Abstract
Pluripotent cells, such as embryonic stem cells (ESCs), divide indefinitely and can differentiate to form mineralised nodules in response to osteogenic supplements. This suggests that they may be used as a cell source for bone replacement strategies. Here, we related the expression of osteogenic and chondrogenic genes in cultures of murine ESCs, marrow stromal cells (MSCs) and calvarial osteoblasts (OBs) differentiating under osteogenic conditions to the biochemical composition and quantity of mineral formed. Mineralisation, measured by calcium sequestration, was >2-fold greater in ESC cultures than in either MSCs or OBs. Micro-Raman spectroscopy and spectral mapping revealed a lower mineral-to-matrix ratio and confirmed a more diffuse pattern of mineralisation in ESCs compared to MSCs and OBs. Baseline expression of chondrogenic and osteogenic genes was between 1 and 4 orders of magnitude greater in MSCs and OBs than in ESCs. Osteogenic differentiation in MSCs and OBs was accompanied by increases in osteogenic gene expression by factors of ~100 compared to only ~10 in ESCs. Consequentially, peak expression of osteogenic and chondrogenic genes was greater in MSCs and OBs than ESCs by factors of 100 - 1000, despite the fact that mineralisation was more extensive in ESCs than either MSCs or OBs. We conclude that the mineralised material observed in cultures of murine ESCs during osteogenic differentiation may accumulate non-specifically, and that thorough characterisation of the tissue formed by ESCs must be achieved before these cells can be considered as a cell source for clinical applications.
Introduction
Bone or joint injury and disease often results in such a severe loss of tissue that restoration of function can only be achieved by its replacement with either transplanted material or a biomaterial prosthesis. The implanted material should provide an environment containing appropriate mechanical and molecular signals conducive to bone formation through the action of endogenous or implanted cells (Giannoudis et al. 2007). Autografting is the preferred, ‘gold standard’ treatment for bone injuries, but the amount of tissue that can be harvested may be limited, and pain and morbidity may result at the donor site (Arrington et al., 1996), problems that are compounded in elderly patients (Giannoudis et al., 2005). Allografts may also be used, but these cadaveric tissues may be of poor quality and have the inherent risk of patient-to-patient disease transmission (Kainer et al., 2004). Many artificial or sterilised tissue-derived biomaterials, often based on recombinant bone morphogenic proteins (BMPs) are used as alternatives to allografts (Place et al., 2009), but these are generally inferior in their ability to promote tissue regeneration, or in some cases may induce aberrant healing and heterotopic bone (Boraiah et al., 2009). To circumvent some of these problems, cells – expanded in vitro and implanted possibly in combination with a physical scaffold – are being considered as an alternative therapy.
There are several candidates as cell sources for bone ‘tissue engineering’. Marrow stromal cells (MSC), discovered in the 1960s (Friedenstein et al., 1970), have a high proliferative potential and contribute to healing bone in animal models (Bruder et al., 1998). It is considered that stem cells, resident in these mixed cell populations, are responsible for both the extended proliferation and multilineage capacity of these cultures. But MSCs do not divide indefinitely. This may limit their use in bone tissue engineering, where large, voluminous constructs may be required to achieve repair. Pluripotent cells, on the other hand, including embryonic stem cells (ESCs) (Evans and Kaufman, 1981; Thomson et al., 1998) and induced pluripotent stem cells (iPSCs) (Takahashi and Yamanaka, 2006), have the ability to both divide indefinitely and differentiate into all cell types found in the adult organism. Several studies have reported osteogenic differentiation of ESCs when cultured with supplements commonly used to induce mineralised nodule formation in primary osteoblast (OB) cultures (Bielby et al., 2004; Buttery et al., 2001; Kawaguchi et al., 2005; Phillips et al., 2001; zur Nieden et al., 2005). However, we recently noted marked differences in the composition of the mineral and the ultrastructure of the nodules formed from ESCs compared to those formed from either MSCs or OBs (Gentleman et al., 2009). This indicates that different mechanisms of differentiation and mineralisation may operate in ESCs compared to the latter cell types. Here, to better characterise the pattern of differentiation in ESCs compared to MSCs or OBs, we expand upon our previous study by directly comparing gene transcription during osteogenic differentiation in the three cell types, and relating them to the morphology and mineral content of nodules. To achieve this, we employed a combination of histological staining and biochemical assay techniques with micro-Raman spectroscopy and quantitative polymerase chain reaction (qPCR). Since it is now established that experimental conditions, cell type or differentiation state (Vandesompele et al., 2002) can affect the expression of ‘housekeeping’ genes (used as normalisers in qPCR for genes of interest), we first evaluated the expression of three potential housekeeping genes in order to establish the most suitable reference gene for comparisons between cell types and experimental conditions. We then tested the hypothesis that, while there are similarities in the material composition of the mineralised tissue that these cells form, the molecular mechanism by which differentiation occurs differs markedly between them.
Methods
Ethics statement
Animals were and sacrificed humanely under Schedule 1 of the Animal Procedures Committee of the United Kingdom Home Office. These procedures do not require specific approval in the UK, but were conducted in accordance with Imperial College’s ethics guidelines under licence PCD 70/2707.
