Abstract
Purpose
Forkhead box K2 (FOXK2) is a member of the forkhead box family of transcription factors. Recently, researchers discovered that overexpression of FOXK2 inhibits the proliferation and metastasis of breast cancer, non-small cell lung cancer, and colorectal cancer, and is related to the clinical prognosis. However, in hepatocellular carcinoma, FOXK2 results in the opposite phenotypes. Currently, the contribution of FOXK2 to glioma pathogenesis is not clear.
Patients and methods
We evaluated the expression of FOXK2 in 151 glioma patients using immunohistochemistry assays. The associations among the expression of FOXK2, clinicopathological parameters, and the prognosis of glioma patients were statistically analyzed. We downregulated and upregulated the level of FOXK2 in glioma cells by transfections with small interfering RNA and plasmids. Then, we investigated the effects on tumor cell behavior in vitro by Cell Counting Kit-8 assays, colony-formation assay, transwell assay, and the epithelial-to-mesenchymal transition (EMT) biomarker levels.
Results
The clinical data showed that expression of FOXK2 gradually decreased with increasing World Health Organization (WHO) grades and a low level of FOXK2 indicates a poor prognosis. FOXK2 expression is negatively correlated with Ki67 expression and the WHO degree but is not correlated with other clinicopathological parameters, including sex, age, Karnofsky Performance Status, tumor diameter, O-6-methylguanine-DNA methyltransferase, and glutathione S-transferase pi. FOXK2 knockdown enhances glioma cell proliferation, migration, invasion, and EMT process, and, in contrast, FOXK2 overexpression inhibits glioma cell proliferation, migration, invasion, and the EMT process.
Conclusion
Expression of FOXK2 gradually decreases with increasing WHO grades. FOXK2 inhibits tumor proliferation, migration, and invasion. FOXK2 is a critical mediator of the EMT process.
Keywords: Forkhead box K2, FOXK2, glioma, oncology
Introduction
Glioma, a tumor derived from glial cells, is the most common primary intracranial tumor. It is a malignant tumor with a high degree of malignancy, a low rate of recovery, and poor prognosis. Glioma formation is a complex process. Although many studies have explored the mechanism and found some therapeutic targets, effective therapeutic targets for glioma have not yet been found. Glioma treatment is still limited to surgical excision, postoperative adjuvant radiotherapy, and chemotherapy, and the prognosis of glioma has not been fundamentally improved.1
Forkhead box K2 (FOXK2), a member of the forkhead transcription factor, is a vital protein that can be phosphorylated by the cyclin complex.2,3 FOXK2 is involved in several signaling pathways, such as the mammalian target of rapamycin, WNT, and AKT signaling pathways. These signaling pathways regulate cell proliferation and death by influencing the expression of important genes through the transcription repressor FOXK2.4,5 Recently, researchers found that FOXK2 plays a crucial role in the occurrence and development of various tumors, and a high expression of FOXK2 is correlated with poor survival.6–8 Chen et al8 found that overexpression of FOXK2 suppresses epithelial-to-mesenchymal transition (EMT) in non-small cell lung cancer, inhibiting the expression of N-cadherin and Snail. However, Lin et al9 found that FOXK2 expression increased in hepatocellular carcinoma. FOXK2 is an oncogene and a promising prognostic biomarker in hepatocellular carcinoma.
Gene expression is regulated by a transcription program. FOXK2, as a transcription factor, regulates a number of genes involved in cell adhesion and motility, metabolism, apoptosis, and cancer tumorigenesis.10–13 Recently, a report from Shan et al14 demonstrated that FOXK2 interacted with polycomb complex molecules and recruited distinct corepressor complexes, such as SIN3 transcription regulator family member A (SIN3A), RE1 silencing transcription factor (REST), nuclear receptor corepressor 2 (SMRT), and histone deacetylase (HDAC), which are important for gene transcription.
Currently, the role of FOXK2 in gliomas has not been studied. In this study, we examined 151 cases of glioma and analyzed the correlation with clinicopathological features. Meanwhile, the expression changes were used to examine the influence of FOXK2 on the glioma cell line.
Materials and methods
Clinical sample
During the period of January 2013–January 2015, 151 patients who were all diagnosed with glioma were randomly selected at the Department of Neurosurgery in Tianjin Huanhu Hospital, China. This research was approved by the Ethics Committee of Tianjin Huanhu Hospital. Written informed consent for this study was obtained from each patient. None of the selected patients received radiotherapy, adjuvant chemotherapy, or other supplementary therapies before the surgeries. After excision, the resected tissue samples were fixed immediately in 4% neutral-buffered formalin and were then processed for paraffin sectioning. To verify the results, another 46 fresh glioma tissues were obtained for real-time polymerase chain reaction (PCR). Two senior neuropathologists evaluated the extent of the pathology by the World Health Organization (WHO) classification system. Clinical follow-up was completed for all patients. The patients who died from other diseases or unexpected reasons were excluded from the evaluation. The expression score of FOXK2 was calculated by multiplying the staining intensity and the positive cells’ percentage scores, ranging from 0 to 12. The staining intensity was scored as follows: 0 (no staining), 1 (weak staining), 2 (moderate staining), and 3 (strongly positive) (Figure S1). The percentage of FOXK2-positive cells was scored as follows: 0 (0%), 1 (1%–25%), 2 (26%–50%), 3 (51%–75%), and 4 (76%–100%).
