Abstract
Cell division and differentiation are two fundamental physiological processes that need to be tightly balanced to achieve harmonious development of an organ or a tissue without jeopardizing its homeostasis. The role played by the centriolar protein STIL is highly illustrative of this balance at different stages of life as deregulation of the human STIL gene expression has been associated with either insufficient brain development (primary microcephaly) or cancer, two conditions resulting from perturbations in cell cycle and chromosomal segregation. This review describes the recent advances on STIL functions in the control of centriole duplication and mitotic spindle integrity, and discusses how pathological perturbations of its finely tuned expression result in chromosomal instability in both embryonic and postnatal situations, highlighting the concept that common key factors are involved in developmental steps and tissue homeostasis.
Facts
STIL is a cell cycle-regulated protein specifically recruited at the mitotic centrosome to promote the duplication of centrioles in dividing cells.
Complete loss of STIL results in no centrosomes, no cilia, and is not compatible with life.
By contrast, residual or increased expression of STIL is viable but alters the centriole duplication process leading to either impaired or excessive centrosome formation.
Genetic mutations in human STIL result in either residual expression or stabilization of STIL at the centrosome both leading to mitotic spindle defects and primary microcephaly (MCPH7).
Abnormally high expression of STIL in differentiated tissues triggers centrosomal amplification and is associated with an increased metastatic potential in multiple cancers.
Open questions
Centrosome amplification is seen both in cancer and in MCPH phenotype. How is the context important in determining the phenotype?
Is presence of STIL in the centrosome important in determining cell fate?
STIL has several binding partners. To what extent is the STIL phenotype due to the independent functions of these binding partners?
Introduction
The developing brain appears particularly sensitive to centrosome dysfunction, which is also associated with a wide range of cancers. The centrosome is a cytoplasmic organelle built around microtubule-based core components called centrioles. This review focuses on the STIL gene that encodes a regulatory protein necessary for centriole biogenesis and is expressed in many cell types. The structure and function of STIL is described followed by an account of two phenotypes that have been associated with STIL dysfunction, autosomal recessive primary microcephaly (MCPH), and cancer. Reasons for these different phenotypes are discussed in relation with centriole duplication and mitotic checkpoints that operate in different cellular contexts to deal with aneuploidy and chromosomal instability.
STIL structure
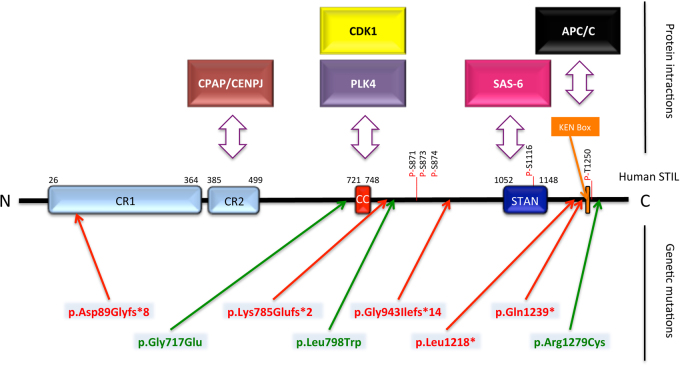
The human STIL gene was initially identified in a common chromosomal rearrangement in T-cell acute lymphoblastic leukemia and named SCL/TAL1 Interrupting Locus (SIL/STIL)1. It is predicted to encode five isoforms. Isoform1 (NP_001041631.1; henceforth referred to as STIL) is a 1288-amino-acid protein; isoform2 has 1287 amino acids (NP_001269865.1) and differs from isoform1 by a missing serine875; isoforms3–5 (NP_001269866.1, NP_001269867.1, NP_001269868.1) have several amino acids missing but their significance is unknown. STIL contains conserved regions and interacts with several proteins (Fig. 1). CR2 (amino acids 385–499) is a proline-rich domain that includes the conserved PRXXPXP motif, which interacts with the Centrosomal P4.1-Associated Protein (CPAP/CENPJ). The coiled-coil domain (amino acids 721–748) is important both for Polo-Like Kinase 4 (PLK4) and Cyclin Dependent Kinase 1 (CDK1 also known as CDC2)/CyclinB binding2,3 and for STIL oligomerization4. The STAN domain (amino acids 1052–1148) mediates the binding of the centriole protein SAS-62. At the C terminus is the KEN box involved in Anaphase-Promoting-Complex/Cyclosome (APC/C)-mediated degradation of STIL5. The C terminus of STIL can also interact with conserved components of the Hedgehog signaling such as Suppressor-of-fused homolog (SUFU)6 and GLI17.
Fig. 1. Conserved regions, functional domains, and genetic mutations in the human STIL protein.
Blue and orange boxes represent regions of the protein that are highly conserved across species (CR conserved region, CC Coiled-Coil domain, STAN STIL/Ana2 domain, KEN Box Conserved Lys-Glu-Asn residues). Double-headed arrows indicate known interactions with STIL. Known phosphorylated residues in 871, 873, 874, 1116, and 1250 are indicated by a red line. Human mutations predicted to truncate the protein are displayed in red and missense mutations are in green
STIL function in centriole duplication
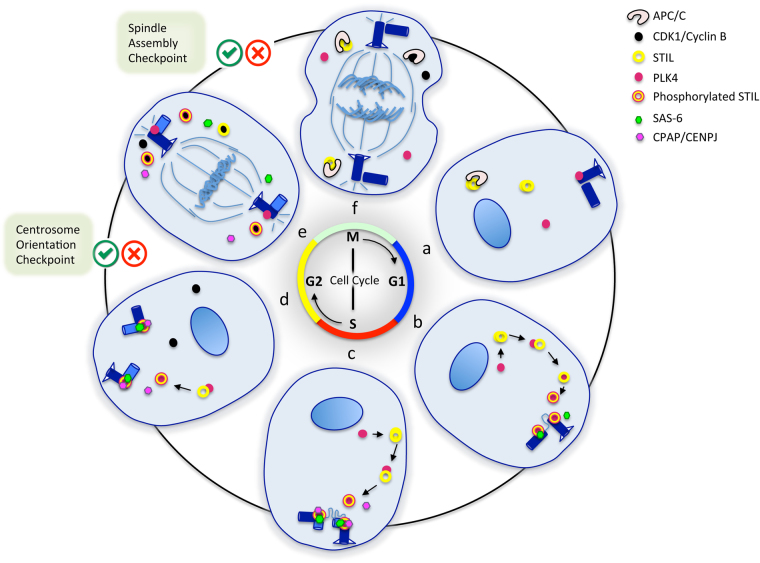