Cell isolation and culture
E14 TG2α ESCs were cultured on gelatin-coated surfaces (0.1% [w/v] in phosphate buffered saline [PBS] for 24 hours prior to culture) in DMEM supplemented with 10% (v/v) fetal bovine serum (FBS), 2 mM L-glutamine, 50 U/ml penicillin, 50 mg/ml streptomycin (Invitrogen, Paisley, UK), 100 μM β-mercaptoethanol (Sigma, Poole, UK), and leukaemia inhibitory factor (LIF) at 1000 U/ml (Chemicon, Chandler’s Ford, UK). Cells were seeded at 6000 – 8000/cm2 and passaged every 3-4 days. MSCs were isolated from the femora of female FVB/N mice by removing the epiphyses and passing growth medium (αMEM supplemented with 15% (v/v) FBS, 50 U/ml penicillin, and 50 mg/ml streptomycin) several times through the marrow cavity of the diaphyses. Marrow cells were collected in tissue culture flasks and allowed to attach for 3 days. Adherent cells were then passaged at 90% confluence and fed every 2 – 3 days with growth medium before use in differentiation experiments. OBs were isolated from the calvaria of neonate CD-1 mice. Soft tissue was removed from the bone and the bone was then rinsed in 500 U/ml penicillin, and 500 mg/ml streptomycin in PBS for 5 minutes. Calvaria were then transferred to a solution containing 0.05% (w/v) trypsin and 0.1% (w/v) collagenase P and were minced finely with dissecting scissors. Bone fragments were subjected to five 15-minute digestions; at the end of each digestion the supernatant was removed, centrifuged, resuspended in growth medium and plated in a 25cm2 flask, while fresh trypsin-collagenase solution was added to the fragments. Cells from digestions III, IV and V were pooled, passaged at confluence and fed every 2 – 3 days with growth medium before use in differentiation experiments. MSCs and OBs were used at passage 1 or 2 in all experiments, and experiments described were conducted on cells obtained from 3 separate cell isolations (for both MSCs and OBs). FBS in all experiments was batch-tested for its ability to support bone nodule formation in differentiating cultures of MC3T3-E1 osteoblasts (FBS reserve number 1222532, batch 07F7260K, Invitrogen, UK).
Differentiation of cells
To induce embryoid body (EB) formation, ESCs were dissociated into clumps of around 20 cells by partial trypsin digestion and transferred to bacteriological-grade 90 mm Petri dishes. EBs were cultured in suspension for 5 days in αMEM supplemented with 15% (v/v) FBS, 50 U/ml penicillin, and 50 mg/ml streptomycin with feeding after 3 days. After five days (referred to in differentiation experiments as d0) EBs were dissociated into single cells with trypsin-EDTA (Invitrogen, Paisley, UK) and seeded at a density of 30 000 cells/cm2 in αMEM supplemented with 10% (v/v) FBS, 50 U/ml penicillin, and 50 mg/ml streptomycin, 280 μM ascorbate, 10 mM β-glycerophosphate and 1 μM dexamethasone (differentiation medium) and were then fed every 2-3 days for a period of up to 28 days. Both MSCs and OBs were seeded at 30,000 cells/cm2 in the same medium and cultured for the same period of time.
Alizarin red S staining and mineral quantification
At d21, cells cultures were washed twice in PBS and fixed for 20 minutes in 10% (v/v) formalin buffered saline. Cells were then washed and incubated in 2% (w/v) alizarin red S (ARS; Sigma, Poole, UK), pH 4.2 for 10 minutes. Cultures were washed in running tap water for 30 minutes, and were then left to air dry. Cells were examined under an inverted epifluorescence microscope at wavelengths of 510-560 nm excitation and >590 nm emission. Calcium content was quantified as previously described (McCullen et al.). After 21 days in culture, cultures were washed with PBS and digested in 0.5M HCl at 4 °C overnight. Calcium content was assessed with a Calcium Colorimetric Assay Kit (BioVision Research Products, Mountain View, CA, USA) based on the formation of a chromogenic complex between 0-cresolphthalein and calcium ions, and quantified by measuring absorbance at 575 nm on a colorimetric plate reader (Molecular Devices, Wokingham, UK).
Raman spectroscopy
Raman spectra of live cell cultures and bone samples were collected in DMEM at 37°C as previously described (Gentleman et al., 2009) with a Renishaw InVia spectrometer connected to a Leica microscope and equipped with a 785 nm line-focus laser. Spectra were recorded at a resolution of ~1-2 cm-1, with 100 s integration time. For mineralised nodules, spectra of live cultures were collected along a line through the centre of the nodule at equally-spaced intervals. Some nodules were also rinsed in PBS and dried in a bell jar desiccator prior to Raman spectroscopic analysis to create spectral maps. These spectra were collected using a Renishaw InVia spectrometer with a 532 nm line-focus doubled Nd:YAG laser connected to a Leica confocal microscope with a motorized stage. An 1800 line/mm grating was used in scanning mode (10s/scan) to collect spectra (350 to 3200 Raman shift cm-1) with ~1 cm-1 resolution. One-second static scans were also completed for each map position with a limited spectral range (900 cm-1 to 1700cm-1) to confirm that no laser damage was observed during the 10s extended scans. For each nodule, spectra were collected in ~40 μm x 50 μm sections with 1 μm raster steps between measurements. Prior to each measurement the system was calibrated for position and intensity using and internal silicon standard. For live cell cultures, Raman spectra were processed and analysed with software developed in-house for use in the Matlab (The Mathworks, Cambridge, UK). Raman spectra were intensity-corrected for instrument response, and the background subtracted using the Modpoly algorithm (5th order polynomial, 1000 iterations) (Lieber and Mahadevan-Jansen, 2003). Individual spectra were lightly smoothed using a 5-point Savitsky-Golay filter (2nd order polynomial) (Savitsky and Golay, 1964), and the wavenumber axis of each spectrum was standardised and aligned to the sharp phenylalanine peak at 1003 cm−1. Prior to analysis, a linear baseline was subtracted from each band envelope. For Raman spectral maps of dried samples, no pre-processing was conducted on the spectra prior to curve fitting. All curve-fitting was completed using Renishaw’s Wire software. Phosphate mineral content was estimated by fitting one (for maps) or two bands to the PO43- ν1stretch at approximately 960 cm-1. To estimate matrix content, two curves were fit to the baseline-corrected 1600–1700 cm−1 spectral region to model the amide I band at ~1650 cm−1, and the C=C stretch of amino acids tyrosine and tryptophan at ~1620 cm−1. Mineral to matrix ratios were computed by dividing the PO43- ν1 band area by the matrix band area. To generate false colour mapping plots of dried samples, the mineral to matrix ratio was calculated at each point in the array. These values were then scaled for each map to represent the intensity range of 5-95% of the maximum signal intensity. A linear interpolation routine was used to smooth the map intensity profile. For mineral to matrix ratio measurements, 80, 53 and 87 Raman spectra from 11, 9, and 13 nodules for ESC, MSC and OB, respectively (obtained from 2 separate cell isolations in the case of MSCs and OBs) were analysed; 44 spectra from 3 native bone samples were also examined.