Immunohistochemistry staining
Formalin-fixed tissue samples were prepared before immunohistochemical staining, and the staining used the avidin–biotin complex method. The sections were blocked with normal goat serum and were then incubated with the following primary antibodies overnight at 4°C: an anti-FOXK2 antibody (1:500); an anti-Ki67 antibody (1:200); an anti-O-6-methylguanine-DNA methyltransferase (MGMT) antibody (1:200); and an anti-glutathione S-transferase pi (GST-pi) antibody (1:200) (Abcam, Cambridge, UK). The secondary antibody was purchased from Zhongshan Biology (Foshan, China) and was diluted at 1:100. The incubation was at 37°C for 1 h and was then incubated in the avidin–biotin complex solution for another 1 h. Finally, the expression of FOXK2 was detected by coloration with a diaminobenzidine buffer, and the procedure was detected as described previously.
Cell culture and transfection
The human glioma cell lines LN229 and U373 were obtained from American Type Culture Collection (Manassas, VA, USA). The cells were cultured in Dulbecco’s Modified Eagle’s Medium (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (Gibco®, Thermo Fisher Scientific, Waltham, MA, USA), 100 μg/mL penicillin (Sigma-Aldrich Co., St Louis, MO, USA), and 100 μg/mL streptomycin (Sigma) under 5% CO2 at 37°C. The glioma cells were seeded and grown in 6-well plates overnight. The transfections with small interfering RNA (siRNA) and the plasmids were performed with a transfection reagent (Hoffman-La Roche Ltd, Basel, Switzerland) according to the manufacturer’s protocol. Forty-eight hours following the transfection, the cells were collected and seeded for assays. The sequence-specific siRNA to FOXK2 (si-FOXK2-1 targeting the following sequence 5′-GAGTTCGAGTATCTGATGA-3′, si-FOXK2-2 targeting the following sequence 5′-GCGAAC ACGTACACTGTCT-3′) and the control siRNA were purchased from RiboBio (Guangzhou, China). A plasmid encoding the FOXK2 protein was purchased from Genscript (cloning ID S57120; Nanjing, China).
Western blot analysis, RNA isolation, and real-time PCR
Antibodies for FOXK2 (1:1,000), E-cadherin (1:1,000), N-cadherin (1:1,000), and vimentin (1:1,000) (Abcam) were used for the Western blot analysis. Total RNA was isolated with TRIzol (Invitrogen, Thermo Fisher Scientific) according to the manufacturer’s protocol. Equal amounts of RNA were converted into cDNA using FOXK2 primers with the PrimeScript RT reagent Kit (Promega Corporation, Fitchburg, WI, USA). The primer sequences for FOXK2 were as follows: forward, 5′-GCTCACCCTGAACGGGATTT-3′; and reverse, 5′-TCCTCCAGAACGAGCCTTTG-3′. The real-time PCR analysis contrasts the relative expression levels of FOXK2.
Cell Counting Kit-8 assay
We used Cell Counting Kit-8 (CCK-8) assays to evaluate the growth rates. After transfection for 48 h, the cells were plated in 96-well plates. After culturing for 24, 48, 72, and 96 h, 20 μL of CCK-8 solution (Beyotime, Shanghai, China) was incubated in each well at 37°C for 4 h. The absorbance was measured via a spectrophotometer at 495 nm. The rate of proliferation was equal to the OD value divided by the average OD value at 24 h.
Transwell migration assay
The ability of cells to migrate was assessed using transwell filters (Corning Incorporated, Corning, NY, USA). The cells were incubated for 48 h at 37°C in 5% CO2. Cell migration and invasion to the underside of the transwell membrane was detected by staining the cells with crystal violet, and then the cells were counted and imaged under an inverted microscope at ×200 magnification (Olympus, Tokyo, Japan). The results are expressed as the average number of invading cells per field.
Transwell invasion assay
The ability of cells to invade was assessed using transwell filters (Corning). A total of 30–50 μL of Matrigel (Corning) was added to the transwell inserts to form a thin gel layer. The cells were incubated for 48 h at 37°C in 5% CO2. Cell migration and invasion to the underside of the transwell membrane was detected by staining the cells with crystal violet, and then the cells were counted and imaged under an inverted microscope at ×200 magnification (Olympus). The results are expressed as the average number of invading cells per field.
