Abstract
The Aurora kinase family (Aurora A, B and C) are crucial regulators of several mitotic events, including cytokinesis. Increased expression of these kinases is associated with tumorigenesis and several compounds targeting Aurora kinase are under evaluation in clinical trials (a.o. AT9283, AZD1152, Danusertib, MLN8054). Here, we demonstrate that the pan-Aurora kinase inhibitor Tozasertib (VX-680 and MK-0457) not only causes cytokinesis defects through Aurora kinase inhibition, but is also a potent inhibitor of necroptosis, a cell death process regulated and executed by the RIPK1, RIPK3 and MLKL signalling axis. Tozasertib’s potency to inhibit RIPK1-dependent necroptosis and to block cytokinesis in cells is in the same concentration range, with an IC50 of 1.06 µM and 0.554 µM, respectively. A structure activity relationship (SAR) analysis of 67 Tozasertib analogues, modified at 4 different positions, allowed the identification of analogues that showed increased specificity for either cytokinesis inhibition or for necroptosis inhibition, reflecting more specific inhibition of Aurora kinase or RIPK1, respectively. These results also suggested that RIPK1 and Aurora kinases are functionally non-interacting targets of Tozasertib and its analogues. Indeed, more specific Aurora kinase inhibitors did not show any effect in necroptosis and Necrostatin-1s treatment did not result in cytokinesis defects, demonstrating that both cellular processes are not interrelated. Finally, Tozasertib inhibited recombinant human RIPK1, human Aurora A and human Aurora B kinase activity, but not RIPK3. The potency ranking of the newly derived Tozasertib analogues and their specificity profile, as observed in cellular assays, coincide with ADP-Glo recombinant kinase activity assays. Overall, we show that Tozasertib not only targets Aurora kinases but also RIPK1 independently, and that we could generate analogues with increased selectivity to RIPK1 or Aurora kinases, respectively.
Introduction
Mitosis is a multi-step process that is tightly regulated by several classes of kinases, like cyclin-dependent kinases (CDKs) and Aurora kinases1. The Aurora serine/threonine kinase family consists of three kinases in mammals: Aurora A, Aurora B and Aurora C2. All three Aurora kinases, although being structurally similar, have different functions and cellular localisations during mitosis2–5. Aurora A and B are expressed in most cell types and play important roles in centrosome maturation, mitotic spindle formation, kinetochore assembly and cytokinesis, the final step of cell division2,6,7. Aurora C, in contrast, is only expressed in testis where it is crucial for spermatogenesis2. Aurora A and B have been described as oncogenes and increased expression or polymorphisms of these kinases have been observed in several types of cancer6, like breast cancer8,9, prostate cancer10,11 and non-small-cell lung carcinoma12. Inhibition of Aurora kinase results in failure of G2/M transition, abnormal spindle formation leading to cytokinesis defects and apoptosis13. Several Aurora kinase inhibitors have been developed, like MLN8054 and MLN8273, which are currently in clinical trials phase I and phase II for the treatment of solid tumours and hematopoietic cancers5,6. In this paper, we will focus on the pan-Aurora kinase inhibitor Tozasertib (VX-680, MK-0457), which has been described as a type I small molecule inhibitor that targets the ATP-binding pocket of Aurora kinases14. Tozasertib retards tumour growth in xenograft models (a.o. HL-60 and HCT116)2,3,15 and was in clinical trial phase II for solid tumours and leukaemia5,6. Although Tozasertib treatment has clear anti-tumour activity, studies were discontinued due to toxic adverse effects5,16. In a broad spectrum drug/kinome-binding study, it was reported that Tozasertib can bind to receptor-interacting-protein kinase 1 (RIPK1) with a Kd of 20 nM17, while the Kd value is 0.6 nM for binding to Aurora A15. RIPK1 is part of the receptor-interacting-protein kinase family that is involved in the regulation of cell death processes and inflammation18–21. Reports are also emerging of its involvement in tumorigenesis22,23. Necroptosis is a cell death process regulated by a kinase signalling cascade in the absence of caspase activation18,21,24–26. TNF-induced necroptosis involves activation of RIPK1 and RIPK3 in a necrosome complex, leading to the activation of the execution protein-mixed lineage kinase domain-like protein (MLKL)18,20,27–29. Here, we characterise Tozasertib as cytokinesis and necroptosis inhibitor in a cellular model system, simultaneously analysing multiple image-based parameters reflecting these phenotypes. A screening of 67 novel Tozasertib analogues suggests that both pathways are independent from each other and that analogues can be identified that have a higher selectivity for either cellular response.
Results
Tozasertib induces cytokinesis defects and inhibits necroptosis at similar dose dependency
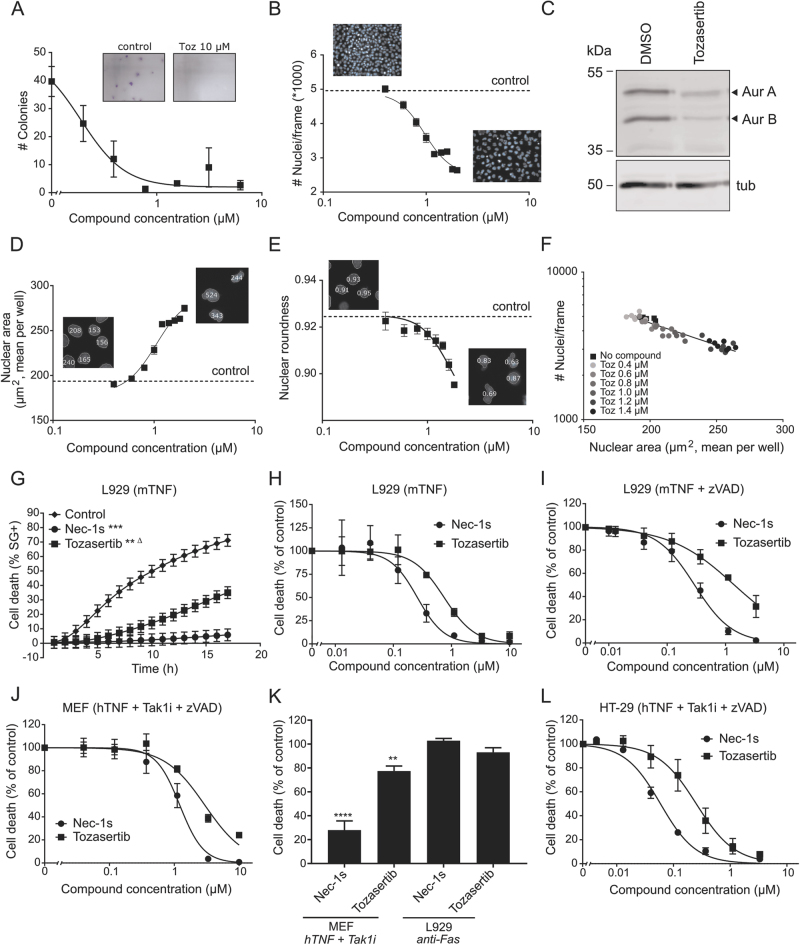
Since Tozasertib is a pan-Aurora kinase inhibitor14, we used the well-established L929 cellular model for TNF-induced necroptosis research30 to confirm inhibitory effects of Tozasertib on cell growth and cytokinesis and to evaluate its effect on cell death. Tozasertib is not toxic on itself in murine L929sAhFas cells at 10 µM or lower (Supplementary Figure 1B), but impairs colony formation in a clonogenic assay from 0.2 µM on (Fig. 1a), illustrating its cytostatic activity. Also, the total number of nuclei per image frame decreased with increasing concentration of Tozasertib (Fig. 1b), reflecting Tozasertib-induced growth arrest (IC50 value of 0.122 µM). Tozasertib-induced inhibition of autophosphorylation of endogenous Aurora A (Aur A) and B (Aur B) confirms that both Aurora kinases are targeted in L929sAhFas cells (Fig. 1c). Using high-content imaging, not only the number of nuclei, but also the nuclear morphology can be assessed31. Both nuclear area and nuclear roundness were analysed as parameter for cytokinesis defects (Fig. 1d–e). With increasing concentration of Tozasertib, nuclear area increased (mean increase to 250 µm2 at highest concentration) and nuclear roundness decreased (mean decrease to 0.80) (Fig. 1d–e). The parameter nuclear area showed a linear correlation with the number of cells or cell growth in function of the concentration of Tozasertib (Fig. 1f). Therefore, cell number and nuclear area will be further used to evaluate cytokinesis inhibition, a consequence of Aurora kinase inhibition5–7 (Fig. 1f). Next, Tozasertib was tested in murine (L929sAhFas, MEF) and human (HT29) cells for inhibition of RIPK1-dependent necroptosis. Indeed, Tozasertib (3 µM) protects against TNF-induced RIPK1-dependent necroptosis over time in L929sAhFas cells (Fig. 1g). This protection is dose dependent with an IC50 value of 0.55 µM (Fig. 1h and Table 1). Although Necrostatin-1s (Nec1s) protects against RIPK1-dependent cell death (Fig. 1g–l), RIPK1 inhibition