Gene expression
At indicated time points, cultures of ESCs, MSCs and OBs were collected, pelleted in microcentrifuge tubes and snap frozen in liquid N2. RNA was extracted using Qiashredder and RNeasy kits (Qiagen, Germany). Sensimix OneStep kit (Quantace, London, UK) was used to synthesize cDNA and to amplify cDNA by PCR in a single step, using SYBR green to detect increases in amplicon concentration. Thermal cycling and SYBR green fluorescence detection were performed using a Corbett Rotorgene (Qiagen, Germany). All primers were designed using Primer Bank free software (http://pga.mgh.harvard.edu/primerbank/). Sequences and cycling conditions of all primers are listed in Supplementary Table 1. For comparison of the expression of housekeeping genes (Gapdh, B2m and Actb) equal masses of RNA, isolated from each cell type, were used in all reaction tubes. Expression of a gene of interest in one selected reference sample (one of the ESC samples; R) was compared to the expression of the same gene in a sample of interest (I) by the equation:
where RE is the relative expression of the gene of interest in I compared to R; E is the efficiency of the primer pair; Ct[I] is the PCR cycle number at which the fluorescent signal reaches a given threshold for a sample of interest; and Ct[R] is the PCR cycle number at which the fluorescent signal reaches a given threshold for the reference sample. Relative gene expression with respect to cell type or time was measured using the Pfaffl method (Pfaffl, 2001) using the equation:
where RE is the expression of a gene at a timepoint of interest (dx) relative to expression at day 0 (d0). EGOI is the efficiency of the primer pair of the gene of interest, ENOR is the efficiency of the normalising gene primer pair, and Ct[dx] and Ct [d0] are the PCR cycle numbers where the fluorescent signal of either the gene of interest or the normalising gene reaches a given intensity threshold. All PCR reactions were performed in duplicate. Efficiencies of all primer pairs were calculated by serial dilutions of either RNA isolated from ESCs (for Nanog and Oct4) or from OBs (for all other genes).
Statistics
Statistical analyses of calcium quantification and real-time RT-PCR data were performed using ANOVA with a post-hoc Tukey test. A Kruskal-Wallis non-parametric ANOVA was used to determine significance (p < 0.05) between mineral to matrix ratios. A Mann-Whitney test with Bonferroni correction was used to test for statistical significance between individual groups. The slopes and correlation coefficients of linear regressions calculated for Gapdh expression as a function of time in culture were compared by calculating the t-statistic and by using Fisher’s method, respectively. Replicate numbers are stated for each experiment in the figure legends and in the Results. Significance was noted at p < 0.05.
Results
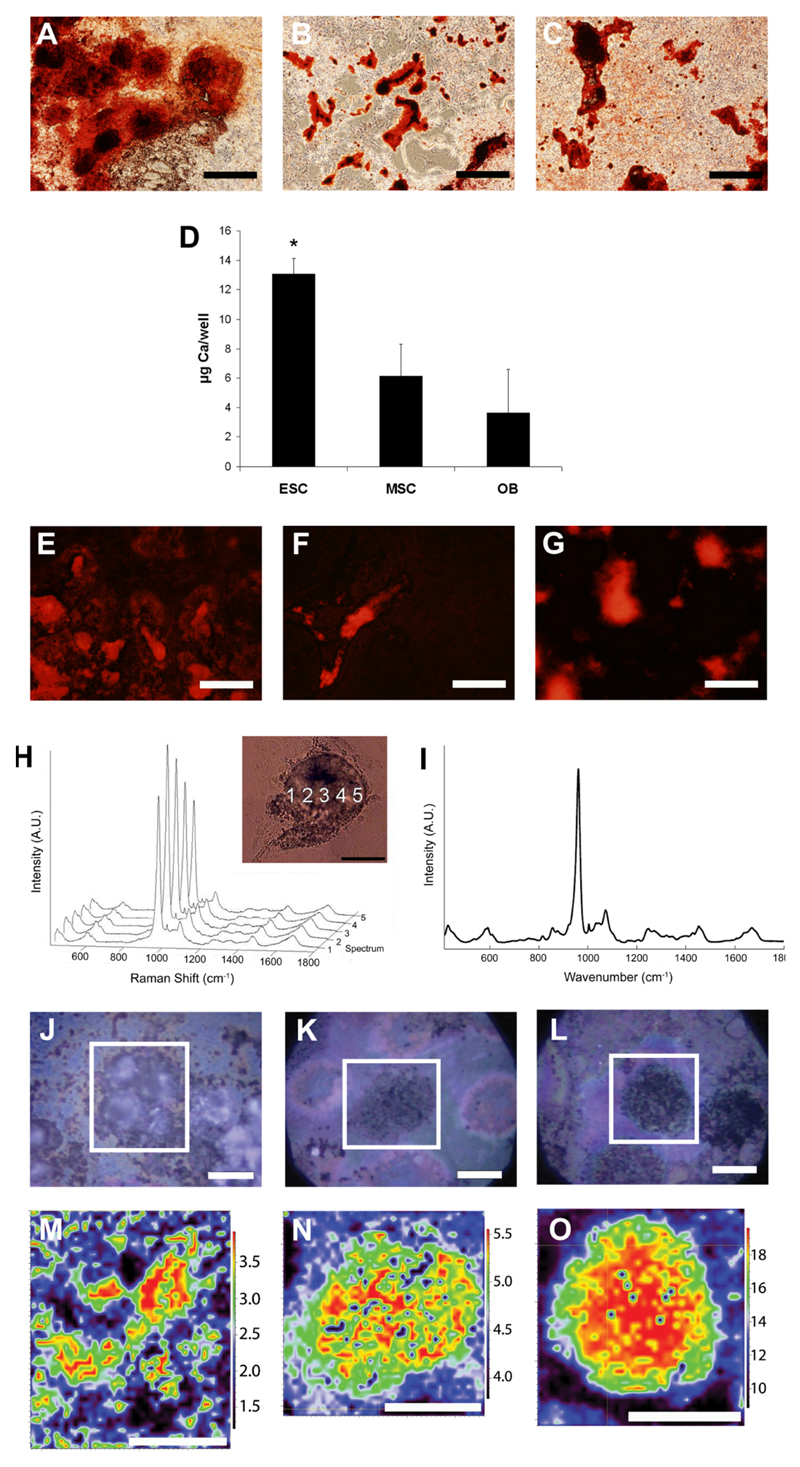