Statistical analysis
The clinicopathological correlations were analyzed using chi-square analysis. Survival curves were plotted with the Kaplan–Meier method, and differences between the survival curves were tested by the log-rank test. Cox’s proportional hazards model was used to identify factors with an independent influence on survival. Student’s t-test was conducted for comparison between two groups. The data are presented as the mean values (±SD). SPSS version 16.0 software (SPSS Inc., Chicago, IL, USA) was used for all data analyses. The differences were considered significant when p<0.05.
Results
FOXK2 is downregulated in high-grade glioma compared to low-grade glioma
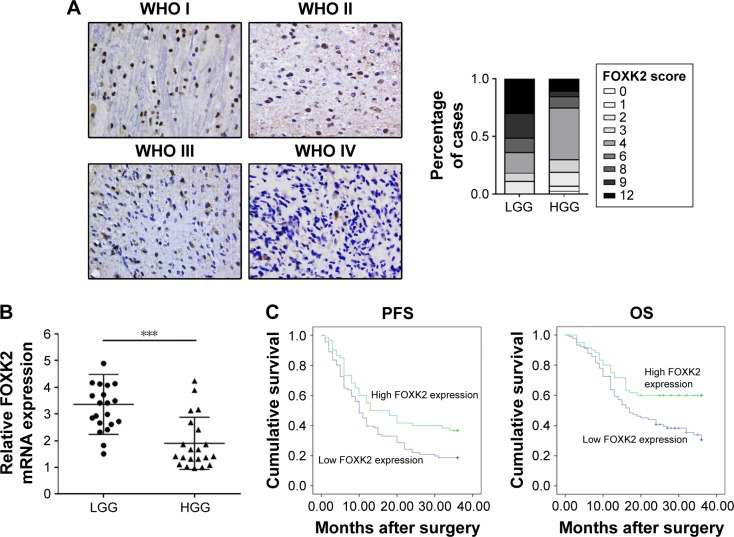
The expression score of WHO grade I gliomas, WHO grade II gliomas, WHO grade III gliomas, and WHO grade IV gliomas were 8.13±3.96, 7.42±3.55, 5.46±3.59, and 4.36±2.63, respectively. The expression score of FOXK2 for high-grade glioma (WHO grade III and IV gliomas) was significantly lower compared to the low-grade glioma (WHO grade I and II gliomas) (p<0.05, Figure 1A). The expression level of FOXK2 was divided into low (score 0–4) and high (score 6–12) expression groups using the mean expression score (5.95) as a cutoff point. The proportion of Ki67-positive cells ranged from 1% to 72%, and a median value of 13.9% was used as the cutoff value for distinguishing between the low and high groups. The proportion of MGMT-positive cells was 52.3%. The proportion of GST-pi-positive cells was 43.7%.
Figure 1.
Expression of FOXK2 in different WHO grade gliomas. (A) Immunohistochemistry staining of FOXK2 in different WHO grade gliomas (original magnification ×200). The histogram shows that the expression score of FOXK2 in high-grade glioma (WHO grade III and IV gliomas) was significantly lower compared to the low-grade glioma (WHO grade I and II gliomas). The staining intensity was scored as follows: 0 (no staining), 1 (weak staining), 2 (moderate staining), and 3 (strong staining) (see Figure S1). The percentage of FOXK2-positive cells was scored as follows: 0 (0%), 1 (1%–25%), 2 (26%–50%), 3 (51%–75%), and 4 (76%–100%). (B) A total of 23 low-grade glioma tissues and 23 high-grade glioma tissues were subjected to quantitative reverse transcription PCR. The results showed that the FOXK2 mRNA level in the high-grade glioma tissues was significantly lower than in the low-grade glioma tissues. ***p<0.001. (C) Kaplan–Meier analysis for the PFS and OS based on the FOXK2 expression in the glioma patients (log-rank test, p<0.01). Blue indicates low FOXK2 expression group; green indicates high FOXK2 expression group.
Abbreviations: FOXK2, Forkhead box K2; HGG, high-grade glioma; LGG, low-grade glioma; OS, overall survival; PCR, polymerase chain reaction; PFS, progression-free survival; WHO, World Health Organization.
To verify the expression of FOXK2 in glioma, 23 low-grade glioma tissues and 23 high-grade glioma tissues were subjected to quantitative reverse transcription PCR. The results showed that the FOXK2 mRNA level in high-grade glioma tissues was significantly lower than that in the low-grade glioma tissues (Figure 1B).