by Nec1s does not result in cytokinesis defects (Supplementary Figure 2). Also, treatment with GSK’963, a more potent and selective RIPK1 inhibitor than Nec-1s32, did not affect cytokinesis in L929sAhFas cells (Supplementary Figure 2), suggesting that RIPK1 kinase activity is not involved in cytokinesis. In sensitising conditions in L929sAhFas cells (mTNF+zVAD.fmk) and in MEF cells, Tozasertib protects against cell death with an IC50 of 1.08 µM (Fig. 1i and Table 1) and 2.56 µM (Fig. 1j and Table 1), respectively. Although Tozasertib inhibits RIPK1-dependent apoptosis in MEF cells, its inhibition on RIPK1-dependent necroptosis in the same cells is remarkably less effective (Fig. 1j, k). This suggests that RIPK1 kinase engagement might be different in apoptotic and necroptotic conditions. In the case of induction of RIPK1-independent apoptosis in L929sAhFas cells by agonistic anti-Fas antibody, no inhibition is observed (Fig. 1k). This indicates that Tozasertib does not affect Fas-mediated apoptosis at the level of FADD/caspase-8 activation, cytochrome c release or the Apaf-1-mediated caspase cascade. Finally, Tozasertib also blocks RIPK1-dependent necroptosis in the human HT29 cell line with an IC50 of 0.26 µM (Fig. 1l and Table 1). Both in sensitising necroptosis conditions in L929sAhFas cells (mTNF+zVAD.fmk) (Supplementary Figure 3A) and in HT29 cells (hTNF+Tak1i+zVAD.fmk) (Supplementary Figure 3B), the protective effect of Tozasertib is partially lost at 10 µM and was excluded for IC50 determination. Overall, Tozasertib-induced growth arrest (IC50 0.97 µM), cytokinesis inhibition (IC50 1.06 µM) and necroptosis inhibition (IC50 0.55 µM) show similar dose dependency in different cellular models (Table 1).
Fig. 1. Tozasertib induces cytokinesis defects and inhibits necroptosis with similar dose dependency.
a L929sAhFas cells were treated with Tozasertib (concentration as indicated) for 24 h. Then, a clonogenic assay was performed to determine colony-forming capacity. Quantification was performed using ImageJ. Data represent mean values ±S.E.M. (n = 3). b, d-f L929sAhFas cells were treated with DMSO (control) or Tozasertib (Toz) (concentrations as indicated) for 24 h (f) or 48 h (b, d–e). Cells were stained with Hoechst (1 µM) and high-content images were acquired using BD pathway Bio-imager and analysed using ColumbusTM software. The dotted control line indicates the real y-axis value corresponding to a normal nucleus (DMSO treatment). Data represent mean values ±S.E.M., n = 6. Nuclear roundness is quantified as with 0 being a straight line and 1 being a perfect circle. Number of nuclei was plotted against the nuclear area (µm2) for all replicates (n = 6). Linear relation between parameters was shown using GraphPad Prism 7. c L929sAhFas cells were treated with DMSO or Tozasertib (10 µM) for 16 h, lysed and immunoblotted with the indicated antibodies. g–I L929sAhFas cells were pre-treated with DMSO (control), Nec1s (3 µM) or Tozasertib (3 µM) (g) or with concentrations as indicated (h-i) for 1 h, followed by mTNF (20 ng/mL) (g, h) (n = 4) or mTNF (20 ng/mL) + zVAD.fmk (20 µM) (i) stimulation (n = 3). * = Significance compared to DMSO, Δ = significance compared to Nec-1s. (see 'Material and methods' for statistical analysis) j MEF cells were pre-treated with DMSO, Nec-1s or Tozasertib (concentration as indicated) for 1 h, followed by stimulation with hTNF (20 ng/mL) + Tak1i (1 µM) + zVAD.fmk (20 µM) for 4 h (n = 3). k MEF (n = 3) and L929sAhFas (n = 3) cells were pre-treated with DMSO (control), Nec1s (10 µM) or Tozasertib (10 µM) for 1 h, followed by stimulation with agonistic anti-Fas antibody (250 ng/mL) or hTNF (20 ng/mL) + Tak1i (1 µM), respectively. One-way ANOVA was performed using Tukey correction for multiple comparison (compared to DMSO control). l HT-29 cells were pre-treated with DMSO, Nec1s or Tozasertib (concentration as indicated) for 1 h, followed by stimulation with hTNF (100 ng/mL) + Tak1i (1 µM) + zVAD.fmk (20 µM) for 16 h. g, l Cells were stained with SytoxGreen (5 µM) and cell death was followed over time using the FluoStar fluorescence detection system. h–k Cells were stained with propidium iodide (PI) (3 µM) and Hoechst (1 µM) for image analysis with BD pathway. Cell death percentage (% PI-positive nuclei) was calculated as percent of control. Data represent mean values ±S.E.M. (g–l). The time kinetic experiment (g) was statistically analysed using repeated measurements data analysis and dose responses (IC50 determination) analysed using probit analysis (see 'Material and methods')
Table 1.
Tozasertib induces cytokinesis defects and inhibits necroptosis with similar potency, while Nec1s inhibits necroptosis without affecting cytokinesis
| Tozasertib | Nec-1s | ||||||
|---|---|---|---|---|---|---|---|
| Compound (pre)treatment time (h) | Necroptosis treatment time (h) | IC50 (µM) | 95% CI (µM) | IC50 (µM) | 95% CI (µM) | ||
| Cytokinesis | Clonogenicity | 24 | – | 0.122 | 0.104–0.139 | NT | NT |
| Cell growth | 48 | – | 0.969 | 0.915–1.025 | NC | NC | |
| Nuclear area! | 48 | – | 1.056 | 1.013–1.101 | NC | NC | |
| Nuclear roundness | 48 | – | 1.639 | 1.521–1.842 | NC | NC | |
| Necroptosis | L929-mTNF | 1 | 5 | 0.554ΔΔΔ | 0.483–0.635 | 0.149 | 0.126–0.174 |
| L929-mTNF+zVAD | 1 | 3 | 1.082ΔΔΔ | 0.923–1.277 | 0.24 | 0.210–0.275 | |
| MEF-hTNF+TAK1i+zVAD | 1 | 4 | 2.561ΔΔΔ | 2.237–2.934 | 0.803 | 0.703–0.914 | |
| HT29-hTNF+TAK1i+zVAD | 1 | 16 | 0.257ΔΔΔ | 0.227–0.291 | 0.077 | 0.067–0.090 | |
Characterisation and quantification of Tozasertib and Nec1s affecting cytokinesis (clonogenicity, cell growth, nuclear area and nuclear roundness as parameters) and necroptosis (L929, MEF and HT29 cell lines as model systems). IC50 values were calculated and compared using dose-response curves of three independent experiments and Probit analysis (see 'Material and methods'). The 95% confidence intervals (CI) for IC50s are also indicated
NT not tested, NC not calculable ! values represent EC50, Δ refers to the significance of the difference relative to the IC50 of Nec-1s
Tozasertib-induced cytokinesis defects and necroptosis inhibition are not correlated
Previous results with Nec1s and GSK’963 suggested that RIPK1 is not involved in cytokinesis (Supplementary Figure 2). In order to further investigate whether Aurora kinase-dependent cytokinesis and RIPK1-dependent necroptosis are interrelated, a small panel of pan-Aurora kinase inhibitors were tested for their ability to inhibit necroptosis in L929sAhFas cells (Table 2). This panel included Tozasertib, Barasertib (more specific for Aurora B), AMG-900, Danusertib and SNS-314 mesylate. Of all Aurora kinase inhibitors tested, only Tozasertib and SNS-314 mesylate were able to inhibit necroptosis with IC50 values of 1.1 µM and 0.4 µM, respectively (Table 2). AMG-900 inhibits cytokinesis with an IC50 in nanomolar range (nuclear area) but cannot inhibit necroptosis (Table 2). These results suggest that cytokinesis and necroptosis are independent processes.