After 21 days, ESCs, OBs and MSCs all formed mineralised nodules that stained positively with the calcium stain ARS. ESC cultures displayed more extensive staining than OB and MSC cultures, however, covering almost the entire culture surface (Figure 1A, B, and C, respectively). A calcium quantification assay confirmed that ESC cultures accumulated significantly more calcium per well than either MSC or OB after 21 days (Figure 1D). We also noticed differences in the staining pattern of ESC nodules as compared to OB and MSC nodules. While nodules formed from all three cell types fluoresced when stained with ARS (Figure 1E, F, G), in ESCs the areas stained with ARS had poorly defined boundaries while, in contrast, the boundaries of mineralised nodules formed from both MSCs and OBs were clearly defined, with a sudden decrease in ARS fluorescence intensity at the nodule edges.
Figure 1. Histological staining and spectroscopic analyses of mineralised nodules formed from ESC, MSC and OB.
Alizarin Red S (ARS) staining of mineralised nodules formed in cultures of (A) ESC, (B) MSC and (C) OB. Notice the extensive staining in ESC compared to MSC and OB. Scale bar = 500 μm. (D) Total calcium per well in cultures of ESC, MSC and OB expressed as μg Ca/well (means ± standard deviation) * indicates ESC cultures contained significantly (p < 0.05) more Ca per well than either MSC or OB. Fluorescence micrographs of typical mineralised nodules formed from (E) ESC, (F) MSC and (G) OB stained with ARS (510-560 nm excitation, >590 nm emission). Scale bar = 500 μm. Note that while OB and MSC nodules fluoresce in discrete areas, the fluorescence in ESC is more diffuse. (H) Representative Raman spectra of a line scan of a typical live mineralised nodule formed from MSC. Inset shows the position at which each of the spectra was collected. (I) Raman spectrum of a fresh bone sample harvested from an adult mouse. False-colour plots of Raman maps created from spectra of dried mineralised nodules formed from ESC (J,M), MSC (K,N) and OB (L,O) with their respective images of individual nodules. Scale bar in all images is 20 μm. False colour images were created by collecting Raman spectra then taking the ratio of the PO43- ν1 area to the area of the Amide I peak and scaling each map to represent the intensity range of 5-95% of the maximum signal intensity. Colours represent the relative amount of mineral at individual points in nodules. Note that ESC nodules lack discrete boundaries.
To better understand these differences in staining patterns, we examined the live cultures with micro-Raman spectroscopy. Raman spectra collected in a line across a typical live mineralised MSC nodule (Figure 1H) were notable for peaks with marked similarities to spectra of native bone (Figure 1I) including strong peaks near 960 cm-1 (resulting from symmetric stretch of the PO43- ν1), 1070 cm-1 (resulting from in-plane vibrations of substituted CO32- ν1), and 1,595–1,720 cm-1 and 1,243–1,269 cm-1 (corresponding to Amide I and III). Spectral maps of dried nodules formed from ESC (Figure 1J,M), OB (K,N) and MSC (L,O), which represent the ratio of the mineral content to the protein content in false colour, revealed discrete mineralisation boundaries in OB and MSC nodules, while ESC nodule boundaries were more diffuse.