FOXK2 expression is negatively correlated with the WHO grade and Ki67, and a high level of FOXK2 improves the prognosis
We investigated the association of FOXK2 expression with the clinicopathological characteristics. FOXK2 expression was negatively correlated with the WHO grade and Ki67 (p<0.01). There was no significant association between FOXK2 expression and the other clinicopathological parameters, including sex, age, Karnofsky Performance Status, tumor diameter, MGMT, and GST-pi (Table 1).
Table 1.
Relationship between FOXK2 expression and clinico-pathological features
| Clinicopathological features | Patients (n) |
FOXK2
|
p-value | |
|---|---|---|---|---|
| Low (n) | High (n) | |||
| Sex | 0.974 | |||
| Male | 76 | 46 | 30 | |
| Female | 75 | 45 | 30 | |
| Age | 0.082 | |||
| <50 years | 86 | 57 | 29 | |
| ≥50 years | 65 | 34 | 31 | |
| WHO grade | <0.01 | |||
| Low (WHO grade I and II) | 57 | 20 | 37 | |
| High (WHO grade III and IV) | 94 | 71 | 23 | |
| Tumor size | 0.796 | |||
| <5 cm | 5 | 30 | 21 | |
| ≥5 cm | 100 | 61 | 39 | |
| Karnofsky Performance Status | 0.152 | |||
| <70 | 114 | 65 | 49 | |
| ≥70 | 37 | 26 | 11 | |
| Ki67 | <0.01 | |||
| <10% | 68 | 33 | 35 | |
| ≥10% | 83 | 58 | 25 | |
| MGMT | 0.385 | |||
| − | 72 | 46 | 26 | |
| + | 79 | 45 | 34 | |
| GST-pi | 0.940 | |||
| − | 85 | 51 | 34 | |
| + | 66 | 40 | 26 | |
Abbreviations: FOXK2, Forkhead box K2; GST-pi, glutathione S-transferase pi; MGMT, O-6-methylguanine-DNA methyltransferase; WHO, World Health Organization.
We investigated the prognostic value of FOXK2 expression and the clinicopathological parameters on the overall survival (OS) of the patients using survival analysis. The result revealed that patients with a high FOXK2 expression had a longer OS (Figure 1C). Cox regression analysis showed that the OS was relative to the expression of FOXK2 (p<0.01). Combined with the earlier results, FOXK2 was an independent prognostic indicator for glioma patients (Table 2).
Table 2.
Univariate and multivariate analyses of prognostic parameters in patients with glioma
| Parameter | Univariate log-rank test p-value | Cox multivariable analysis p-value | Relative risk |
|---|---|---|---|
| Age (<50 years vs ≥50 years) | 0.274 | – | – |
| Sex (male vs female) | 0.286 | – | – |
| WHO grade (low vs high) | <0.01 | <0.01 | 112.772 |
| Karnofsky Performance Status (<70 vs ≥70) | 0.998 | – | – |
| Tumor size (<5 cm vs ≥5 cm) | 0.165 | – | – |
| Ki67 (<10% vs ≥10%) | <0.01 | 0.266 | 1.467 |
| MGMT (− vs +) | 0.122 | – | – |
| GST-pi (− vs +) | 0.234 | – | – |
| FOXK2 (low vs high) | <0.01 | 0.023 | 1.794 |
Abbreviations: FOXK2, Forkhead box K2; GST-pi, glutathione S-transferase pi; MGMT, O-6-methylguanine-DNA methyltransferase; WHO, World Health Organization.
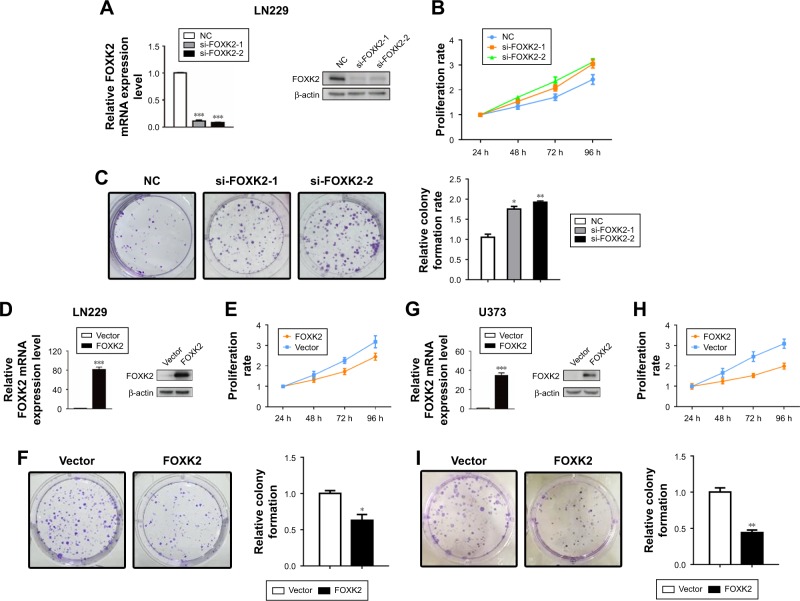
FOXK2 inhibits glioma cell proliferation
To identify the role of FOXK2, we downregulated the level of FOXK2 in the glioma cell line LN229. The transfection efficiency was validated by the Western blot analysis and real-time PCR analysis (Figure 2A). The CCK-8 assays revealed that downregulated FOXK2 expression promoted the proliferation of LN229 cells (Figure 2B). The colony-formation assay confirmed that the proliferation rate was increased by FOXK2 knockdown (Figure 2C). Then, we upregulated the level of FOXK2 in the glioma cell lines LN229 and U373. The transfection efficiency was validated by Western blot analysis and real-time PCR analysis (Figure 2D and 2G). The CCK-8 assays revealed that increased FOXK2 expression inhibited the proliferation of the glioma cells (Figure 2E and 2H). The colony-formation assay confirmed that the proliferation rate was inhibited by FOXK2 knockdown (Figure 2F and 2I).