Table 2.
Tozasertib-induced cytokinesis defects and necroptosis inhibition are not correlated
| Inhibitor | Target | Cytokinesis | Necroptosis | ||
|---|---|---|---|---|---|
| EC50 | 95% CI for EC50 | IC50 | 95% CI for IC50 | ||
| Nec-1s | RIPK1 | NC | NC | 0.367 | 0.187–0.739 |
| Tozasertib | pan-Aurora | 0.591 | 0.230–1.581 | 1.143 | 1.405–7.749 |
| AMG-900 | pan-Aurora | 0.018 | 0.006–0.093 | NC | NC |
| Barasertib | Aurora B | 1.264 | 0.444–3.181 | NC | NC |
| Danusertib | pan-Aurora | 2.528 | 1.143–4.223 | 13.92 | 6.088–47.84 |
| SNS-314 | pan-Aurora | 0.073 | 0.018–0.320 | 0.399 | 0.129–1.424 |
L929sAhFas cells were pre-treated with the indicated Aurora inhibitors (dose-response from 10 µM–0.0022 µM) for 1 h, followed by stimulation with mTNF (20 ng/mL) for 5 h. EC50 and IC50 values (µM) were calculated for, respectively, nuclear area and inhibition of necroptosis using nonlinear regression in GraphPad Prism 7. The 95% confidence intervals (CI) for IC50s are also indicated
NC not calculable
Dissection of cytokinesis inhibition and necroptosis inhibition by SAR of Tozasertib analogues
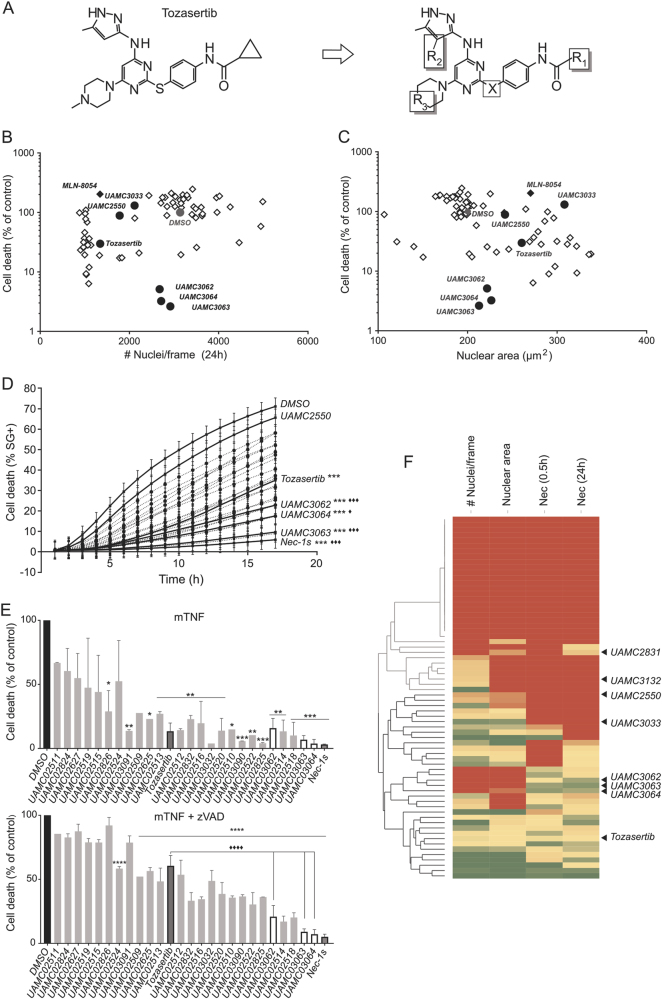
In order to analyse the required chemical structures that define induction of cytokinesis and necroptosis inhibition in more detail and to design therapeutic Tozasertib analogues that would target RIPK1 with high specificity and potency, 67 Tozasertib analogues were developed (Hofmans S. et al., manuscript under review) and tested for both cytokinesis and necroptosis inhibition (PI positivity with both 0.5 h and 24 h pre-treatment) (Fig. 2). These analogues were designed by changing 4 different positions of Tozasertib (Fig. 2a, Hofmans S. et al., manuscript under review). Scatter plots of cell growth or nuclear area in relation to the necroptosis-inhibiting capacity indicate that Tozasertib inhibits both cellular processes (Fig. 2b, c). Several Tozasertib analogues lose their potency to inhibit these cellular processes, while others preferably still inhibit cytokinesis and proliferation without affecting necroptosis (UAMC2550 and UAMC3033), or vice versa (UAMC3062, UAMC3063 and UAMC3064) (Fig. 2b, c). Also, the Aurora A inhibitor MLN-8054 was included in the screening as benchmark of cytokinesis inhibition without affecting RIPK1-dependent cell death (Fig. 2b, c). These data confirm the previous results (Table 2) that Aurora kinase inhibition has no contribution to the necroptosis inhibition. All necroptosis inhibitors have been validated using a kinetic cell death assay (Fig. 2d)33. Both after mTNF and mTNF+zVAD.fmk (sensitising condition) stimulation in L929sAhFas cells, a similar ranking of the Tozasertib analogues was obtained, with UAMC3062, UAMC3063 and UAMC3064 being more potent necroptosis inhibitors than Tozasertib (Fig. 2d, e). Although analogues were identified with better specificity to necroptosis inhibition compared to Tozasertib, they were not more efficient than Nec1s (Fig. 2d, e). Next, hierarchical clustering of all analogues using cytokinesis and cell death IC50 values resulted in the identification of different classes of Tozasertib analogues (Fig. 2f). This Ward’s method clustering identified analogues with increased selectivity for one or the other phenotype (Fig. 2f). Compared to Tozasertib, the analogues UAMC3062, UAMC3063 and UAMC3064 were more selective for necroptosis protection and UAMC3033 and UAMC2550 were more selective for cytokinesis defects (Fig. 2f and Supplementary Figure 4). Overall, based on the characterisation of the cellular phenotypes, analogues were identified with higher selectivity to either Aurora kinases or RIPK1 but not higher potency compared to Tozasertib and Nec1s, respectively.
Fig. 2. Dissection of cytokinesis inhibition and necroptosis inhibition by SAR of Tozasertib analogues.
