(2):349-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burger G, Abu-Hanna A, de Keizer N, Cornet R. Natural language processing in pathology: a scoping review. J Clin Pathol. 2016;jclinpath-2016-203872. [DOI] [PubMed] [Google Scholar]
- 27.Denny JC, Choma NN, Peterson JF, et al. Natural language processing improves identification of colorectal cancer testing in the electronic medical record. Med Decis Making. 2012;32(1):188-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hripcsak G, Friedman C, Alderson PO, DuMouchel W, Johnson SB, Clayton PD. Unlocking clinical data from narrative reports: a study of natural language processing. Ann Intern Med. 1995;122(9):681-688. [DOI] [PubMed] [Google Scholar]
- 29.Ye JJ. Pathology report data extraction from relational database using R, with extraction from reports on melanoma of skin as an example. J Pathol Inform. 2016;7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Healthinformatics. NegEx Algorithm. 2017. https://healthinformatics.wikispaces.com/NegEx+Algorithm. Accessed Jan 23, 2017.
- 31.Gianinazzi ME, Rueegg CS, Zimmerman K, Kuehni CE, Michel G; Swiss Paediatric Oncology Group . Intra-rater and inter-rater reliability of a medical record abstraction study on transition of care after childhood cancer. PLoS One. 2015;10(5):e0124290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Attanoos RL, Bull AD, Douglas-Jones AG, Fligelstone LJ, Semararo D. Phraseology in pathology reports: a comparative study of interpretation among pathologists and surgeons. J Clin Pathol. 1996;49(1):79-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kho AN, Pacheco JA, Peissig PL, et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med. 2011;3(79):79re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peissig PL, Rasmussen LV, Berg RL, et al. Importance of multi-modal approaches to effectively identify cataract cases from electronic health records. J Am Med Inform Assoc. 2012;19(2):225-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eAppendix 1. Skin Biopsy Identification and Melanoma Identification Using Corresponding HCPCS/CPT 4, ICD-9, and ICD-O-3 Codes
eAppendix 2. Additional Details on NLP Methods and Health Plan SEER Data
eAppendix 3. Diagnosis at patient level at the index biopsy and when the patient record was followed forward to 90 days and 365 days to evaluate follow-up biopsy results. Results are shown overall and stratified by sex.