Figure 2.
FOXK2 inhibits glioma cell proliferation. (A) The transfection efficiency of siRNA was validated by Western blot analysis and real-time PCR analysis. (B) The CCK-8 assays revealed that downregulated FOXK2 expression promoted proliferation of LN229 cells. (C) A colony-formation assay confirmed that the proliferation rate was increased by FOXK2 knockdown. (D) The transfection efficiency of the plasmid encoding the FOXK2 protein was validated by Western blot analysis and real-time PCR analysis in LN229 cells. (E) The CCK-8 assays revealed that increased FOXF2 expression inhibited proliferation of LN229 cells. (F) A colony-formation assay confirmed that the proliferation rate of LN229 cells was inhibited by FOXK2 knockdown. (G) The transfection efficiency of the plasmid encoding the FOXK2 protein was validated by Western blot analysis and real-time PCR analysis in U373 cells. (H) The CCK-8 assays revealed that increased FOXF2 expression inhibited the proliferation of U383 cells. (I) A colony-formation assay confirmed that the proliferation rate of U373 cells was inhibited by FOXK2 knockdown. *p<0.05; **p<0.01; and ***p<0.001.
Abbreviations: CCK-8, Cell Counting Kit-8; FOXK2, Forkhead box K2; NC, negative control; PCR, polymerase chain reaction; si, small interfering.
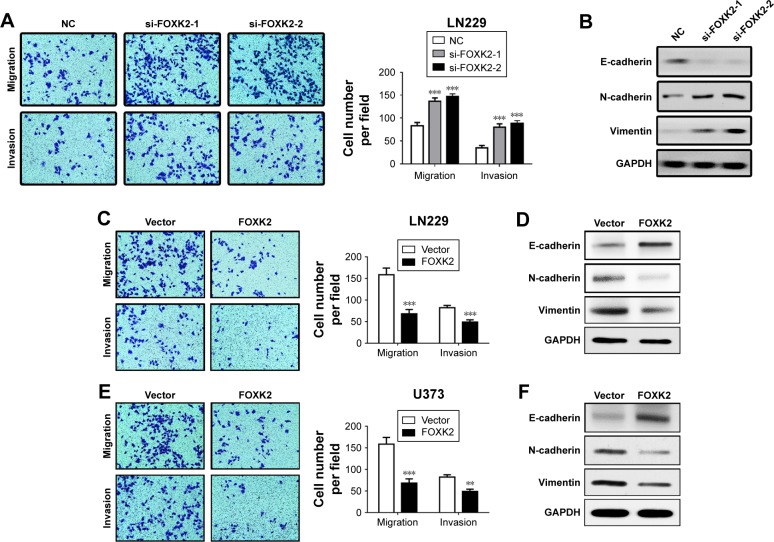
FOXK2 inhibits glioma cell migration and invasion
Downregulation of FOXK2 efficiently increased the cell migration and invasion of the LN229 cells compared to the control cells (Figure 3A). To explain the reason for the increased cell migration and invasion, we measured the EMT biomarker levels. The expression of E-cadherin was decreased by knockdown of FOXK2 in the LN229 cells, and conversely, the expression of vimentin and N-cadherin increased (Figure 3B). Upregulation of FOXK2 efficiently reduced the cell migration and invasion of the LN229 and U373 cells compared to the control cells (Figure 3C and 3E). We also measured the EMT biomarker levels. The expression of E-cadherin increased with upregulation of FOXK2 in the LN229 and U373 cells, and, conversely, the expression of vimentin and N-cadherin decreased (Figure 3D and F).
Figure 3.