a Structure of Tozasertib and an illustration of the four different positions that have been altered to the molecule resulting in 67 Tozasertib analogues (aliphatic substituents (R1), pyrazole moieties (R2), cyclohexyl moieties (R3) and S<>N linker (X)). b–c The 67 Tozasertib analogues were screened for both cytokinesis defects and inhibition of necroptosis in one cellular screening assay. Scatter plots of cell growth (amount of nuclei/frame) or nuclear area in relation to necroptosis-inhibiting capacity are shown. L929sAhFas cells were pre-treated with Tozasertib or its analogues (3 µM) for 0.5 h or 24 h. Next, cells were left unstimulated or stimulated with mTNF (40 ng/mL) + zVAD.fmk (10 µM) for 3 h. Proliferation, nuclear area and cell death were quantified by staining the cells with propidium iodide (PI) (3 µM) and Hoechst (1 µM) for image analysis with BD pathway. Percentage of cell death (% PI-positive nuclei) was calculated as percent of control (DMSO). d The selection of Tozasertib analogues that inhibited necroptosis was validated in a fluorometric cell death assay over time using SytoxGreen (SG) (5 µM). L929sAhFas cells were pre-treated with Nec1s, Tozasertib or its analogues (3 µM) for 0.5 h, followed by mTNF (20 ng/mL) stimulation. Significance was analysed using repeated measurements data analysis. Data represent mean values ±S.E.M. (n = 4). * refers to significance compared to DMSO, ♦ refers to significance compared to Tozasertib. e Ranking of validated Tozasertib analogues according to their potential to inhibit necroptosis. Nec1s, Tozasertib or its analogues (3 µM) were ranked for two conditions in L929sAhFas cells: mTNF (20 ng/mL) and mTNF (20 ng/mL) + zVAD.fmk (10 µM)-induced necroptosis, the latter being a sensitising condition for induction of necroptosis. Cells were stimulated for 7 h with mTNF and 4 h with mTNF + zVAD.fmk. Data represent mean values ±S.E.M. (n = 2). One-way ANOVA was performed using Tukey correction for multiple comparison (compounds compared to both DMSO control (*) and Tozasertib (♦)). f Heat map of screening data (b–c). Dose responses were included in the screening (b–c) assay conditions. IC50 values were determined for cell growth (amount of nuclei/frame), nuclear area and necroptosis protection after 0.5 h or 24 h pre-treated with compound. Hierarchical clustering (row dendrogram using Ward’s hierarchical clustering method with normalisation using Z-score calculation) of the IC50 values was then applied on these results in order to create phenotypic clusters and allow SAR analysis of Tozasertib and its analogues (Tibco Spotfire software). Selected analogues for further characterisation are indicated with arrows on the heat map
Potency and specificity ranking of Tozasertib analogues is confirmed by in vitro kinase activity profiling and validated in a human and mouse cell line
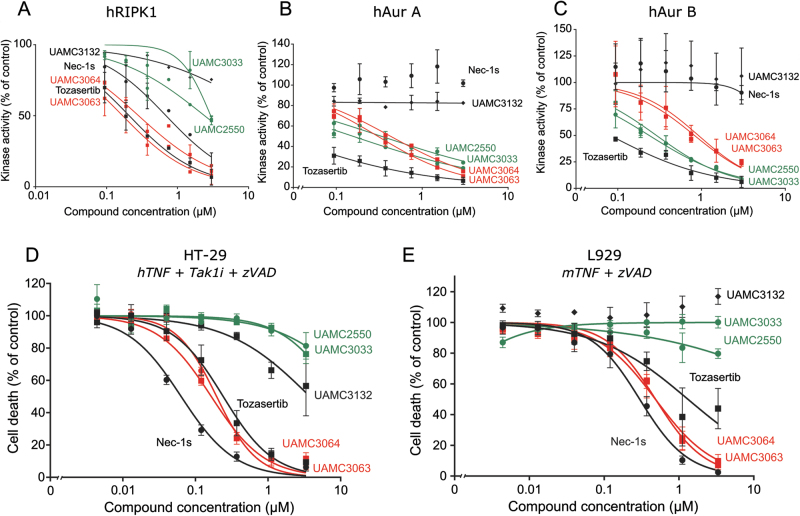
Since all data so far suggested that Tozasertib is a RIPK1 inhibitor and Tozasertib was shown to bind to RIPK117, we performed ADP-Glo kinase activity assays using recombinant hRIPK1, hAurora A (hAur A) and hAurora B (hAur B) (Fig. 3a–c). In order to confirm specificity, IC50 values were determined for all three recombinant kinases. Indeed, Tozasertib inhibits hAur A and hAur B kinase activity with an IC50 value of 0.030 µM and 0.068 µM, respectively, but it also inhibits hRIPK1 kinase activity with an IC50 of 0.18 µM (Fig. 3a–c and Supplementary Table 1). Inhibition of necroptosis by Tozasertib seems to act specifically through RIPK1 inhibition, since Tozasertib is not able to inhibit recombinant mRIPK3 kinase activity (Supplementary Figure 5). Although IC50 values for the cellular phenotypes were similar, the IC50 values for inhibition of recombinant kinase activity differs between the Aurora kinases and RIPK1 (Table 1 and Fig. 3a–c). The analogues that showed proliferation and cytokinesis defects without inhibition of necroptosis (UAMC3033 and UAMC2550) inhibit hAur A and hAur B kinase activity, albeit with lower potency than Tozasertib (IC50s between 0.2 and 0.4 µM), whereas limited inhibition of hRIPK1 kinase activity is observed (Fig. 3a–c and Supplementary Table 1). On the other hand, the analogues with higher specificity for necroptosis inhibition compared to Tozasertib (UAMC3063 and UAMC3064) could inhibit hRIPK1 kinase activity with a similar dose response as Tozasertib. Although no cytokinesis and proliferation defects were detected based on the cellular parameters analysed, these analogues are still able to inhibit hAur A and hAur B kinase activity, albeit with a >10-times lower potency than Tozasertib (Fig. 3a–c and Supplementary Table 1). Due to redundancy in the function of Aurora A and B, loss in potency of both Aurora A and B inhibition, may give rise to the more specific cell death phenotype for these analogues. Also, for Aurora kinase inhibition, IC50 values between recombinant kinase assays and cellular phenotypes differ significantly from nanomolar range for the recombinant kinase assays to micromolar range for cellular phenotypes (Table 1 and Supplementary Table 1). This may be due to timing and parameter chosen for the cellular assay, or to poor membrane permeability. Therefore, small changes in the enzymatic IC50 values could result in significant changes in cellular phenotype. Nec1s only inhibits recombinant hRIPK1 and the inactive analogue UAMC3132 lost the ability to inhibit all three recombinant proteins (Fig. 3a–c). Overall, these data indicate that the inhibition of Aurora kinases and RIPK1 by Tozasertib and its analogues is correlated to the proliferation and cytokinesis defects and necroptosis protection, respectively.
Fig. 3. Potency and specificity ranking of Tozasertib analogues is confirmed by in vitro kinase activity profiling and validated in a human and mouse cell line.
a–c An in vitro ADP-Glo kinase assay using recombinant human (h)RIPK1 (100 nM) (a), hAur A kinase (25 nM) (b) or hAur B kinase (25 nM) (c) was performed. Recombinant protein was incubated with the selected analogues, according to potency and specificity (selection indicated), at concentrations as indicated. Also, Nec1s and Tozasertib were included. Data represent mean value ±S.E.M. (n = 2). d Human HT-29 cells were pre-treated with Nec1s, Tozasertib and the Tozasertib analogues (concentration as indicated) for 1 h, followed by stimulation with hTNF (100 ng/mL) + Tak1i (1 µM) + zVAD.fmk (20 µM) for 17 h. Cells were stained with SytoxGreen (5 µM) and cell death was measured using the FluoStar fluorescence detection system. e Murine L929sAhFas cells were pre-treated with Nec1s, Tozasertib and the Tozasertib analogues (concentration as indicated) for 1 h, followed by stimulation with mTNF (20 ng/mL) for 3 h. Percentage cell death (% PI-positive nuclei) was determined as percent of control. d–e Data represent mean value ±S.E.M. (n = 3)
In a final step, the selected analogues with increased specificity were also tested in a cell death assay in the human HT29 (Fig. 3d) and the mouse L929sAhFas (Fig. 3e) cell lines (sensitised condition). After stimulation with hTNF+Tak1i+zVAD.fmk to induce necroptosis in HT29, UAMC3063-UAMC3064 could protect with similar IC50 values as Tozasertib (0.1–0.2 µM), while UAMC2550-UAMC3033 (more specific for Aurora kinase) and the inactive analogue UAMC3132 could not protect (Fig. 3d and Supplementary Table 2). Similar results were obtained in mouse L929sAhFas cells stimulated with mTNF+zVAD.fmk, a sensitising necroptotic condition (Fig. 3e and Supplementary Table 2). Overall, these data illustrate that Tozasertib targets RIPK1, but not RIPK3, and that Tozasertib analogues were designed with increased specificity for Aurora kinase or RIPK1 as characterised with human and mouse cellular phenotypes and with direct inhibition of recombinant kinases.