The centriole is an evolutionarily conserved structure consisting of a ninefold symmetric cartwheel with microtubule triplets, and thought to have been present in the Last Eukaryotic Common Ancestor8. The centrosome has two centrioles orthogonal to each other, surrounded by proteins constituting the pericentriolar material (PCM). Both the centriolar core and the PCM nucleate microtubules, which are important for positioning the mitotic spindles and imparting polarity and asymmetry to the cell. During cell division the centrioles duplicate only once; STIL and its interactors are central to this process, ensuring the fidelity of this unique duplication, thereby minimizing chromosome instability9. STIL transiently associates with PLK4 and SAS-6 to form the core module for centriole duplication10. PLK4 is related to the Polo-kinase family of serine/threonine kinases and initiates centriole formation11,12, while SAS-6, a coiled-coil protein that self-assembles into the ninefold symmetry gives the carthweel its shape13,14. STIL and PLK4 are initially maintained at low levels in non-dividing and differentiated cells15: STIL forms dimers or tetramers through its CC domain and is degraded in the cytoplasm by APC/C and its co-activator CDC20, through recognition of the KEN motif4,5. STIL level in the cytoplasm rises in G1 because its degradation by APC/C is prevented by the absence of CDC205. PLK4 recruits STIL at the base of the parental centriole through the CC domain and phosphorylates the STAN domain at the late G1/G1-S transition16–18. Phosphorylated STIL then promotes its binding to the C-terminal region of SAS-6 and recruits it to the outside wall of the centriole2,17. This core module along with CPAP, also recruited by STIL to the centriole, starts the assembly of the cartwheel and centriole duplication19,20. At early S phase, the centrosome-associated protein ROTATIN was recently shown to associate with STIL and contribute to building full-length centrioles21. At S–G2 transition, the centrioles are duplicated but remain close to each other, potentially preventing reduplication. Remarkably, CDK1, transiently expressed at late G2 and during M phase, competes with PLK4 for binding to the CC domain of STIL, preventing PLK4 from recruiting STIL until mitosis is completed3,16,22. The spindle assembly checkpoint, a highly conserved mechanism also called mitotic checkpoint, prevents the degradation of CDK1 during mitosis by the inhibition of APC/C-CDC20 until all chromosomes are properly segregated23. CDK1 is thought to trigger progressive dissociation of STIL and SAS-6 from early mitotic centrosomes, thereby initiating cartwheel disassembly during M phase and ensuring that centriole biogenesis occurs only once before cell divides5. Upon activation, APC/C-CDC20 degrades CDK1 for entry into anaphase and again marks STIL for degradation until next the G1 phase. Thus, by prometaphase there is no STIL at the centrosome and by anaphase there is no STIL in the cytoplasm either until upon mitotic exit APC/C is blocked and STIL levels builds up once again in G1 (Fig. 2).
Fig. 2. STIL regulation during the cell cycle. Six phases of the cell cycle are represented.
a Early G1 phase: STIL levels are low in the cytoplasm and STIL is absent at the centrosome. b, c G1–S and S phases: STIL levels are high in the cytoplasm and STIL starts being associated with the centrosome. PLK4 interacts and phosphorylates STIL. STIL recruits SAS-6 and CPAP to the centrosome, contributing to the assembly of the cartwheel. The procentriole starts elongating in S phase. d G2 phase: the two centrosomes begin to move apart. CDK1/CYCLIN B is active. Centrosome orientation checkpoint. e The nuclear envelope breakdown occurs, CDK1 binds to STIL and moves it to the cytoplasm, there is no more STIL at the centrosome, cartwheel disassembles. Spindle assembly checkpoint. f Anaphase, APC/C is fully active, cytoplasmic STIL is degraded. The cell nucleus or chromosomes with their mitotic spindle are represented in blue. Centrioles are shown in dark blue. STIL is represented by a yellow donut surrounded by a red circle when phosphorylated. PLK4 is represented by a red dot, CDK1/CYCLIN B by a black dot, SAS-6 by a small green hexagon, CPAP/CENPJ by a small pink hexagon, and APC/C by a black and white C-shape. Centrosome orientation and spindle assembly checkpoints are indicated by green/red tick boxes
STIL function in SHH signal transduction
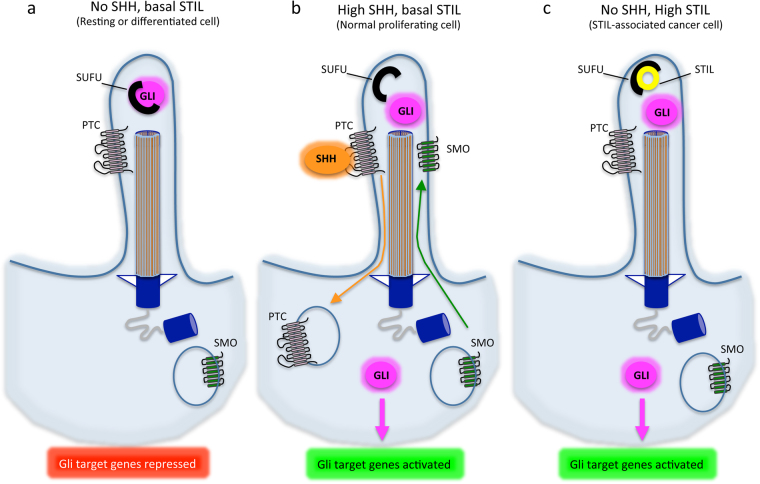
SHH is a morphogen involved in patterning, proliferation, and survial of neural stem cells during development24. SHH binds to its receptor Patched thereby relieving its inhibition on Smoothened and resulting in the activation of GLI transcription factors. That GLI proteins are the main downstream effectors for SHH signaling is borne out by the fact that activated GLI can rescue the loss-of-function of SHH and lead to proliferation and increased cell survival25. The initial findings that STIL participates in the control of SHH signaling came from Stil−/− mouse embryos, which showed a marked reduction of Patched and Gli1 expression26. Interestingly, Stil−/− mutants also lack primary cilia27, a structure present in almost all cell types, which is assembled beneath the plasma membrane, by the protrusion of the microtubule-based axoneme. During interphase, the mother centriole and its appendages constitute the basal body, from which the axoneme elongates28,29. STIL requirement for cilia formation likely stems from its function in supporting centriole biogenesis and stability. Receptors for SHH are abundant on cilia membranes allowing cilia to act as receivers of signals and as platforms where downstream effectors can be modified30. Thus, cilia, whose presence depends on STIL, are required for activity of the SHH pathway31.
STIL also interferes with the SHH pathway in a direct manner, by interacting with SUFU. SUFU acts as a negative regulator of SHH by tethering GLI1 in the cytoplasm. STIL binds to SUFU in the cytoplasm of pancreatic cancer cells, thus releasing GLI1 for transcription of SHH-downstream genes6. In PC12 cells, STIL interacts with the SUFU/GLI1 complex, and its downregulation results in a decrease in both SHH signaling and cell proliferation7. A similar correlation has been evidenced in zebrafish retina cells, further suggesting that STIL plays a role in cell proliferation through the SHH pathway32 (Fig. 3).
Fig. 3. STIL and SHH signaling at the cilium.