FOXK2 inhibits glioma cell migration and invasion. (A) Downregulation of FOXK2 efficiently increased the cell migration and invasion of LN229 cells compared to the control cells (original magnification ×40). (B) Expression of E-cadherin was decreased by FOXK2 knockdown in LN229 cells, and conversely, expression of vimentin and N-cadherin increased. (C) Upregulation of FOXK2 efficiently reduced the cell migration and invasion of LN229 cells compared to the control cells (original magnification ×40). (D) Expression of E-cadherin was increased by upregulation of FOXK2 in LN229 cells, and conversely, expression of vimentin and N-cadherin decreased. (E) Upregulation of FOXK2 efficiently reduced the cell migration and invasion of U373 cells compared to the control cells (original magnification ×40). (F) Expression of E-cadherin was increased by upregulation of FOXK2 in U373 cells, and conversely, expression of vimentin and N-cadherin decreased. **p<0.01 and ***p<0.001.
Abbreviations: FOXK2, Forkhead box K2; NC, negative control; si, small interfering.
Discussion
Growing evidence shows that FOXK2 plays a crucial role in cancer development, which makes FOXK2 a valuable biomarker for the treatment of cancer.7–9,15 Overexpression of FOXK2 inhibits the proliferation and metastasis of breast cancer, non-small cell lung cancer, and colorectal cancer and is related to the clinical prognosis.7,8,15 Liu et al15 found that FOXK2 might act as a negative regulator of ERa and suppressed the proliferation of ERa-positive breast cancer cells, and a high expression of FOXK2 was associated with poor clinical outcomes. Chen et al8 found that Foxk2 inhibits non-small cell lung cancer cell growth by regulating cyclin D1 and CDK4, and foxk2 also affects the non-small cell lung cancer EMT process; reducing foxk2 expression is related to better OS. Qian et al7 suggested that the oncogene Sox9 activates foxk2 and participates in the pathogenesis of colorectal cancer, which may be a new target for the treatment of colorectal cancer, and the patients with a high FOXK2 expression had a poor OS. However, in hepatocellular carcinoma, FOXK2 resulted in the opposite phenotypes. Lin et al9 found that high expression of FOXK2 promoted tumor cell growth and migration via activation of the phosphoinositide 3-kinase/AKT pathway and was associated with a poor prognosis. The molecular mechanism of how FOXK2 impacts cancer cell development remains incompletely clear. Shan et al14 found that FOXK2 inhibits the proliferation and invasion of breast cancer cells and suppresses the growth and metastasis of breast cancer. However, Marais et al16 found that knocking down FOXK2 decreased BrdU incorporation and H3 phosphorylation and increased caspase 3 activity in NmuMG cells, suggesting that tumor proliferation was weakened and apoptosis was increased. The effect of FOXK2 on cell proliferation and survival is associated with Bcl-2.16 Moreover, Wang et al5 found that disheveled (DVL) proteins, a common essential component of Wnt/β-catenin, were activated by FOXK1/2 and, thus, may contribute to intestinal tumorigenesis. Therefore, the role of FOXK2 may not be the same in different tumor cell lines or under different conditions.
The forkhead family of transcription factors contains more than 100 members, and all of these contain a special DNA binding domain.17 FOXK2, as one of the members, affects a variety of gene expression through sequence-specific DNA binding. In breast cancer cells, Shan et al14 found that FOXK2 interacted with transcription corepressor complexes containing HDAC activities, such as NCoR/SMRT, SIN3A, nucleosome remodeling and deacetylase (NuRD), and REST/CoREST, to repress the amount of genes, such as hypoxia-inducible factor 1-beta (HIF1β) and enhancer of zeste 2 poly-comb repressive complex 2 subunit (EZH2), and to regulate several signaling pathways, such as hypoxia, Notch, blood vessel development, and cell metabolism. Nestal de Moraes et al6 found that FOXK2 combined with the FOXO3a promoter. In breast cancer cells, FOX3a is a crucial mediator of the cytotoxic and antiproliferative effect of chemotherapeutic agents. Ji et al12 found that FOXK2 recruited BAP1, an important tumor suppressor protein that promotes local histone deubiquitination and causes changes in target gene activity, to the target gene. Bowman et al18 found that FOXK1/2 recruited Sin3A–HDAC complexes, causing the decreased acetylation of histone H4, which decreased the expression of autophagy genes. At the present time, the mechanism of FOXK2 in different cancers is complicated, and the role of FOXK2 in gliomas has not been explored.