Discussion
Tozasertib, also known as VX-68034, is a small molecule type I kinase inhibitor targeting the Aurora family of serine/threonine protein kinases15. This kinase family is crucial for proper proliferation and cell cycle progression3,4,7,35. Increased expression or gene amplification of Aurora A/B has been observed in a number of cancers6,8,36,37 and its oncogenic potential has been illustrated by the fact that Aurora A overexpression induces transformation of mammalian fibroblasts38. Tozasertib-induced inhibition of Aurora kinase activity results in cell proliferation defects, disruption of bipolar spindle formation, polyploidy and finally cell death3,6,39–43. These phenotypic parameters are associated with defective mitosis and cytokinesis. Tozasertib has entered phase I/II clinical trials for the treatment of solid tumours44, chronic myeloid leukaemia and acute lymphocytic leukaemia with T351I BCR-ABL mutations45–47. Although Tozasertib treatment has clear anti-tumour activity, studies were discontinued due to toxic adverse effects including febrile neutropenia, anaemia and thrombocytopenia5,16. Type I kinase inhibitors are known to have more than one possible target or binding partner. Also, Tozasertib has been shown to bind other kinases than Aurora. Other kinase targets that have been described for Tozasertib are the oncogenic kinases Flt-3 and Abl (both wild-type Abl kinase and imatinib-resistant Abl mutant (T315I)) and JAK 2 kinase, with the latter playing a role in imatinib resistance in chronic myelogenous leukaemia3,5,14,15,45,48. Davis et al. performed an interaction study consisting of 72 kinase inhibitors and 442 kinases, covering more than 80% of the human catalytic protein kinome17. This study showed that Tozasertib binds to RIPK1 with a Kd of 20 nM, suggesting that Tozasertib could inhibit TNF-induced necroptosis, a RIPK1-dependent pathway19,21,49,50. In our study, we show that Tozasertib not only targets Aurora kinases, resulting in proliferation and cytokinesis defects, but also inhibits TNF-induced necroptosis and that its potency is similar for both cellular phenotypes. Both in murine (L929sAhFas, MEF cells) and human (HT-29) cell lines, Tozasertib inhibits TNF-induced necroptosis at concentrations below 10 µM (IC50 HT-29 is 0.3 µM and IC50 L929 is 0.7 µM). Targeting the active site of kinases, like the competition of Tozasertib with ATP for binding to the active site14,51, remains a challenge due to the fact that ATP-binding pockets are very similar between kinases14. This specificity issue for kinase inhibitors is a.o. illustrated by a recent report about PERK inhibitors that are also very potent RIPK1 inhibitors52. Even when sequence identity with Aurora kinase is less than 50%, which is the case for Flt3 and Abl, kinase inhibition by Tozasertib is still observed3, indicating that other factors (a.o. structural properties) may also influence interaction of the kinases with Tozasertib.
In order to study the possible relation between Aurora kinase inhibition and RIPK1-dependent necroptosis inhibition, other known Aurora kinase inhibitors (AMG-900, Barasertib, Danusertib, SNS-314 mesylate) were tested for inhibition of TNF-induced necroptosis. Except for SNS-314 mesylate, these Aurora kinase inhibitors were not able to protect against TNF-induced necroptosis, illustrating that Aurora and necroptosis inhibition are independent processes. Nec1 and Nec1s were the first RIPK1 inhibitors discovered and are now used in research as benchmark for RIPK1 inhibition19,49,53,54. Other necrostatins have also been developed49,55–57, but although they had proper selectivity, pharmacokinetic properties of all necrostatins were moderate58. This indicates the need for better therapeutic RIPK1 inhibitors. Recently, GSK developed a type II/III RIPK1 inhibitor (GSK2982772) that is currently in clinical trial phase IIa for the treatment of psoriasis, rheumatoid arthritis and ulcerative colitis59,60. Since necrostatins and the GSK RIPK1 inhibitor are classified as type II or III kinase inhibitors, and Tozasertib has structural elements of a type I kinase inhibitor (targeting the ATP-binding pocket of the kinase), this created the opportunity to develop type I RIPK1 inhibitors based on Tozasertib structure. Therefore, 67 Tozasertib analogues were developed (Hofmans S. et al, manuscript under review) and tested for cytokinesis defects and necroptosis protection in order to design novel type I RIPK1 inhibitors with higher potency and selectivity than Tozasertib. In a second step, analogues that inhibited necroptosis were validated using a real-time fluorometric method33. After detailed analysis and clustering of IC50 values, Tozasertib analogues were selected that had increased specificity for either cytokinesis defects (UAMC3033 and UAMC2550) or necroptosis protection (UAMC3063 and UAMC3064). These selected Tozasertib analogues have been tested for their potency to inhibit recombinant kinases Aur A, Aur B and RIPK1. These data confirmed our observation in the cellular assay that the selected analogues have similar potency for Aurora and/or RIPK1, but show increased specificity. Although analogues specific for necroptosis (RIPK1) inhibition in cellular assays still inhibited recombinant Aurora kinases, the more than tenfold decrease in potency is enough to reduce significantly the cytokinesis defects in the cellular assay.
It is not clear how Tozasertib and its analogues bind RIPK1. Each of the Tozasertib analogues can have a very different binding and interaction with both Aurora A, B and RIPK1. Indeed, kinases can have two different conformations, the ‘DFG-in’ active conformation and the ‘DFG-out’ inactive conformation61. RIPK1 has a DLG motif instead of the well-known DFG motif, resulting in different organisation of the amino acids during catalysis. Necrostatins (Nec-1a, Nec-3a and Nec-4) can stabilise RIPK1 in an inactive conformation that has structural similarities with the inactive conformation of the protein PKA (with DFG motif)55. Many therapeutic kinase inhibitors bind the closed conformation (Imatinib, GW572016 and A-770041) by making hydrophobic interactions with the enzyme14,62–64, but Tozasertib has a different way to interact with Aurora kinases. Tozasertib is described as a type I kinase inhibitor that binds at the ATP site without penetrating the allosteric pocket and therefore does not depend on specific kinase conformation for its binding65,66. Full activation of Aurora A kinase is only achieved when it binds to its co-factor targeting protein for Xklp2 (TPX2). Binding of TPX2 causes the activation loop to fold away from the active site, providing the DFG-in ‘open’, active kinase conformation67,68. Similar interaction of Aurora B with its co-factor inner centromere protein (INCENP) has been described69. Depending on the presence of the co-factor, Tozasertib can bind different conformations. Tozasertib was shown to bind to the closed conformation of Aurora A in the absence of TPX214,70,71. On the other hand, in the presence of TPX2 it binds to the open, active conformation of Aurora A51,67. Tozasertib also binds to the DFG-in ‘open’, active conformation of the imatinib-resistant form of Abl kinase containing H396P mutation65. This shows that prediction of the binding mode of Tozasertib, and especially its analogues, to RIPK1 (and Aurora kinase) will be difficult due to the high flexibility of these kinases (switching between inactive–active conformations) and needs further investigation.
Tozasertib is able to reduce tumour growth in vivo in xenograft models of leukaemia and colon cancer (HL-60 and HCT-116)15. With its clear anti-cancer activity15,39,45 and apoptosis-inducing capacity42,43, it is surprising that it at the same time inhibits TNF-induced necroptotic cell death at concentrations below 10 µM. But, Tozasertib is not the only chemotherapeutic agent that inhibits necroptosis. Other kinase inhibitors have been described to inhibit RIPK1 and/or RIPK3: ponatinib72,73, dabrafenib74, pazopanib72 and Sorafenib75. The fact that so many chemotherapeutic kinase inhibitors have been selected and turned out to have necroptosis inhibition properties may suggest that necroptosis targeting paradoxically may be part of the anti-tumour mechanism. It could also be that it just reflects the high chance for resemblance of ATP pocket targeting kinase inhibitors, especially small molecule type I inhibitors designed to insert into the ATP pocket, a feature that can have high similarity among kinases. However, the idea that proteins of the core pathway of necroptotic cell death (RIPK1/3 and MLKL) can promote tumour growth has been recently published22,23,76,77. During necroptosis, the cell explodes and releases damage-associated molecular patterns (DAMPs)78. These events may happen in the necrotic core22. This continuous presence of necrotic dying cells and the released DAMPs could create a pro-tumorigenic inflammatory environment that enhances the growth of the tumour mass. This could explain why targeting RIPK1 could contribute to the anti-cancer activities of these chemotherapeutics. Overall, we show that Tozasertib directly targets RIPK1 and therefore protects against TNF-induced necroptosis, and that Tozasertib analogues were designed with increased specificity for Aurora kinases or RIPK1 kinase.