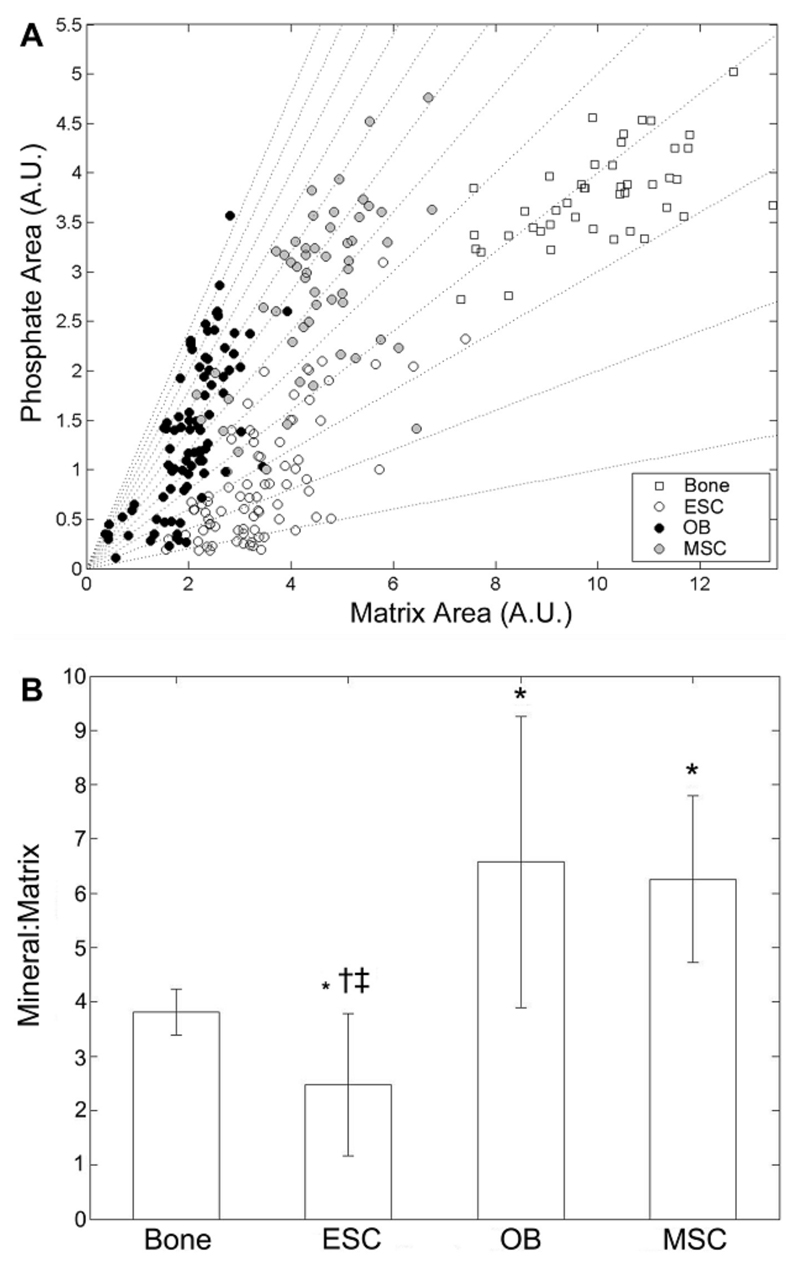
In addition to differences in the distribution of mineral in ESCs compared to MSCs or OBs, we also found that the mineral-to-matrix ratio, an average measure of the relative amount of phosphate mineral present in nodules as compared to the amount of protein, was significantly lower in ESC than in either OB, MSC or native bone samples (Figure 2). We found a broad range of mineral peak areas for given matrix areas (Figure 2A, which depicts the scatter of the mineral and matrix peak areas for ESC, OB and MSC nodules and native bone), but the trend was for increasing mineral content with increasing matrix content for all samples. ESC had the lowest phosphate peak area among all the groups, which when combined with their intermediate matrix peak area yielded the lowest mineral to matrix ratios (Figure 2B). Conversely, while OB and MSC nodules had similar matrix levels to ESC, they had higher mineral peak areas, yielding significantly higher mineral to matrix ratios. Bone samples yielded the highest mineral and matrix peak areas and therefore their spectra clustered in the upper right portion of the scatter plot, with a mineral to matrix ratio significantly greater than ESC nodules.
Figure 2. Univariate peak analysis of mineral-to-matrix ratios in ESC, MSC and OB nodules, and native bone.
Mineral-to-matrix was calculated by measuring the area of the PO43- ν1 peak divided by the area of the Amide I band. In (A) the data scatter is plotted with dotted lines representing constant mineral to matrix ratios. (B) Mean mineral to matrix ratio (± standard deviation). * represents a statistically significant difference between the given sample and bone. † significantly different from OB; ‡ significantly different from MSC.
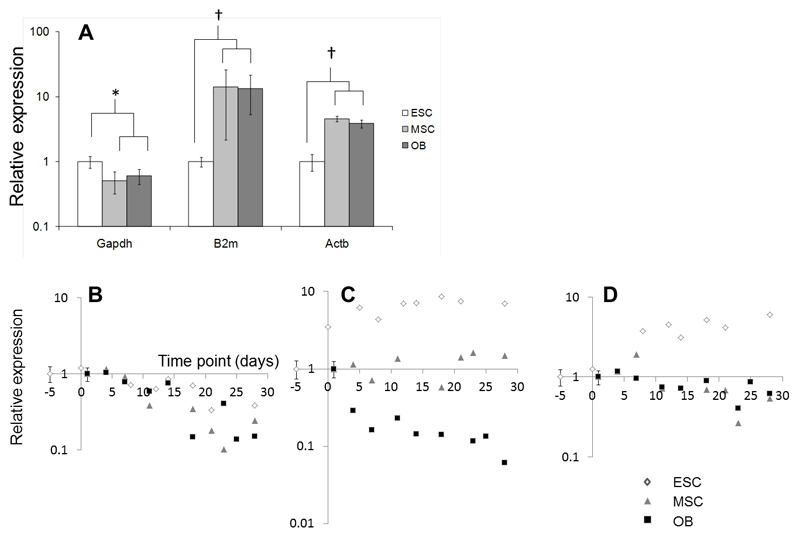
To attempt to explain the differences in ARS uptake and Raman spectra of bone nodules, we next tested for differences in the molecular mechanisms of osteogenic differentiation by comparing and quantifying gene expression in ESCs, MSCs and OBs. To compare gene expression between cell types, the selection of a normalising housekeeper gene that is expressed similarly both in different cell types and under different conditions is critical. We compared three commonly used housekeeping genes: Gapdh (glyceraldehyde-3-phosphate dehydrogenase; an enzyme necessary for glycolysis), B2m (β2-microglobulin; a component of the major histocompatibility complex (MHC) present on all nucleated cells) and Actb (β-actin; a component of the cytoskeleton) (Figure 3). At day 0 (i.e. when comparing undifferentiated ESCs, MSCs and OBs) Gapdh expression was significantly lower in both MSCs and OBs than in ESCs (p < 0.05), while both B2m and Actb expression was significantly higher (p < 0.01) (Figure 3A). There was no significant difference in the expression of Gapdh when comparing MSCs and OBs with d5 embryoid bodies (corresponding to d0 in our ESC osteogenic differentiation experiments; Supplementary Figure 1A)). During the course of a differentiation experiment we could not detect differences in the expression of Gapdh between ESCs, MSCs and OBs, with no significant differences in either the slopes of linear regressions (p > 0.1 for all comparisons) calculated for the expression data as a function of time in culture, or their correlation coefficients (p > 0.5) (Figure 3B). This was also true when Gapdh expression was normalised to that in EBs (d0 in ESC osteogenic differentiation experiments; Supplementary Figure 1B). In contrast, both B2m and Actb increased in ESCs over the course of the differentiation experiment by factors of 6 – 10 fold, while declining or remaining unchanged in both MSCs and OBs (Figure 3C and 3D). As Gapdh expression did not differ between cell types during osteogenic differentiation, we chose this gene as a normaliser to compare genes of interest between cell types.