In our research, the level of FOXK2 was evaluated in different grades of glioma tissues. We found that expression of FOXK2 was lower in high-grade (WHO III and IV) glioma compared with low-grade (WHO I and II) glioma. The correlation analysis of the clinical data showed that FOXK2 negatively correlated with the WHO grade and Ki67 but was not correlated with MGMT and GST-pi. Ki67 is an indicator of cell proliferation in clinical work.19 MGMT and GST-pi are recognized as resistance genes of the antineoplastic alkylating agent.20–23 Therefore, our study showed that FOXK2 may promote the proliferation of glioma but is not involved in the drug resistance mechanisms of glioma. The Kaplan–Meier analysis and Cox regression analyses revealed that the high level of FOXK2 means a better prognosis, and FOXK2 may serve as a potential therapeutic target in glioma and as a prognostic marker for glioma patients.
We decreased the expression of FOXK2 in LN229 cells using siRNA. We found that the proliferation, migration, and invasion of the cells were inhibited via the CCK-8 experiments, plate cloning experiments, and transwell experiments. Then, we upregulated the level of FOXK2 in the glioma cell lines LN229 and U373. We found that the proliferation, migration, and invasion of cells were enhanced via the CCK-8 experiments, plate cloning experiments, and transwell experiments. These experiments proved that FOXK2 promoted cell proliferation, migration, and invasion.
Recently, EMT in glioma has attracted much attention. Glioma, like other types of cancer, has a high plasticity, includ ing changes in epithelial and mesenchymal phenotypes.24 Some researchers propose the term “glial-to-mesenchymal transition (GMT)” or an EMT-like process.25 EMT is a critical step in cancer metastasis. During the EMT process, epithelial cells obtain the characteristics of mesenchymal phenotypes and gain the ability to migrate and contribute to tumor metastasis. The major changes in the EMT process are the continuous decreased E-cadherin expression level and the increased N-cadherin expression level.26 In our study, we found that FOXK2 inhibited the process of EMT in glioma, which increased invasion and migration. FOXK2 is a critical mediator of the EMT process.
Conclusion
In summary, our study suggests that FOXK2 may play an important role in glioma. FOXK2 inhibited glioma cell proliferation, migration, and invasion. FOXK2 was a critical mediator of the EMT process. However, the exact molecular mechanisms by which FOXK2 inhibits glioma cell development require further study.
Supplementary material
Quantification of immunohistochemistry staining of Forkhead box K2 (FOXK2). Note: Original magnification ×200.
Acknowledgments
We thank all surgeons from the Neurosurgery Department of Tianjin Huanhu Hospital for their help in gathering the tumor biopsies. This work was supported by the Chinese National Natural Science Foundation (grant number 81671380), Key Project of Tianjin National Natural Science Foundation (grant number 17JCZDJC35900), The Basic and Advanced Technology Research Foundation (grant number 152300410162), and the Science and Technology Development Foundation (grant number 172102310103) from the Science and Technology Department of Henan Province.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Weller M, Wick W, Aldape K, et al. Glioma. Nature Reviews Disease Primers. 2015;1:15017. doi: 10.1038/nrdp.2015.17. [DOI] [PubMed] [Google Scholar]
- 2.Fujii Y, Nakamura M. FOXK2 transcription factor is a novel G/T-mismatch DNA binding protein. J Biochem. 2010;147(5):705–709. doi: 10.1093/jb/mvq004. [DOI] [PubMed] [Google Scholar]
- 3.Katoh M, Katoh M. Human FOX gene family (review) Int J Oncol. 2004;25(5):1495–1500. [PubMed] [Google Scholar]
- 4.van der Heide LP, Wijchers PJ, von Oerthel L, Burbach JP, Hoekman MF, Smidt MP. FoxK2 is required for cellular proliferation and survival. J Cell Physiol. 2015;230(5):1013–1023. doi: 10.1002/jcp.24828. [DOI] [PubMed] [Google Scholar]
- 5.Wang W, Li X, Lee M, et al. FOXKs promote Wnt/β-catenin signaling by translocating DVL into the nucleus. Dev Cell. 2015;32(6):707–718. doi: 10.1016/j.devcel.2015.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nestal de Moraes G, Khongkow P, Gong C, et al. Forkhead box K2 modulates epirubicin and paclitaxel sensitivity through FOXO3a in breast cancer. Oncogenesis. 2015;4:e167. doi: 10.1038/oncsis.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qian Y, Xia S, Feng Z. Sox9 mediated transcriptional activation of FOXK2 is critical for colorectal cancer cells proliferation. Biochem Biophys Res Commun. 2017;483(1):475–481. doi: 10.1016/j.bbrc.2016.12.119. [DOI] [PubMed] [Google Scholar]