Materials and methods
Cell culture, cytokines and reagents
L929 cells stably transfected with hFas and designated as L929sAhFas were generated as previously described30. All L929 cell lines and MEF cells were cultured in DMEM supplemented with 10% (v/v) FCS and l-glutamine (0.03%). HT-29 cells were cultured in EMEM medium supplemented with 10% (v/v) FCS and l-glutamine (0.03%). Following cytokines were used: recombinant (rec) human and murine TNF-α were produced at VIB protein Service Facility (Ghent, Belgium) with a specific biological activity of 3 × 107 IU/mg and 1.27 × 108 IU/mL, respectively. Following reagents were purchased as indicated: Nec1s (synthesised by the Laboratory of Medicinal Chemistry; University of Antwerp), GSK’963 (Aobious, AOB9775, Gloucester, MA), Tozasertib (LC Laboratories, T-2304), Tozasertib analogues (synthesised by the Laboratory of Medicinal Chemistry; University of Antwerp), Tak1 kinase inhibitor NP-009245 (AnalytiCon Discovery GmbH, Potsdam, Germany), z-VAD-fmk (Bachem, Bubendorf, Switzerland and R&D systems) and agonistic anti-human Fas (clone 2R2, Cell Diagnostica, Munster, Germany).
Antibodies and immunoblotting
Antibodies used were anti-phospho-Aurora A (Thr288)/Aurora B (Thr232)/Aurora C (Thr198) (D13A11) (Cell Signaling, #2914, Danvers, MA, USA), anti-tubulin-HRP (abcam, ab21058, Cambridge, UK). After treatments, cells were washed twice with PBS and lysed with 2× Laemmli buffer directly. These samples were used for SDS-PAGE and immunoblotting. Tubulin was used as loading control.
Growth arrest and cell death analysis
Proliferation, cytokinesis defects and cell death were analysed using a BDPathway 855 high-content screening instrument (BD Biosciences)31,79. Ten thousand (1 h assay) or five thousand (24 h assay) cells were seeded in a black-clear 96-well plate. The next day, the cells were pre-treated with the indicated compounds for 1 or 24 h and then stimulated with mTNF, hTNF or agonistic anti-Fas Ab (concentration as indicated) in the presence of 3 µM propidium iodide (Sigma-Aldrich) and 1 µM Hoechst (Sigma-Aldrich). Per condition, 25 image frames were taken with a ×20 objective containing a minimum of 1000 cells. Data were analysed using the Columbus software package (PerkinElmer). The image algorithm identifies objects as nuclei using the Hoechst channel and categorises cells as life or dead using the PI channel. The percentage of PI+ nuclei is calculated as read-out for cell death. The total number of nuclei after 24 or 48 h treatment reflects cell growth and nuclear area/nuclear roundness are the parameters that measure Aurora kinase inhibition and therefore growth arrest and cytokinesis defects. For time kinetics and when indicated, cell death was analysed on a FLUOSTAR Omega (BMG Labtech, Offenburg, Germany). Cells were stained with 5 µM SytoxGreen (Life Technologies) and 33 µM ac-DEVD-amc (PeptaNova GmbH, Sandhausen, Germany) together with treatment as indicated. Maximal cell death was obtained by permeabilizing all cells with Triton X-100 (0.05%). Cell death and caspase activity was determined as described previously33.
Clonogenic assay
L929sAhFas cells were treated with Tozasertib (concentration as indicated) for 24 h. Next, treatment was removed and 50 cells/condition were seeded. After 10 days, cells were fixed with 4% PFA for 10 min. After fixation, cells were stained with a 0.01% crystal violet solution for 1 h. Cells were washed with PBS, dried overnight and images were taken. Colonies were counted using ImageJ.
In vitro ADP-Glo kinase assay
Compounds (concentrations as indicated) were co-incubated with recombinant hRIPK1 (100 nM) (produced in our laboratory), hAurora kinase A (25 nM) or hAurora kinase B (25 nM) (ADP-GloTM kinase assay + Aurora A/Aurora B kinase enzyme system, Promega, V9081 and V9181). For hRIPK1, the kinase assay buffer (2×) contains 50 mM HEPES pH 7.5, 30 mM MgCl2, 50 mM NaCl, 0.5 mg/mL BSA, 0.02% CHAPS and 1 mM DTT. For the Aurora kinases, the kinase assay buffer was used from the promega assay (V9081, V9181). Next, the in vitro ADP-GloTM kinase assay (Promega, V6930) was used to determine kinase activity. The assay was performed according to the manufacturer’s protocol. The primary kinase reaction was carried out for 4 h at room temperature and a 2:2:1 ratio of kinase reaction volume to ADP-Glo reagent volume to kinase detection reagent volume was used.
Statistics
Two or three independent experiments were performed (as indicated in Figure legends) and data were analysed using GraphPad Prism 7 (when indicated). Values represent the mean values ± standard error of the mean, unless indicated otherwise. IC50 values were calculated using nonlinear curve fitting (four parameters) with fixed top and bottom in GraphPad Prism 7 (Table 2). All parameters, in which the compounds are inhibitory, are indicated with IC50 values. Only for nuclear area, the values indicate EC50, as Tozasertib (and analogues) enhances nuclear area. The definition of IC50 and EC50 is also indicated in the legends of the Tables. When indicated, one-way ANOVA was performed using Tukey correction for multiple comparison. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, with *, Δ and ♦ referring to significance compared to respectively DMSO control, Nec-1s and Tozasertib.
Repeated measurements data analysis (FIG1G and FIG2D)
Percentages of cell death were analysed as repeated measurements using the residual maximum likelihood (REML) approach as implemented in Genstat v18 (Genstat Reference Manual (Release 18), Part 3 Procedures. 2015. VSN International, Hemel Hempstead, UK). Briefly, a linear mixed model with compound, time and compound × time interaction as fixed terms, and subject.time used as residual term, was fitted to data. Times of measurement were set at equal intervals and an autoregressive correlation structure of order 1 (AR1) with equal variances across time, was selected as best model fit in all cases, based on the Aikake information coefficient. Significances of the fixed terms and pairwise comparisons of compounds across time were assessed by an F-test.
Probit analysis
Probit analysis, as implemented in Genstatv18 (Genstat Reference Manual (Release 18), Part 3 Procedures. 2015. VSN International, Hemel Hempstead, UK), was used to model the relationship between the doses of compounds and the cell death response or kinase activity. Doses were transformed to logarithms and IC50 values for each compound were calculated. The dispersion parameter was fixed to 1 and used when calculating standard errors of the IC50 values. The significance of differences between EC50 values was assessed on a log-scale using a t test, before back transforming to the original scale.
Electronic supplementary material
Acknowledgements
This research was financed by the fund for Scientific Research Flanders (FWO) under grant agreement no G078713N. S.M. was supported by the ‘Institute for the promotion of Innovation by Science and Technology in Flanders’ (IWT). V.G. has been supported by Methusalem grant (BOF09/01M00709 and BOF16/MET_V/007). Research in the group of Prof. P. Vandenabeele is supported by grants from the Vlaams Instituut voor Biotechnologie (VIB), from Ghent University (MRP, GROUP-ID consortium), grants from the ‘Foundation against Cancer’ (2012-188 and FAF-F/2016/865), grants from the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (FWO) (FWO G.0787.13N, FWO G.0C37.14N, FWO G.0C31.14N, FWO G.0E04.16N), grants from the Flemish Government (Methusalem BOF09/01M00709 and BOF16/MET_V/007), a grant from the Belgian science policy office (BELSPO)(IAP 7/32). This research is part of the Cancer Research Institute Ghent (CRIG) consortium. S.H. is paid by the University of Antwerp as a researcher to work in the Laboratory of Medicinal Chemistry. The Laboratory of Medicinal Chemistry is partner of the Antwerp Drug Discovery Network (www.addn.be). This research was also funded by a FWO PhD fellowship under grant agreement no. 11Z815N and 11Z817N appointed to L.D.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by B. Turk
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41419-017-0245-7).