A cilium and its axonemal structure are represented in three different contexts of expression of SHH and STIL. SMO = SMOOTHENED receptor, PTC = PATCHED receptor. a In resting or differentiated cells, there is no SHH and low STIL expressed in the cilium. The PTC receptor is expressed at the membrane while the SMO receptor is degraded in the cytoplasm. SUFU binds and tether GLI proteins in the cytoplasm, blocking GLI-dependent transcription. b When SHH is expressed, it binds to its receptor PTC inducing its internalization and degradation in the cytoplasm. This allows SMO expression at the membrane of the cilium and leads to GLI derepression. c When STIL is abnormally highly expressed during interphase, it becomes abundant in the cilium where it binds to SUFU, releasing GLI proteins and thus activating GLI-mediated transcription even in the absence of SHH
Consequences of STIL deregulation
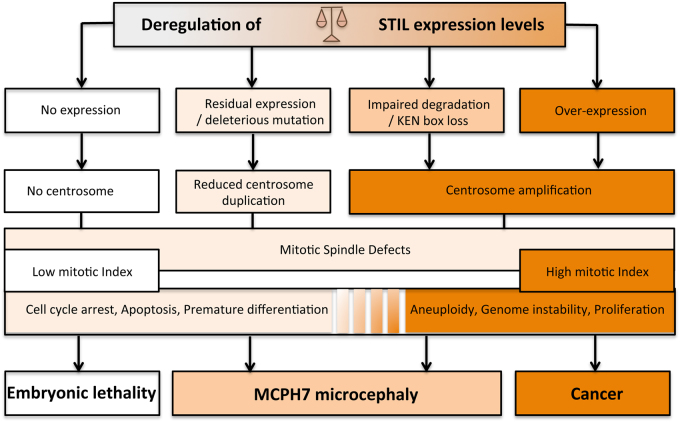
Given the centrality of STIL in centrosome duplication and cilium biogenesis, deregulation of STIL is predictably profound. Stil−/− embryos die at midgestation with axial midline defects, resulting from aberrant SHH signaling26. MEFs derived from Stil−/− embryos show a marked decrease in mitotic index, and Stil knockdown results in the absence of identifiable centrosomes during interphase and multiple spindle poles with disrupted γ-tubulin signals during mitosis33, as well as in the loss of primary cilia27. Conversely, STIL overexpression causes centrosome amplification, resulting in a star-like pattern around the parental centriole, consistent with a role of STIL in the onset of procentriole formation. Accordingly, this star-like structure is positive for the centriolar proteins CP110 and centrin20,34. Mutations within the various domains of STIL give predictable phenotypes. When the CR2 domain or the CC domain or the STAN domain is deleted, there is no centrosomal duplication16,18, whereas removal of the KEN box leads to centrosome amplification due to the inability of STIL to be degraded5. However, both overduplication and lack of duplication of centrosomes lead to abnormal mitotic spindle assembly and consequently increase the chances of abnormal chromosomal segregation and aneuploidy.
Another consequence of STIL deregulation is the indirect effect on interacting partners. STIL binding to PLK4 supresses PLK4 auto-inhibition, thereby allowing its trans-phosphorylation and protecting activated PLK4 from degradation35. Conversely, depletion of STIL leads to a marked accumulation of PLK4 in an inactive conformation. Therefore, changes in STIL levels have immediate consequences on PLK4 activation levels10,17. STIL also negatively regulates Chfr, an E3 ligase that blocks mitotic entry in response to mitotic stress. No direct interaction between STIL and Chfr has been shown, but STIL expression results in an increase of Chfr auto-ubiquitination, thereby promoting its proteasomal degradation and proper progression of cells through mitosis33. Data support a role of Chfr in defective mitotic progression associated with reduced activation of CDK1/CYCLIN B and centrosomal abnormalities caused by the lack of STIL33. Thus, STIL deregulation likely impacts the activation level of CDK1/CYCLIN B and the timing of entry into mitosis, through its regulation of Chfr.
STIL expression during human brain development
STIL is expressed in both fetal and adult tissues. However, its expression levels fluctuate with the cell cycle, making it difficult to detect in whole tissue, especially if the cells are not synchronized. Indeed, it is detected more easily in cancer cell lines and tissue where it is overexpressed36,37. The BrainSpan Atlas (http://www.brainspan.org/) and the BrainCloud database (http://braincloud.jhmi.edu) show its expression pattern during brain development. The association of STIL with cell proliferation is borne out by its expression pattern during fetal stages. At 15 postconceptional weeks STIL is strongly expressed in the ventricular and subventricular zones of the forebrain, the ganglionic eminence and the rostral migratory stream, and less expressed in intermediate zone, subplate, cortical plate, marginal zone, and subgranular layer of the forebrain. This pattern persists at 21 postconceptional weeks, although the expression is reduced in the subventricular zone. The expression of PLK4, SAS-6, and CPAP also broadly shows this pattern of expression, although the expression of SAS-6 and STIL diverges in some regions of the cortical plate. In the cerebellum, STIL, PLK4, and SAS-6 but not CPAP are expressed in the external granule layer and regions of the rhombic lip. However, none of these genes are expressed in the transient Purkinje cell cluster, the ventricular matrix zone of the cerebellum or in the migratory streams of the hindbrain. Microdissection experiments at different developmental stages confirmed that STIL expression is high in the ventricular zone and low in the subplate and cortical zones (http://www.brainspan.org/). Further, microdissected diencephalon did not have STIL (http://www.blueprintnhpatlas.org/). This suggests that (i) the structural components of the cartwheel STIL-SAS-6-PLK4 is by no means obligatory for centrosome duplication in all cells and (ii) STIL is present in specific populations of dividing cells.