Figure 3. Comparison of housekeeping gene expression in cultures of ESCs, MSCs and OBs.
In undifferentiated cells, Gapdh was expressed at levels significantly lower in both MSCs and OBs than in ESCs, while both B2m and Actb were expressed at significantly higher levels in MSCs and OBs than in ESCs (A; * indicates p < 0.05, † indicates p < 0.01; n = 5). During a time course of differentiation, Gapdh declined in all cells at a similar rate (B). B2m (C) and Actb (D) increased markedly in cultures of ESCs over a time period of 28 days. In OBs, Actb declined during the same time period, while B2m declined in OBs, but remained constant in MSCs.
We next investigated compared the expression of genes expected to change during osteogenic differentiation of the three cell types (Nanog, Oct4, Sox9, Col2a1, Bglap, Sparc, Akp2, Spp1, Runx2 and Col1a1; see Supplementary Table 1 for explanation of gene functions), by comparing expression in undifferentiated ESCs, cells derived from d5 EBs (corresponding to ‘d0’ in our differentiation experiments for ESCs), undifferentiated MSCs, and undifferentiated OBs. In all cases Gapdh was chosen as the normalising gene due to the results described above. As might be expected, at baseline timepoints (d-5 [undifferentiated cells] or d0 [embryoid bodies] in ESCs or at d1 in MSCs or OBs), markers of osteogenic and chondrogenic differentiation were substantially and significantly greater in MSCs and OBs than in ESCs (Sox9, Col2a1, Bgalp1, Spp1, Akp2, Sparc, Runx2, Col1a1), while the opposite was true of genes such as Nanog and Oct4 (Supplementary Figure 2). We obtained similar results by normalising gene expression to the amount of input RNA in our PCR reactions (Supplementary Table 2 and Supplementary Figure 3). All genes remained detectable in all cell types (See Supplementary Table 2 for details of Ct values for each gene in each cell type).
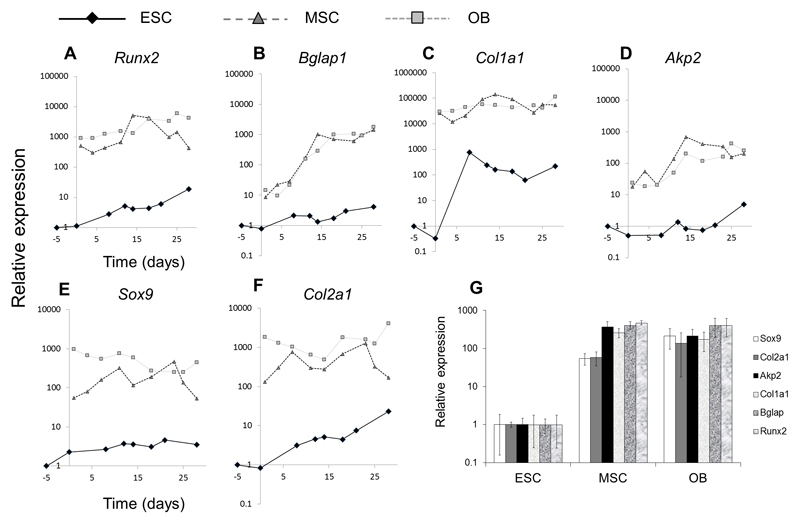
Having established baseline comparisons of gene expression between undifferentiated cell types, we next investigated the expression of selected chondrogenic and osteogenic genes during the course of osteogenic differentiation in ESCs, OBs and MSCs. To directly and quantitatively compare gene expression between cell types during the time-course of differentiation, we normalised expression to the baseline comparisons obtained in our experiments above. Figure 4 shows that during osteogenic differentiation of all cell types, expression of osteogenic and chondrogenic genes was always greater in both MSCs than in OBs relative to either MSCs or OBs (despite the comparative amounts of mineral that formed in ESCs, MSCs and OBs, as shown in Figure 1). Runx2 and Bgalp1 expression increased modestly in ESCs during osteogenic differentiation (by factors of ~5-10; Figure 4A and B) but remained between 100-1000 lower than in MSCs or OBs even at peak expression levels (Figure 4G). In addition, Bglap1 expression increased in both MSCs and OBs by a factor of ~100 during a time-course of differentiation, while increasing by a factor of only ~4 in ESCs. The expression of type I collagen increased by ~100-1000 times in ESCs during a time course of differentiation, while remaining relatively unchanged in MSCs and OBs (Figure 4C). But due to the significantly higher baseline expression of this gene in the latter cell types, Col1a1 was expressed ~100-1000 times lower in ESCs than MSCs or OBs at peak expression levels (Figure 4G). Akp2 expression increased by a factor of ~10 in MSCs and OBs while remaining relatively unchanged in ESCs (Figure 4D), with a peak expression ~100 times greater in MSCs or OBs than in ESCs. Similar to osteogenic genes, the expression of chondrogenic genes Sox9 and Col2a1 increased modestly in ESCs during differentiation, while fluctuating or showing biphasic patterns of expression in either MSCs or OBs (Figure 4E and F). Again, due to significantly higher baseline expression of Sox9 and Col2a1 in the latter cell types, peak expression of these genes remained ~100 higher than in ESCs (Figure 4G).