References
- 1.Pitts TM, Davis SL, Eckhardt SG, Bradshaw-Pierce EL. Targeting nuclear kinases in cancer: development of cell cycle kinase inhibitors. Pharmacol. Ther. 2014;142:258–269. doi: 10.1016/j.pharmthera.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 2.Marumoto T, Zhang D, Saya H. Aurora-A - a guardian of poles. Nat. Rev. Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- 3.Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 4.Ducat D, Zheng Y. Aurora kinases in spindle assembly and chromosome segregation. Exp. Cell Res. 2004;301:60–67. doi: 10.1016/j.yexcr.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 5.Falchook GS, Bastida CC, Kurzrock R. Aurora kinase inhibitors in oncology clinical trials: current state of the progress. Semin. Oncol. 2015;42:832–848. doi: 10.1053/j.seminoncol.2015.09.022. [DOI] [PubMed] [Google Scholar]
- 6.Yan M, et al. Aurora-A kinase: a potent oncogene and target for cancer therapy. Med. Res. Rev. 2016;36:1036–1079. doi: 10.1002/med.21399. [DOI] [PubMed] [Google Scholar]
- 7.Afonso O, Figueiredo AC, Maiato H. Late mitotic functions of Aurora kinases. Chromosoma. 2017;126:93–103. doi: 10.1007/s00412-016-0594-5. [DOI] [PubMed] [Google Scholar]
- 8.Xu J, et al. Aurora-A identifies early recurrence and poor prognosis and promises a potential therapeutic target in triple negative breast cancer. PLoS ONE. 2013;8:e56919. doi: 10.1371/journal.pone.0056919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Assoro AB, et al. The mitotic kinase Aurora--a promotes distant metastases by inducing epithelial-to-mesenchymal transition in ERalpha(+) breast cancer cells. Oncogene. 2014;33:599–610. doi: 10.1038/onc.2012.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park K, et al. Prostate cancer with Paneth cell-like neuroendocrine differentiation has recognizable histomorphology and harbors AURKA gene amplification. Hum. Pathol. 2014;45:2136–2143. doi: 10.1016/j.humpath.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buschhorn HM, et al. Aurora-A over-expression in high-grade PIN lesions and prostate cancer. Prostate. 2005;64:341–346. doi: 10.1002/pros.20247. [DOI] [PubMed] [Google Scholar]
- 12.Takeshita M, et al. Aurora-B overexpression is correlated with aneuploidy and poor prognosis in non-small cell lung cancer. Lung Cancer. 2013;80:85–90. doi: 10.1016/j.lungcan.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 13.Bavetsias V, Linardopoulos S. Aurora kinase inhibitors: current status and outlook. Front. Oncol. 2015;5:278. doi: 10.3389/fonc.2015.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheetham GM, Charlton PA, Golec JM, Pollard JR. Structural basis for potent inhibition of the Aurora kinases and a T315I multi-drug resistant mutant form of Abl kinase by VX-680. Cancer Lett. 2007;251:323–329. doi: 10.1016/j.canlet.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Harrington EA, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat. Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 16.Cheung CH, Sarvagalla S, Lee JY, Huang YC, Coumar MS. Aurora kinase inhibitor patents and agents in clinical testing: an update (2011 - 2013) Expert Opin. Ther. Pat. 2014;24:1021–1038. doi: 10.1517/13543776.2014.931374. [DOI] [PubMed] [Google Scholar]
- 17.Davis MI, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 18.Newton K, Manning G. Necroptosis and Inflammation. Annu. Rev. Biochem. 2016;85:743–763. doi: 10.1146/annurev-biochem-060815-014830. [DOI] [PubMed] [Google Scholar]
- 19.Degterev A, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho YS, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat. Rev. Mol. Cell Biol. 2010;11:700–714. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- 22.Liu X, et al. Key roles of necroptotic factors in promoting tumor growth. Oncotarget. 2016;7:22219–22233. doi: 10.18632/oncotarget.7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu XY, et al. RIP1 kinase is an oncogenic driver in melanoma. Cancer Res. 2015;75:1736–1748. doi: 10.1158/0008-5472.CAN-14-2199. [DOI] [PubMed] [Google Scholar]
- 24.He S, et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell. 2009;137:1100–1111. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 25.Newton K, et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science. 2014;343:1357–1360. doi: 10.1126/science.1249361. [DOI] [PubMed] [Google Scholar]
- 26.Zhang DW, et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332–336. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- 27.Dondelinger Y, et al. MLKL compromises plasma membrane integrity by binding to phosphatidylinositol phosphates. Cell Rep. 2014;7:971–981. doi: 10.1016/j.celrep.2014.04.026. [DOI] [PubMed] [Google Scholar]
- 28.Chen X, et al. Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res. 2014;24:105–121. doi: 10.1038/cr.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, et al. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol. Cell. 2014;54:133–146. doi: 10.1016/j.molcel.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Vercammen D, et al. Inhibition of caspases increases the sensitivity of L929 cells to necrosis mediated by tumor necrosis factor. J. Exp. Med. 1998;187:1477–1485. doi: 10.1084/jem.187.9.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadaie M, et al. Cell-based screen for altered nuclear phenotypes reveals senescence progression in polyploid cells after Aurora kinase B inhibition. Mol. Biol. Cell. 2015;26:2971–2985. doi: 10.1091/mbc.E15-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berger SB, et al. Characterization of GSK'963: a structurally distinct, potent and selective inhibitor of RIP1 kinase. Cell Death Discov. 2015;1:15009. doi: 10.1038/cddiscovery.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grootjans S, et al. A real-time fluorometric method for the simultaneous detection of cell death type and rate. Nat. Protoc. 2016;11:1444–1454. doi: 10.1038/nprot.2016.085. [DOI] [PubMed] [Google Scholar]
- 34.Bebbington D, et al. The discovery of the potent aurora inhibitor MK-0457 (VX-680) Bioorg. Med. Chem. Lett. 2009;19:3586–3592. doi: 10.1016/j.bmcl.2009.04.136. [DOI] [PubMed] [Google Scholar]
- 35.Bischoff JR, Plowman GD. The Aurora/Ipl1p kinase family: regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 1999;9:454–459. doi: 10.1016/S0962-8924(99)01658-X. [DOI] [PubMed] [Google Scholar]
- 36.de Paula Careta F, et al. TheAurora A and B kinases are up-regulated in bone marrow-derived chronic lymphocytic leukemia cells and represent potential therapeutic targets. Haematologica. 2012;97:1246–1254. doi: 10.3324/haematol.2011.054668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guan Z, et al. Aurora-A, a negative prognostic marker, increases migration and decreases radiosensitivity in cancer cells. Cancer Res. 2007;67:10436–10444. doi: 10.1158/0008-5472.CAN-07-1379. [DOI] [PubMed] [Google Scholar]
- 38.Bischoff JR, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin YG, et al. Targeting aurora kinase with MK-0457 inhibits ovarian cancer growth. Clin. Cancer Res. 2008;14:5437–5446. doi: 10.1158/1078-0432.CCR-07-4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, et al. The effect of Aurora kinases on cell proliferation, cell cycle regulation and metastasis in renal cell carcinoma. Int. J. Oncol. 2012;41:2139–2149. doi: 10.3892/ijo.2012.1633. [DOI] [PubMed] [Google Scholar]
- 41.Li Y, et al. VX680/MK-0457, a potent and selective Aurora kinase inhibitor, targets both tumor and endothelial cells in clear cell renal cell carcinoma. Am. J. Transl. Res. 2010;2:296–308. [PMC free article] [PubMed] [Google Scholar]
- 42.Gizatullin F, et al. The Aurora kinase inhibitor VX-680 induces endoreduplication and apoptosis preferentially in cells with compromised p53-dependent postmitotic checkpoint function. Cancer Res. 2006;66:7668–7677. doi: 10.1158/0008-5472.CAN-05-3353. [DOI] [PubMed] [Google Scholar]