STIL mutations and primary microcephaly
Microcephaly (small brain size) is indirectly diagnosed by a head circumference smaller than the age-specific and gender-adjusted mean by more than 2 standard deviations (S.D.s) at birth. Primary microcephaly refers to hereditary microcephalies already detectable in utero. Most of them are autosomal recessive and include (i) isolated forms called MicroCephaly Primary Hereditary (MCPH), (ii) forms associated with growth retardation, called microcephalic dwarfism. Most of STIL mutations identified in patients are associated with a MCPH phenotype and STIL is recognized as MCPH738–40. However, a few patients exhibit criteria of microcephalic dwarfism, as short stature has been reported41. So far, eight mutations in STIL have been described in 37 patients all showing severe microcephaly (−4 to −10 S.D.). Five mutations are splice42,43, deletion, nonsense43, or duplication44 that predict the production of truncated proteins, while three mutations are missense substitutions44–46 (Fig. 1). Despite the different kinds of mutations, the various domains affected (Table 1), and the report of lobar holoprosencephaly and partial agenesis of the corpus callosum in addition to microcephaly in some cases42,44,45, no clear genotype–phenotype correlation has come out so far. However, the fact that these mutations trigger a phenotype compatible with life (although very severe) is surprising since genetic ablation of STIL is embryonically lethal early during embryogenesis in mice and fish26,47. This suggests that STIL mutations in human do not result in a complete loss-of-function, or are partially compensated by other genes. The first hypothesis is supported by the findings that two C-terminal nonsense mutations (p.Val1219* and p.Gln1239*), which result in the loss of the KEN box43, do not affect the centrosomal localization of STIL nor its functionality but rather abolish its degradation by APC/C in late mitosis5. Removal of the KEN box thus causes a strong accumulation of the mutant protein, resulting in centrosome amplification5. Alternatively, microcephaly in MCPH7 patients can result from a decrease of STIL levels. This was illustrated by rescue experiments in U2OS cells showing that the pGly717Glu mutation induces a reduced but non-null activity of STIL on centriole duplication45. Another mutation, inducing exon 5 skipping and frameshift in the STIL sequence, is likely close to a null mutation as all domains are predicted to be lost42. However exon skipping was only partial in the patient analyzed, suggesting some residual activity of STIL, and providing an explanation for the fact that this mutation is compatible with life and the idea that MCPH7 microcephaly can result from a decreased STIL activity. Therefore, both accumulation and impairment of STIL protein levels during cell cycle affect centriole regulation and result in microcephaly (Fig. 4).
Table 1.
List of mutations identified in the human STIL gene, and their functional consequence
| Mutation (nt) | Mutation (aa) | Domain affected (protein) | Functional consequence | Head OFC (S.D.) | Brain MRI data | References |
|---|---|---|---|---|---|---|
| c.453 + 5 G > A (splice donor) | p.Asp89Glyfs*8 (truncating) | All domains | No functional protein | −9 to −10 | Short cc, simplified gyration, lobar HPE | 42 |
| c.2150 G > A | p.Gly717Glu (missense) | CC domain | Presumably inability of STIL to oligomerize and interact with PLK4—decreased centriole duplication capacity | −7 to −8 | Partial cc agenesis, simplified gyration, lobar HPE | 4,45 |
| c.2354_2355dupGA | p.Lys785Glufs*2 (truncating) | Phosphorylation sites, STAN domain, KEN box | Removes critical PLK4 phosphorylation sites | 26 cm at birth | Cc agenesis, simplified gyration, lobar HPE | 44 |
| c.2392 T > G | p.Leu798Trp (missense) | — | ND | NA | NA | 46 |
| c.2826 + 1 G > A (splice donor) | p.Gly943Ilefs*14 (likely truncating) | STAN domain, KEN box | Not able to duplicate centrosomes since they cannot recruit SAS6 | −5 | NA | 27,43 |
| c.3655delG | p.Val1219* (initially reported as p.Leu128*; truncating) | KEN box | Resistance to APC/C-mediated degradation | −4 to −10 | NA | 5,18,27,43 |
| c.3715 C > T | p.Gln1239* (truncating) | KEN box | Resistance to APC/C-mediated degradation | −7 to −8 | NA | 18,27,43,95 |
| c.3835 C > T | p.Arg1279Cys (missense) | C terminus | ND | 26 cm at birth | Cc agenesis, simplified gyration, lobar HPE | 44 |
Fig. 4. Consequences of deregulating STIL expression levels.
STIL expression levels are finely regulated both during development and in differentiated tissues. While total absence of STIL is lethal during development, high STIL levels due to random somatic mutations in differentiated tissues result in aneuploidy and/or high GLI-mediated transcription that can lead to cancer. Congenital deregulation of STIL (both a too high and a too weak expression) leads to microcephaly by either reduced or amplified centrosome duplication, highlighting the importance of a tightly regulated STIL expression
STIL deregulation and cancer
Centrosomes are essential for chromosomal stability, and abnormalities of their number, or structure affect cell division22. Considering the central role of STIL in maintaining centrosome integrity in highly proliferating cells, STIL has been found upregulated in several cancers of bad prognosis, including lung cancer, colon carcinoma, prostate adenocarcinoma36, and ovarian cancers48. Moreover, STIL expression is associated with an increased metastatic potential in multiple cancers49. As expected, STIL upregulation is associated with a high histopathological mitotic index in tumors36 and presumably affects the formation of mitotic spindles, as well as SHH signaling and the function of its interactors.
Although cells can proliferate and microtubule can nucleate in their absence, centrosomes are obligatory for controlling spindle orientation50. Loss of this control leads to spindle defects, especially in highly polarized cells, where misoriented spindles influence daughter cell positioning, possibly causing a disruption in tissue morphology51. Several tumor suppressor genes such as APC, or VHL, are important for stabilizing microtubules and maintaining spindle orientation51. Similarly, overexpression of genes such as STIL could act as oncogenes and lead to cancer by promoting spindle defects, although direct evidence of a link between oncogene activation and spindle misorientation is lacking. STIL overexpression, which results in supernumerary centrosomes, could lead to cancer in inducing chromosomal instability10,52.
The role of STIL in promoting SHH signaling could be another pathway to account for its association with cancer. Following the initial observation that SHH overexpression results in the development of basal cell carcinomas in mice53, a large body of literature has implicated the SHH pathway in development of various cancers including a subset of medulloblastomas54,55. STIL overexpression in pancreatic adenocarcinoma was shown to de-repress GLI1 from SUFU-mediated control and this phenotype was reversed when STIL was downregulated6. Thus, during the process of carcinogenesis, the increased expression of STIL promotes the transcriptional activity of GLI1, which is no longer regulated quantitatively. GLI1 upregulates genes that promote sustained proliferation, cell death resistance, stemness, angiogenesis, and genomic instability, which are hallmarks of cancer56. Therefore, increase in STIL expression leading to uncontrolled GLI1 de-repression likely represents a crucial step toward cancer progression.
MCPH and cancer—two conditions associated with abnormal centrosome duplication
STIL loss-of-function results in embryonic lethality in mice, fish, and most likely in human as well. MCPH7 phenotype reflects that perturbing the tuning of STIL expression levels is compatible with life but triggers severe patterning phenotypes during development, including brain growth. Thus, microcephaly caused by STIL mutations can result from either too less centrosome duplication or too much centrosome amplification (Fig. 4). In both cases, the integrity of the mitotic spindle is compromised and cell divisions have a greater chance of chromosomal instability. Similarly, the association of STIL upregulation with various cancers reflects a deregulation of STIL expression levels in specific tissues contributing to genomic instability. What results in MCPH rather than cancer could be the difference in mitotic checkpoints in different contexts13,57. Centrosome orientation checkpoint ensures that the centrosomes are duplicated and appropriately positioned before entry into mitosis and several centrosomal proteins such as CNN and SAS-6 are part of this checkpoint58. This checkpoint may not be present in all cells at all developmental stages but rather in those cases where the cell division plane has important outcomes59. In germline stem cells, for example, the division plane determines the distribution of cell fate determinants in daughter cells, helding cells at the G2–M phase until their mitotic spindles are properly oriented58. STIL downregulation results in disorganized mitotic spindles resulting in a loss of spindle orientation control47. In cortical development, STIL mutations leading to spindle orientation defects likely result in a depletion of cortical progenitors due to cell death or premature differentiation and give rise to a MCPH phenotype57 (Fig. 4). PLK4 overexpression also results in centrosome amplification and aneuploidy leading to a reduction in brain size due to cell death60. Apoptosis inhibition in this background causes the accumulation of aneuploid cells unable to proliferate efficiently, leading to premature neuronal differentiation60, whereas in a P53−/− context, PLK4 overexpression results in aneuploidy and skin cancer61.