Figure 4. Time course expression of osteogenic (A-D) or chondrogenic (E,F) or genes in cultures of ESCs, MSCs or OBs respectively and peak expression comparison of genes (G).
For each gene, values are given as the relative expression levels for each of the three cell types at baseline timepoints (i.e. in undifferentiated cells - d1 in MSCs and OBs and d-5 in ESCs), thus charts A-F compare the expression of each gene between cell types. Time course plots are from a single differentiation experiment and are representative of data from two separate cell isolations. G compares the peak expression of selected genes normalised to ESCs (by taking the means of the three highest data points in each time-course) illustrating a significantly lower expression in ESCs – by factors of ~100 to ~1000 - of all genes compared to either MSCs or OBs (results shown as + standard deviation; n = 3).
Discussion
Stem cells, such as ESCs and MSCs, are considered as potential sources of cells for use in regenerative medicine and tissue engineering strategies for the skeleton. Before they can be used for these purposes, however, it is necessary to demonstrate that they are able to function in the same way as the cells and tissues they are intended to replace. Here, we investigated the morphology and biochemical composition of nodules formed by ESCs and MSCs, and compared them to those formed by primary osteoblasts. We then related this to the quantity of mineral formed by each cell type and the magnitude of the expression of genes involved in osteogenic differentiation.
As in previous publications, we found that ESCs, MSCs and OBs form ARS-positive mineralised areas after prolonged culture in media containing osteogenic supplements (Buttery et al., 2001; Karp et al., 2006; Sottile et al., 2003, zur Nieden et al., 2003, zur Nieden et al., 2005). Similarly, in agreement with a previous study, we found that whole ESC cultures accumulate more calcium-containing mineral than OB or MSC (Shimko et al., 2004). But we also observed that the morphology of mineralised nodules was subtly different in in ESC cultures compared to in OB and MSC cultures, and that both the ratio of mineral-to-matrix and the absolute phosphate concentration in individual ESCs nodules was lower compared to nodules in MSCs, OBs or in native bone. This indicates a more diffuse but more extensive mineral accumulation in ESC cultures, and suggests that a different mechanism may mediate calcium deposition in the former cultures as compared to the latter. Therefore, we next thoroughly investigated the gene transcription profiles of ESCs, MSCs and OB cultures over the differentiation period, comparing expression of both osteogenic and chondrogenic genes. We found that many genes involved in osteogenic and chondrogenic differentiation were expressed at levels several orders of magnitude lower in ESCs than in MSCs or OBs, despite the high levels of mineralisation in the former cell cultures.
To allow us to compare gene expression between the three cell types, we first had to validate a suitable housekeeping gene to act as normaliser, and chose to investigate three commonly used housekeeping genes. In all cases, we observed no difference in any housekeeping gene when comparing MSCs with OBs. This is perhaps due to their similar morphology and shared cell lineage. In contrast there were significant differences in the expression of all housekeeping genes when comparing undifferentiated ESCs to either MSCs or OBs. Gapdh, for instance, was expressed at slightly higher levels in undifferentiated ESCs than in either undifferentiated MSCs or OBs, but was not significantly different after EB formation (5 days post differentiation, and referred to as d0 in our differentiation experiments). This is probably because cells of the early embryo, including embryonic stem cells, have fewer mitochondria than more differentiated cells (Facucho-Oliveira and St John, 2009) and rely to a greater extent on glycolysis rather than aerobic respiration for ATP generation (Cho et al., 2006; Facucho-Oliveira et al., 2007; Powers et al., 2008; St John et al., 2005). B2m and Actb, on the other hand were expressed at higher levels in MSCs and OBs compared to ESC, supporting previous observations where Tian et al were unable to detect MHC class I molecules in ESCs by FACS (Tian et al., 1997) and Dressel et al also noted low B2m in ESC lines (Dressel et al., 2009). The low expression of cytoskeletal gene β-actin in ESCs compared to MSCs or OBs may reflect the fact that ESCs have a poorly defined cytoskeleton, and grow as rounded colonies rather than flat, spread cells (as is the case in MSCs and OBs).
During differentiation, expression of Gapdh declined in all cell types at similar rates, with no significant difference in expression between the three cell types, a fact that allowed us to use this housekeeping gene as a normaliser between cell types. The decline we observed, however, may reflect the increase in mitochondria and decreased reliance on glycolysis that accompanies ESC differentiation, and also with the observation that during the course of osteogenic differentiation, osteoblasts also decrease their reliance on glycolysis and increase their reliance on oxidative metabolism (Komarova et al., 2000). The variations in the expression of housekeeping genes between cell types and during differentiation illustrates that careful validation of housekeeping gene expression should be performed before choosing normalisers to analyse gene expression in such experiments.