- 43.Huang XF, et al. Aurora kinase inhibitory VX-680 increases Bax/Bcl-2 ratio and induces apoptosis in Aurora-A-high acute myeloid leukemia. Blood. 2008;111:2854–2865. doi: 10.1182/blood-2007-07-099325. [DOI] [PubMed] [Google Scholar]
- 44.Traynor AM, et al. Phase I dose escalation study of MK-0457, a novel Aurora kinase inhibitor, in adult patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2011;67:305–314. doi: 10.1007/s00280-010-1318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giles FJ, et al. MK-0457, a novel kinase inhibitor, is active in patients with chronic myeloid leukemia or acute lymphocytic leukemia with the T315I BCR-ABL mutation. Blood. 2007;109:500–502. doi: 10.1182/blood-2006-05-025049. [DOI] [PubMed] [Google Scholar]
- 46.Giles FJ, et al. MK-0457, an Aurora kinase and BCR-ABL inhibitor, is active in patients with BCR-ABL T315I leukemia. Leukemia. 2013;27:113–117. doi: 10.1038/leu.2012.186. [DOI] [PubMed] [Google Scholar]
- 47.Seymour JF, et al. A phase 2 study of MK-0457 in patients with BCR-ABL T315I mutant chronic myelogenous leukemia and philadelphia chromosome-positive acute lymphoblastic leukemia. Blood Cancer J. 2014;4:e238. doi: 10.1038/bcj.2014.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samanta AK, Lin H, Sun T, Kantarjian H, Arlinghaus RB. Janus kinase 2: a critical target in chronic myelogenous leukemia. Cancer Res. 2006;66:6468–6472. doi: 10.1158/0008-5472.CAN-06-0025. [DOI] [PubMed] [Google Scholar]
- 49.Degterev A, et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 50.Tonnus W, Linkermann A. The in vivo evidence for regulated necrosis. Immunol. Rev. 2017;277:128–149. doi: 10.1111/imr.12551. [DOI] [PubMed] [Google Scholar]
- 51.Zhao B, et al. Modulation of kinase-inhibitor interactions by auxiliary protein binding: crystallography studies on Aurora A interactions with VX-680 and with TPX2. Protein Sci. 2008;17:1791–1797. doi: 10.1110/ps.036590.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rojas-Rivera D, et al. When PERK inhibitors turn out to be new potent RIPK1 inhibitors: critical issues on the specificity and use of GSK2606414 and GSK2656157. Cell Death Differ. 2017;24:1100–1110. doi: 10.1038/cdd.2017.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi N, et al. Necrostatin-1 analogues: critical issues on the specificity, activity and in vivo use in experimental disease models. Cell Death Dis. 2012;3:e437. doi: 10.1038/cddis.2012.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Declercq W, Vanden Berghe T, Vandenabeele P. RIP kinases at the crossroads of cell death and survival. Cell. 2009;138:229–232. doi: 10.1016/j.cell.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 55.Xie T, et al. Structural basis of RIP1 inhibition by necrostatins. Structure. 2013;21:493–499. doi: 10.1016/j.str.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 56.Wang K, et al. Structure-activity relationship analysis of a novel necroptosis inhibitor, necrostatin-5. Bioorg. Med. Chem. Lett. 2007;17:1455–1465. doi: 10.1016/j.bmcl.2006.11.056. [DOI] [PubMed] [Google Scholar]
- 57.Zheng W, Degterev A, Hsu E, Yuan J, Yuan C. Structure-activity relationship study of a novel necroptosis inhibitor, necrostatin-7. Bioorg. Med. Chem. Lett. 2008;18:4932–4935. doi: 10.1016/j.bmcl.2008.08.058. [DOI] [PubMed] [Google Scholar]
- 58.Teng X, et al. Structure-activity relationship study of novel necroptosis inhibitors. Bioorg. Med. Chem. Lett. 2005;15:5039–5044. doi: 10.1016/j.bmcl.2005.07.077. [DOI] [PubMed] [Google Scholar]
- 59.Harris PA, et al. DNA-encoded library screening identifies benzo[b][1,4]oxazepin-4-ones as highly potent and monoselective receptor interacting protein 1 kinase inhibitors. J. Med. Chem. 2016;59:2163–2178. doi: 10.1021/acs.jmedchem.5b01898. [DOI] [PubMed] [Google Scholar]
- 60.Harris PA, et al. Discovery of a first-in-class receptor interacting protein 1 (RIP1) kinase specific clinical candidate (GSK2982772) for the treatment of inflammatory diseases. J. Med. Chem. 2017;60:1247–1261. doi: 10.1021/acs.jmedchem.6b01751. [DOI] [PubMed] [Google Scholar]
- 61.Muller S, Chaikuad A, Gray NS, Knapp S. The ins and outs of selective kinase inhibitor development. Nat. Chem. Biol. 2015;11:818–821. doi: 10.1038/nchembio.1938. [DOI] [PubMed] [Google Scholar]
- 62.Buchdunger E, et al. Inhibition of the Abl protein-tyrosine kinase in vitro and in vivo by a 2-phenylaminopyrimidine derivative. Cancer Res. 1996;56:100–104. [PubMed] [Google Scholar]
- 63.Burchat A, et al. Discovery of A-770041, a src-family selective orally active lck inhibitor that prevents organ allograft rejection. Bioorg. Med. Chem. Lett. 2006;16:118–122. doi: 10.1016/j.bmcl.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 64.Wood ER, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 65.Young MA, et al. Structure of the kinase domain of an imatinib-resistant Abl mutant in complex with the Aurora kinase inhibitor VX-680. Cancer Res. 2006;66:1007–1014. doi: 10.1158/0008-5472.CAN-05-2788. [DOI] [PubMed] [Google Scholar]
- 66.Treiber DK, Shah NP. Ins and outs of kinase DFG motifs. Chem. Biol. 2013;20:745–746. doi: 10.1016/j.chembiol.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 67.Bayliss R, Sardon T, Vernos I, Conti E. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell. 2003;12:851–862. doi: 10.1016/S1097-2765(03)00392-7. [DOI] [PubMed] [Google Scholar]
- 68.Kufer TA, et al. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol. 2002;158:617–623. doi: 10.1083/jcb.200204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elkins JM, Santaguida S, Musacchio A, Knapp S. Crystal structure of human aurora B in complex with INCENP and VX-680. J. Med. Chem. 2012;55:7841–7848. doi: 10.1021/jm3008954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheetham GM, et al. Crystal structure of aurora-2, an oncogenic serine/threonine kinase. J. Biol. Chem. 2002;277:42419–42422. doi: 10.1074/jbc.C200426200. [DOI] [PubMed] [Google Scholar]
- 71.Nowakowski J, et al. Structures of the cancer-related Aurora-A, FAK, and EphA2 protein kinases from nanovolume crystallography. Structure. 2002;10:1659–1667. doi: 10.1016/S0969-2126(02)00907-3. [DOI] [PubMed] [Google Scholar]
- 72.Fauster A, et al. A cellular screen identifies ponatinib and pazopanib as inhibitors of necroptosis. Cell Death Dis. 2015;6:e1767. doi: 10.1038/cddis.2015.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Najjar M, et al. Structure guided design of potent and selective ponatinib-based hybrid inhibitors for RIPK1. Cell Rep. 2015;10:1850–1860. doi: 10.1016/j.celrep.2015.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li JX, et al. The B-Raf(V600E) inhibitor dabrafenib selectively inhibits RIP3 and alleviates acetaminophen-induced liver injury. Cell Death Dis. 2014;5:e1278. doi: 10.1038/cddis.2014.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martens S, et al. Sorafenib tosylate inhibits directly necrosome complex formation and protects in mouse models of inflammation and tissue injury. Cell Death Dis. 2017;8:e2904. doi: 10.1038/cddis.2017.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seifert L, et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature. 2016;532:245–249. doi: 10.1038/nature17403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Strilic B, et al. Tumour-cell-induced endothelial cell necroptosis via death receptor 6 promotes metastasis. Nature. 2016;536:215–218. doi: 10.1038/nature19076. [DOI] [PubMed] [Google Scholar]
- 78.Kaczmarek A, Vandenabeele P, Krysko DV. Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity. 2013;38:209–223. doi: 10.1016/j.immuni.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 79.Vanden Berghe T, et al. Determination of apoptotic and necrotic cell death in vitro and in vivo. Methods. 2013;61:117–129. doi: 10.1016/j.ymeth.2013.02.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.