The spindle assembly checkpoint is the next checkpoint once cells have entered mitosis and formed spindles. In the absence of bipolar spindles, cells are held-up in prometaphase and do not proceed to anaphase58, which results in increased apoptosis62. However, cells can increase their time in mitosis, thus adapting to this checkpoint and can assemble bipolar spindles in the absence of centrosomes or in the presence of supernumerary centrosomes. First in the absence of centrosome duplication cells manage to have bipolar spindles even though there is no centriole attached to one of the poles34. Second is a phenomena of centrosome clustering, wherein multiple centrosomes are clustered to enable the formation of bipolar spindles, a phenomenon whose efficiency could be tissue-specific60. These divisions often result in aneuploidy. When centrosome integrity is compromised, cells usually arrest in G1 and do not re-enter cell cycle after a prolonged cytokinesis to avoid chromosomal instability22 and in many cases proliferation in the absence of centrioles additionally requires the suppression of P5317. Upregulation of STIL giving rise to aneuploidy could play a major role in providing an evolving genome to help the cells adapt to the changing environment of the cancer and escape normal checkpoints. In the developing cortex, abnormal mitotic spindles due to STIL mutations would result in cell fate switches rather than cell hyperproliferation. However, that patients whose STIL mutation is clearly associated with centrosome amplification could be at risk of developing cancer cannot be ruled out.
Seventeen MCPH loci have been described so far40, several of which also play a role in cancer. MCPH1 is an early DNA damage response (DDR) protein and MCPH1 deletions have been identified as a risk for breast cancer63. Centrosome amplification is commonly seen following DNA damage and can arise as part of the DDR64. The serine-threonine kinase CHK1 plays a major role in DDR by halting cell division in G2/M until repair proteins are recruited at lesion sites. In this context, MCPH1 deficiency potentiates CHK1 activity and increases centrosome amplification65. Thus, compromising mitotic checkpoints results in cell division in the presence of abnormal centrosomes63,65. How CHK1 overactivation leads to centrosome amplification is not fully understood, but it is thought to activate CDK2, a cyclin-dependent kinase required for centrosome duplication, likely through activating its phosphorylation66,67. WD repeat-containing protein62 (WDR62), whose deficiency is associated with MCPH268–70, is required for maintaining spindle and centrosome integrity. Its overexpression, coincident with centrosome amplification, is also seen in lung adenocarcinomas and ovarian cancers71,72. WDR62 interacts physically during the cell cycle with the abnormal spindle-like microcephaly-associated protein (ASPM), whose deficiency causes the most frequent primary microcephaly (MCPH5)73. Here too, increased ASPM levels cause tumor growth and are seen in medulloblastomas while its reduction causes a decrease in tumor proliferation74. CDK5RAP2 (MCPH3) is also involved in the DDR; it functions by arresting cells before mitotic entry due to imparied centrosomes but also interacts with BUB1 and MAD2, which are important in spindle activation checkpoint75,76. Interestingly, a correlation between centrosomal abnormalities, aneuploidy, and cytogenic risk profile is seen in acute myeloid leukemia, where gene expression profiling has revealed the differential expression of genes encoding centrosomal and mitotic spindle proteins77. Among these are pericentrin, a scaffold protein that anchors many other proteins at the centrosome78 and NuMA, which associates with dynein and microtubules to create localized pulling forces, thus regulating the correct assembly and positioning of the mitotic spindle79. The role of such factors points to mitotic centrosomal abnormalities as important component of cancer progression.
STIL could also have an indirect effect on cancer, as a downstream effector of PLK4. Increased PLK4 expression has been reported in malignancies such as colorectal cancer80, pediatric medulloblastoma81, and breast tumors82. Its transient overexpression leads to centrosome overduplication and in a P53−/− background that inhibits apoptosis, it results in aneuploidy and spontaneous skin cancer61. Another recent study has convincingly shown that PLK4 overexpression leads to spontaneous tumors in several organs83. It remains to be seen whether this phenotype is dependent on STIL expression. STIL binds to PLK4 in the cytoplasm and therefore STIL expression levels could impact PLK4 cytoplasmic activity17, which functions to remodel the cytoskeleton and may be important for cancer invasion and metastasis as its depletion is correlated with an increase in E-cadherin expression and less metastasis84. CYCLIN B is also frequently found elevated in primary breast cancer, esophageal squamous cell carcinoma, laryngeal squamous cell carcinoma, and colorectal carcinoma85–89 and its expression indicates a bad prognosis and is correlated with the malignancy of gynecological cancers90. Downregulation of STIL decreases CDK1/CYCLIN B activity, prevents G2–M transition, and causes inhibition of tumor growth in vivo91. Conversely, increasing STIL could promote CDK1/CYCLIN B activity and indirectly participate in CYCLIN B-dependent proliferation in tumor cells. Absence of STIL also results in an upregulation of Chfr, and lowers PLK1, resulting in the activation of the CDC25c phosphatase. This pathway could thus control the entry of cells into mitosis independent of its obligatory role in centriole duplication33.
Conclusion
STIL mutations in MCPH show that centrosomes and cilia are essential for normal brain development. During evolution, one way that the cortex has undergone expansion is likely linked to mechanisms controlling spindle orientation and keeping the balance between asymmetric and symmetric cell divisions92,93. A change in the nature of spindle orientation control has been proposed to account for the population of subventricular zone progenitors in human development and based on the strong expression of STIL in the ventricular zone, one may speculate that STIL is important for controlling spindle orientation in giving rise to subventricular zone’s cells94. Deficiency of several centrosomal proteins results in MCPH, and centrosomal proteins may thus be a key to discovering the mechanism of expansion of cortical area38. In addition, the involvement of STIL in cancer shows that the result of spindle defects can be vastly different depending on the stage of development and the tissue involved that probably is partly due to differential responses to mitotic checkpoint mechanisms. STIL intersects several pathways, and lowering its level could block the effects of PLK4 overexpression, CDK1/CYCLIN B activation, and GLI1 signaling, and reduce proliferation even in the presence of aneuploidy. Thus, centrosomal proteins may be a new class of targets for cancer.