- 8.Chen S, Jiang S, Hu F, Xu Y, Wang T, Mei Q. Foxk2 inhibits non-small cell lung cancer epithelial-mesenchymal transition and proliferation through the repression of different key target genes. Oncol Rep. 2017;37(4):2335–2347. doi: 10.3892/or.2017.5461. [DOI] [PubMed] [Google Scholar]
- 9.Lin MF, Yang YF, Peng ZP, et al. FOXK2, regulted by miR-1271-5p, promotes cell growth and indicates unfavorable prognosis in hepatocellular carcinoma. Int J Biochem Cell Biol. 2017;88:155–161. doi: 10.1016/j.biocel.2017.05.019. [DOI] [PubMed] [Google Scholar]
- 10.Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer. 2013;13(7):482–495. doi: 10.1038/nrc3539. [DOI] [PubMed] [Google Scholar]
- 11.Ji Z, Donaldson IJ, Liu J, Hayes A, Zeef LA, Sharrocks AD. The forkhead transcription factor FOXK2 promotes AP-1-mediated transcriptional regulation. Mol Cell Biol. 2012;32(2):385–398. doi: 10.1128/MCB.05504-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji Z, Mohammed H, Webber A, et al. The forkhead transcription factor FOXK2 acts as a chromatin targeting factor for the BAP1-containing histone deubiquitinase complex. Nucleic Acids Res. 2014;42(10):6232–6242. doi: 10.1093/nar/gku274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okino Y, Machida Y, Frankland-Searby S, Machida YJ. BRCA1-associated protein 1 (BAP1) deubiquitinase antagonizes the ubiquitin-mediated activation of FoxK2 target genes. J Biol Chem. 2015;290(3):1580–1591. doi: 10.1074/jbc.M114.609834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shan L, Zhou X, Liu X, et al. FOXK2 Elicits massive transcription repression and suppresses the hypoxic response and breast cancer carcinogenesis. Cancer Cell. 2016;30(5):708–722. doi: 10.1016/j.ccell.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Ao X, Jia Z, et al. FOXK2 transcription factor suppresses ERα-positive breast cancer cell growth through down-regulating the stability of ERα via mechanism involving BRCA1/BARD1. Sci Rep. 2015;5:8796. doi: 10.1038/srep08796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marais A, Ji Z, Child ES, Krause E, Mann DJ, Sharrocks AD. Cell cycle-dependent regulation of the forkhead transcription factor FOXK2 by CDK·cyclin complexes. J Biol Chem. 2010;285(46):35728–35739. doi: 10.1074/jbc.M110.154005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7(11):847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 18.Bowman CJ, Ayer DE, Dynlacht BD. Foxk proteins repress the initiation of starvation-induced atrophy and autophagy programs. Nat Cell Biol. 2014;16(12):1202–1214. doi: 10.1038/ncb3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulus W. GFAP, Ki67 and IDH1: perhaps the golden triad of glioma immunohistochemistry. Acta Neuropathol. 2009;118(5):603–604. doi: 10.1007/s00401-009-0600-6. [DOI] [PubMed] [Google Scholar]
- 20.Calatozzolo C, Gelati M, Ciusani E, et al. Expression of drug resistance proteins Pgp, MRP1, MRP3, MRP5 and GST-pi in human glioma. J Neurooncol. 2005;74(2):113–121. doi: 10.1007/s11060-004-6152-7. [DOI] [PubMed] [Google Scholar]
- 21.von Deimling A, Korshunov A, Hartmann C. The next generation of glioma biomarkers: MGMT methylation, BRAF fusions and IDH1 mutations. Brain Pathol. 2011;21(1):74–87. doi: 10.1111/j.1750-3639.2010.00454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song T, Li H, Tian Z, Xu C, Liu J, Guo Y. Disruption of NF-κB signaling by fluoxetine attenuates MGMT expression in glioma cells. Onco Targets Ther. 2015;8:2199–2208. doi: 10.2147/OTT.S85948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scaringi C, Enrici RM, Minniti G. Combining molecular targeted agents with radiation therapy for malignant gliomas. Onco Targets Ther. 2013;6:1079–1095. doi: 10.2147/OTT.S48224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iser IC, Pereira MB, Lenz G, Wink MR. The epithelial-to-mesenchymal transition-like process in glioblastoma: an updated systematic review and in silico investigation. Medicinal Research Reviews. 2017;37(2):271–313. doi: 10.1002/med.21408. [DOI] [PubMed] [Google Scholar]
- 25.Mahabir R, Tanino M, Elmansuri A, et al. Sustained elevation of Snail promotes glial-mesenchymal transition after irradiation in malignant glioma. Neuro Oncol. 2014;16(5):671–685. doi: 10.1093/neuonc/not239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skrypek N, Goossens S, De Smedt E, Vandamme N, Berx G. Epithelial-to-mesenchymal transition: epigenetic reprogramming driving cellular plasticity. Trends Genet. 2017;33(12):943–959. doi: 10.1016/j.tig.2017.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantification of immunohistochemistry staining of Forkhead box K2 (FOXK2). Note: Original magnification ×200.