Acknowledgments
This work was supported by the Institut National pour la Santé et la Recherche Médicale (INSERM), the Centre National de la Recherche Scientifique (CNRS), the Université Paris 7, the DHU PROTECT, the Indo-French Centre for the Promotion of Advanced Research “CEFIPRA” (grant no 4903-02 to S.M., V.E.G., P.G., and S.P.), and grants from the French National Research Agency (ANR-13-RARE-0007-01 to S.P., P.G., and V.E.G. (ERA-NET E-Rare 2013, EuroMicro project), and ANR-16-CE16-0024- 01 to V.E.G. (PRC GENERIQUE 2016, MicroGol project)). S.M. was supported by a grant from Curadev Pharma.
Competing interests
The authors declare that they have no competing financial interests.
Footnotes
Patwardhan Dhruti, Mani Shyamala, Gressens Pierre, and El Ghouzzi Vincent contributed equally to this work.
Edited by A. Verkhratsky
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Aplan PD, et al. Involvement of the putative hematopoietic transcription factor SCL in T-cell acute lymphoblastic leukemia. Blood. 1992;79:1327–1333. [PubMed] [Google Scholar]
- 2.Ohta M, et al. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat. Commun. 2014;5:5267. doi: 10.1038/ncomms6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zitouni S, et al. CDK1 prevents unscheduled PLK4-STIL complex assembly in centriole biogenesis. Curr. Biol. 2016;26:1127–1137. doi: 10.1016/j.cub.2016.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.David A, et al. Molecular basis of the STIL coiled coil oligomerization explains its requirement for de-novo formation of centrosomes in mammalian cells. Sci. Rep. 2016;6:24296. doi: 10.1038/srep24296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arquint C, Nigg EA. STIL microcephaly mutations interfere with APC/C-mediated degradation and cause centriole amplification. Curr. Biol. 2014;24:351–360. doi: 10.1016/j.cub.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 6.Kasai K, Inaguma S, Yoneyama A, Yoshikawa K, Ikeda H. SCL/TAL1 interrupting locus derepresses GLI1 from the negative control of suppressor-of-fused in pancreatic cancer cell. Cancer Res. 2008;68:7723–7729. doi: 10.1158/0008-5472.CAN-07-6661. [DOI] [PubMed] [Google Scholar]
- 7.Sun L, et al. Characterization of the human oncogene SCL/TAL1 interrupting locus (Stil) mediated Sonic hedgehog (Shh) signaling transduction in proliferating mammalian dopaminergic neurons. Biochem. Biophys. Res. Commun. 2014;449:444–448. doi: 10.1016/j.bbrc.2014.05.048. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. Evolution: tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 2011;194:165–175. doi: 10.1083/jcb.201011152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Funk LC, Zasadil LM, Weaver BA. Living in CIN: mitotic infidelity and its consequences for tumor promotion and suppression. Dev. Cell. 2016;39:638–652. doi: 10.1016/j.devcel.2016.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arquint C, Nigg EA. The PLK4-STIL-SAS-6 module at the core of centriole duplication. Biochem. Soc. Trans. 2016;44:1253–1263. doi: 10.1042/BST20160116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bettencourt-Dias M, et al. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 12.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 13.Kitagawa D, et al. Spindle positioning in human cells relies on proper centriole formation and on the microcephaly proteins CPAP and STIL. J. Cell Sci. 2011;124(Pt 22):3884–3893. doi: 10.1242/jcs.089888. [DOI] [PubMed] [Google Scholar]
- 14.van Breugel M, et al. Structures of SAS-6 suggest its organization in centrioles. Science. 2011;331:1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- 15.Bauer M, Cubizolles F, Schmidt A, Nigg EA. Quantitative analysis of human centrosome architecture by targeted proteomics and fluorescence imaging. EMBO J. 2016;35:2152–2166. doi: 10.15252/embj.201694462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kratz AS, Barenz F, Richter KT, Hoffmann I. Plk4-dependent phosphorylation of STIL is required for centriole duplication. Biol. Open. 2015;4:370–377. doi: 10.1242/bio.201411023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moyer TC, Clutario KM, Lambrus BG, Daggubati V, Holland AJ. Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J. Cell Biol. 2015;209:863–878. doi: 10.1083/jcb.201502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vulprecht J, et al. STIL is required for centriole duplication in human cells. J. Cell Sci. 2012;125(Pt 5):1353–1362. doi: 10.1242/jcs.104109. [DOI] [PubMed] [Google Scholar]
- 19.Cottee MA, et al. Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly. eLife. 2013;2:e01071. doi: 10.7554/eLife.01071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang CJ, et al. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 2011;30:4790–4804. doi: 10.1038/emboj.2011.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen HY, et al. Human microcephaly protein RTTN interacts with STIL and is required to build full-length centrioles. Nat. Commun. 2017;8:247. doi: 10.1038/s41467-017-00305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pihan GA. Centrosome dysfunction contributes to chromosome instability, chromoanagenesis, and genome reprograming in cancer. Front. Oncol. 2013;3:277. doi: 10.3389/fonc.2013.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sivakumar S, Gorbsky GJ. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat. Rev. Mol. Cell Biol. 2015;16:82–94. doi: 10.1038/nrm3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choudhry Z, et al. Sonic hedgehog signalling pathway: a complex network. Ann. Neurosci. 2014;21:28–31. doi: 10.5214/ans.0972.7531.210109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cayuso J, Ulloa F, Cox B, Briscoe J, Marti E. The Sonic hedgehog pathway independently controls the patterning, proliferation and survival of neuroepithelial cells by regulating Gli activity. Development. 2006;133:517–528. doi: 10.1242/dev.02228. [DOI] [PubMed] [Google Scholar]
- 26.Izraeli S, et al. The SIL gene is required for mouse embryonic axial development and left-right specification. Nature. 1999;399:691–694. doi: 10.1038/21429. [DOI] [PubMed] [Google Scholar]
- 27.David A, et al. Lack of centrioles and primary cilia in STIL(−/−) mouse embryos. Cell Cycle. 2014;13:2859–2868. doi: 10.4161/15384101.2014.946830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishikawa H, Marshall WF. Ciliogenesis: building the cell’s antenna. Nat. Rev. Mol. Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- 29.Tanos BE, et al. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 2013;27:163–168. doi: 10.1101/gad.207043.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scholey JM, Anderson KV. Intraflagellar transport and cilium-based signaling. Cell. 2006;125:439–442. doi: 10.1016/j.cell.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 31.Bangs F, Anderson KV. Primary Cilia and Mammalian Hedgehog Signaling. Cold Spring Harb. Persp. Biol. 2017;9:pii: a028175. doi: 10.1101/cshperspect.a028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun L, et al. Transcription of the SCL/TAL1 interrupting Locus (Stil) is required for cell proliferation in adult Zebrafish Retinas. J. Biol. Chem. 2014;289:6934–6940. doi: 10.1074/jbc.M113.506295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castiel A, et al. The Stil protein regulates centrosome integrity and mitosis through suppression of Chfr. J. Cell Sci. 2011;124(Pt 4):532–539. doi: 10.1242/jcs.079731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arquint C, Sonnen KF, Stierhof YD, Nigg EA. Cell-cycle-regulated expression of STIL controls centriole number in human cells. J. Cell Sci. 2012;125(Pt 5):1342–1352. doi: 10.1242/jcs.099887. [DOI] [PubMed] [Google Scholar]
- 35.Arquint, C. et al. STIL binding to Polo-box 3 of PLK4 regulates centriole duplication. Elife4 (2015). [DOI] [PMC free article] [PubMed]
- 36.Erez A, et al. Sil overexpression in lung cancer characterizes tumors with increased mitotic activity. Oncogene. 2004;23:5371–5377. doi: 10.1038/sj.onc.1207685. [DOI] [PubMed] [Google Scholar]
- 37.Izraeli S, et al. Expression of the SIL gene is correlated with growth induction and cellular proliferation. Cell Growth Differ. 1997;8:1171–1179. [PubMed] [Google Scholar]
- 38.Kaindl AM, et al. Many roads lead to primary autosomal recessive microcephaly. Prog. Neurobiol. 2010;90:363–383. doi: 10.1016/j.pneurobio.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 39.Passemard S, et al. Expanding the clinical and neuroradiologic phenotype of primary microcephaly due to ASPM mutations. Neurology. 2009;73:962–969. doi: 10.1212/WNL.0b013e3181b8799a. [DOI] [PubMed] [Google Scholar]
- 40.Verloes, A., Drunat, S., Gressens, P., Passemard, S. Primary autosomal recessive microcephalies and seckel syndrome spectrum disorders. In: Pagon R.A., Adam M.P., Ardinger H.H., Wallace S.E., Amemiya A., Bean L.J.H., et al. (eds). GeneReviews(R): Seattle (WA) (1993). [PubMed]
- 41.Darvish H, et al. A clinical and molecular genetic study of 112 Iranian families with primary microcephaly. J. Med. Genet. 2010;47:823–828. doi: 10.1136/jmg.2009.076398. [DOI] [PubMed] [Google Scholar]
- 42.Kakar N, et al. STIL mutation causes autosomal recessive microcephalic lobar holoprosencephaly. Hum. Genet. 2015;134:45–51. doi: 10.1007/s00439-014-1487-4. [DOI] [PubMed] [Google Scholar]
- 43.Kumar A, Girimaji SC, Duvvari MR, Blanton SH. Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am. J. Hum. Genet. 2009;84:286–290. doi: 10.1016/j.ajhg.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bennett H, et al. A prenatal presentation of severe microcephaly and brain anomalies in a patient with novel compound heterozygous mutations in the STIL gene found postnatally with exome analysis. Pediatr. Neurol. 2014;51:434–436. doi: 10.1016/j.pediatrneurol.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 45.Mouden C, et al. Homozygous STIL mutation causes holoprosencephaly and microcephaly in two siblings. PLoS ONE. 2015;10:e0117418. doi: 10.1371/journal.pone.0117418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Papari E, et al. Investigation of primary microcephaly in Bushehr province of Iran: novel STIL and ASPM mutations. Clin. Genet. 2013;83:488–490. doi: 10.1111/j.1399-0004.2012.01949.x. [DOI] [PubMed] [Google Scholar]
- 47.Pfaff KL, et al. The zebra fish cassiopeia mutant reveals that SIL is required for mitotic spindle organization. Mol. Cell Biol. 2007;27:5887–5897. doi: 10.1128/MCB.00175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rabinowicz N, et al. Targeting the centriolar replication factor STIL synergizes with DNA damaging agents for treatment of ovarian cancer. Oncotarget. 2017;8:27380–27392. doi: 10.18632/oncotarget.16068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 50.Yamashita YM, Fuller MT. Asymmetric centrosome behavior and the mechanisms of stem cell division. J. Cell Biol. 2008;180:261–266. doi: 10.1083/jcb.200707083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pease JC, Tirnauer JS. Mitotic spindle misorientation in cancer--out of alignment and into the fire. J. Cell Sci. 2011;124(Pt 7):1007–1016. doi: 10.1242/jcs.081406. [DOI] [PubMed] [Google Scholar]
- 52.Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- 53.Oro AE, et al. Basal cell carcinomas in mice overexpressing sonic hedgehog. Science. 1997;276:817–821. doi: 10.1126/science.276.5313.817. [DOI] [PubMed] [Google Scholar]
- 54.Kool M, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123:473–484. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Northcott PA, et al. Medulloblastomics: the end of the beginning. Nat. Rev. Cancer. 2012;12:818–834. doi: 10.1038/nrc3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kasai K. GLI1, a master regulator of the hallmark of pancreatic cancer. Pathol. Int. 2016;66:653–660. doi: 10.1111/pin.12476. [DOI] [PubMed] [Google Scholar]
- 57.Wang G, Jiang Q, Zhang C. The role of mitotic kinases in coupling the centrosome cycle with the assembly of the mitotic spindle. J. Cell Sci. 2014;127(Pt 19):4111–4122. doi: 10.1242/jcs.151753. [DOI] [PubMed] [Google Scholar]
- 58.Venkei ZG, Yamashita YM. The centrosome orientation checkpoint is germline stem cell specific and operates prior to the spindle assembly checkpoint in Drosophila testis. Development. 2015;142:62–69. doi: 10.1242/dev.117044. [DOI] [PubMed] [Google Scholar]
- 59.Chen C, Inaba M, Venkei ZG, Yamashita YM. Klp10A, a stem cell centrosome-enriched kinesin, balances asymmetries in Drosophila male germline stem cell division. eLife. 2016;5:pii:e20977. doi: 10.7554/eLife.20977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marthiens V, et al. Centrosome amplification causes microcephaly. Nat. Cell Biol. 2013;15:731–740. doi: 10.1038/ncb2746. [DOI] [PubMed] [Google Scholar]
- 61.Sercin O, et al. Transient PLK4 overexpression accelerates tumorigenesis in p53-deficient epidermis. Nat. Cell Biol. 2016;18:100–110. doi: 10.1038/ncb3270. [DOI] [PubMed] [Google Scholar]
- 62.Novorol C, et al. Microcephaly models in the developing zebrafish retinal neuroepithelium point to an underlying defect in metaphase progression. Open Biol. 2013;3:130065. doi: 10.1098/rsob.130065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mantere T, et al. Targeted next-generation sequencing identifies a recurrent mutation in MCPH1 associating with hereditary breast cancer susceptibility. PLoS Genet. 2016;12:e1005816. doi: 10.1371/journal.pgen.1005816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mullee LI, Morrison CG. Centrosomes in the DNA damage response--the hub outside the centre. Chromosome Res. 2016;24:35–51. doi: 10.1007/s10577-015-9503-7. [DOI] [PubMed] [Google Scholar]
- 65.Antonczak AK, et al. Opposing effects of pericentrin and microcephalin on the pericentriolar material regulate CHK1 activation in the DNA damage response. Oncogene. 2016;35:2003–2010. doi: 10.1038/onc.2015.257. [DOI] [PubMed] [Google Scholar]
- 66.Bourke E, Brown JA, Takeda S, Hochegger H, Morrison CG. DNA damage induces Chk1-dependent threonine-160 phosphorylation and activation of Cdk2. Oncogene. 2010;29:616–624. doi: 10.1038/onc.2009.340. [DOI] [PubMed] [Google Scholar]
- 67.Bourke E, et al. DNA damage induces Chk1-dependent centrosome amplification. EMBO Rep. 2007;8:603–609. doi: 10.1038/sj.embor.7400962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bilguvar K, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nicholas AK, et al. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat. Genet. 2010;42:1010–1014. doi: 10.1038/ng.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu TW, et al. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat. Genet. 2010;42:1015–1020. doi: 10.1038/ng.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shinmura K, et al. WDR62 overexpression is associated with a poor prognosis in patients with lung adenocarcinoma. Mol. Carcinog. 2017;56:1984–1991. doi: 10.1002/mc.22647. [DOI] [PubMed] [Google Scholar]
- 72.Zhang Y, et al. Overexpression of WDR62 is associated with centrosome amplification in human ovarian cancer. J. Ovarian Res. 2013;6:55. doi: 10.1186/1757-2215-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jayaraman D, et al. Microcephaly proteins Wdr62 and Aspm define a mother centriole complex regulating centriole biogenesis, apical complex, and cell fate. Neuron. 2016;92:813–828. doi: 10.1016/j.neuron.2016.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Williams SE, et al. Aspm sustains postnatal cerebellar neurogenesis and medulloblastoma growth in mice. Development. 2015;142:3921–3932. doi: 10.1242/dev.124271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barr AR, Kilmartin JV, Gergely F. CDK5RAP2 functions in centrosome to spindle pole attachment and DNA damage response. J. Cell Biol. 2010;189:23–39. doi: 10.1083/jcb.200912163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang X, et al. CDK5RAP2 is required for spindle checkpoint function. Cell Cycle. 2009;8:1206–1216. doi: 10.4161/cc.8.8.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Neben K, et al. Gene expression patterns in acute myeloid leukemia correlate with centrosome aberrations and numerical chromosome changes. Oncogene. 2004;23:2379–2384. doi: 10.1038/sj.onc.1207401. [DOI] [PubMed] [Google Scholar]
- 78.Delaval B, Doxsey SJ. Pericentrin in cellular function and disease. J. Cell Biol. 2010;188:181–190. doi: 10.1083/jcb.200908114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.di Pietro F, Echard A, Morin X. Regulation of mitotic spindle orientation: an integrated view. EMBO Rep. 2016;17:1106–1130. doi: 10.15252/embr.201642292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Macmillan JC, Hudson JW, Bull S, Dennis JW, Swallow CJ. Comparative expression of the mitotic regulators SAK and PLK in colorectal cancer. Ann. Surg. Oncol. 2001;8:729–740. doi: 10.1007/s10434-001-0729-6. [DOI] [PubMed] [Google Scholar]
- 81.Sredni ST, Tomita T. The polo-like kinase 4 gene (PLK4) is overexpressed in pediatric medulloblastoma. Child’s Nervous Syst. 2017;33:1031. doi: 10.1007/s00381-017-3452-8. [DOI] [PubMed] [Google Scholar]
- 82.Marina M, Saavedra HI. Nek2 and Plk4: prognostic markers, drivers of breast tumorigenesis and drug resistance. Front. Biosci. 2014;19:352–365. doi: 10.2741/4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Levine MS, et al. Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Dev. Cell. 2017;40:313–322. doi: 10.1016/j.devcel.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kazazian K, et al. Plk4 promotes cancer invasion and metastasis through Arp2/3 complex regulation of the actin cytoskeleton. Cancer Res. 2017;77:434–447. doi: 10.1158/0008-5472.CAN-16-2060. [DOI] [PubMed] [Google Scholar]
- 85.Dong Y, Sui L, Watanabe Y, Sugimoto K, Tokuda M. Clinical relevance of cyclin B1 overexpression in laryngeal squamous cell carcinoma. Cancer Lett. 2002;177:13–19. doi: 10.1016/S0304-3835(01)00770-4. [DOI] [PubMed] [Google Scholar]
- 86.Hassan KA, et al. Cyclin B1 overexpression and resistance to radiotherapy in head and neck squamous cell carcinoma. Cancer Res. 2002;62:6414–6417. [PubMed] [Google Scholar]
- 87.Li JQ, et al. Cyclin B1, unlike cyclin G1, increases significantly during colorectal carcinogenesis and during later metastasis to lymph nodes. Int. J. Oncol. 2003;22:1101–1110. [PubMed] [Google Scholar]
- 88.Takeno S, et al. Prognostic value of cyclin B1 in patients with esophageal squamous cell carcinoma. Cancer. 2002;94:2874–2881. doi: 10.1002/cncr.10542. [DOI] [PubMed] [Google Scholar]
- 89.Winters ZE, et al. Subcellular localisation of cyclin B, Cdc2 andp21(WAF1/CIP1) in breast cancer. association with prognosis. Eur. J. Cancer. 2001;37:2405–2412. doi: 10.1016/S0959-8049(01)00327-6. [DOI] [PubMed] [Google Scholar]
- 90.Androic I, et al. Targeting cyclin B1 inhibits proliferation and sensitizes breast cancer cells to taxol. BMC Cancer. 2008;8:391. doi: 10.1186/1471-2407-8-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Erez A, et al. The SIL gene is essential for mitotic entry and survival of cancer cells. Cancer Res. 2007;67:4022–4027. doi: 10.1158/0008-5472.CAN-07-0064. [DOI] [PubMed] [Google Scholar]
- 92.Florio M, Huttner WB. Neural progenitors, neurogenesis and the evolution of the neocortex. Development. 2014;141:2182–2194. doi: 10.1242/dev.090571. [DOI] [PubMed] [Google Scholar]
- 93.Lancaster MA, Knoblich JA. Spindle orientation in mammalian cerebral cortical development. Curr. Opin. Neurobiol. 2012;22:737–746. doi: 10.1016/j.conb.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.LaMonica BE, Lui JH, Hansen DV, Kriegstein AR. Mitotic spindle orientation predicts outer radial glial cell generation in human neocortex. Nat. Commun. 2013;4:1665. doi: 10.1038/ncomms2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Feinstein M, et al. VPS53 mutations cause progressive cerebello-cerebral atrophy type 2 (PCCA2) J. Med. Genet. 2014;51:303–308. doi: 10.1136/jmedgenet-2013-101823. [DOI] [PubMed] [Google Scholar]