During osteogenic differentiation of both OBs and MSCs, the substantial upregulation of Bglap1, the gene for osteocalcin, and of Akp2, the gene for tissue nonspecific alkaline phosphatase (TNAP), with more subtle increases in Runx2 and Col1a1, are reflect the differentiation of osteogenic cells, and accompany the formation of bone matrix and the deposition of mineral in vitro (Aubin et al., 1995). Accordingly, we concurrently observed formation of raised, ARS-positive nodules in culture, which we have previously shown to exhibit characteristics similar to those of native bone tissue. In ESCs, however, there were only modest increases in Bglap, Akp2, and Runx2 and when peak expression of these genes in ESCs was compared with peak expression in MSCs and OBs, we found that all osteogenic genes were expressed at factors of 200 – 500 higher in the latter cells. This would seem to indicate that osteogenic differentiation is less efficient in ESCs than in MSCs or OBs. Considering that ESCs, as uncommitted cells, may undergo non-specific differentiation to other lineages in parallel with osteogenic differentiation, which is less true of MSCs or OBs, this finding alone is not surprising. But coupled with observation that mineralisation, measured by ARS uptake, was higher in ESCs than in the other cell types, our results seem paradoxical. We therefore think it likely that much of the calcium ion accumulation, measured by ARS staining, that occurs in differentiating ESCs in these conditions is not osteoblast-dependent. One possible mechanism for this is that mineral deposition may be promoted by other isoforms and/or homologues of alkaline phosphatase – ALP is known to be expressed at high levels in undifferentiated ESCs. In previous experiments, however, we found that alkaline phosphatase activity (measured biochemically) was no greater in ESCs than in MSCs or OBs (Gentleman et al., 2009). A more likely explanation is that dystrophic calcifications may occur in necrotic cells in culture. Necrotic tissue is known to promote inappropriate mineralisation both in vivo and in vitro due to the accumulation of insoluble salts containing calcium and phosphate (Gadeau et al., 2001; Hussmann et al., 1995; Zhao et al., 2009), and in preliminarly experiments we have observed significant cell death in ESC nodules by ethidium homodimer-1 uptake (Supplementary Figure 4). ESC nodules are thick multi-layered structures, and it is likely that nutrient depletion at the centre of such structures led to necrosis. We did not observe significant ARS staining of ESC cultures in the absence of β-glycerophosphate (not shown), indicating that the presence of phosphate was necessary for the dystrophic mineralisation we observed.
That Sox9 and Col2a1 were expressed at higher levels in OBs compared to in MSCs was surprising. During the normal course of development, the parietal and frontal skull bones of mice (from which OBs are derived) form by intramembanous ossification, with no cartilage intermediate. However, this process has been shown to be perturbed by mechanical force (Shapiro, 2008), growth factor exposure (Govindarajan and Overbeek, 2006) and tissue location (Leucht et al., 2008) and thus we suggest that the chemical milieu of the cell culture environment may promote chondrogenic gene expression in OB. This is supported by our previous observations of localisation of collagen type II by immunostaining and Raman spectroscopy in both OB and MSC (Gentleman et al., 2009). The rise in Col2a1 gene expression that accompanied osteogenic differentiation in MSCs suggests (but does not prove) that, at least in part, osteogenic differentiation proceeds via a cartilage intermeditary.
Our results certainly do not exclude the possibility that it is possible to produce osteogenic cells from embryonic stem cells for tissue engineering applications with greater efficiency. While most of the published papers on osteogenic differentiation describe using only osteogenic supplements such as β-glycerophosphate, ascorbate, dexamethasone (Buttery et al., 2001; Karp et al., 2006; Sottile et al., 2003), BMPs (zur Nieden et al., 2005), and vitamin D (zur Nieden et al., 2003) to promote osteogenesis, Kawaguchi et al did not observe induction of osteogenesis without stimulation of EBs with retinoic acid (Kawaguchi et al., 2005) (others have also noted increased osteogenesis by adding this molecule (Phillips et al., 2001; Yamashita et al., 2005; zur Nieden et al., 2007)). Notably, in this study the authors observed upregulation of Bgalp by a factor of ~100, which indicates a much greater efficiency of osteogenic differentiation than in our study, where maximal increases in the expression of this gene were only ~4 times greater than baseline. Here the authors suggest that early exposure to retinoic acid promotes either the differentiation of neural crest (which gives rise to the bones of the skull) or redirects paraxial mesoderm to form mesenchymal tissues. Further studies may seek to address the poorly-understood mechanisms of osteogenesis by examining the osteogenic potential of subsets of cells that arise in differentiating ESC cultures, and the effect of molecules such as retinoic acid in modulating this process.
Other recent studies have described protocols for deriving and selecting cells with properties of MSCs from cultures of differentiating ESCs (Gruenloh et al., 2011; Lai et al., 2011; Olivier et al., 2006). We did not test whether ESC-derived MSC-like cells expressed osteogenic genes at similar levels to either adult derived MSCs or OBs, but we suggest that future experiments on ESC-derived osteogenic cells should be compared directly with control, adult cells of known osteogenic potential. To our knowledge, there have been no demonstrations to date that ESC-derived osteogenic cells contribute to new, functional bone tissue in an animal model of fracture healing, a necessary test before ESC-derived osteoblasts can be used as a therapy in humans, and safety concerns over the potential of ESCs to cause teratomas will have to be overcome before these cells can be considered for therapeutic use.
We conclude that mineralisation of ESCs cultured under conditions commonly used to promote osteogenic differentiation occurs without either the characteristic high expression or upregulation of genes observed in MSCs and OBs, suggesting a non-specific accumulation of mineral in ESC cultures.
Supplementary Material
Acknowledgements
The authors would like to thank the EPSRC for funding. N.D.E. gratefully acknowledges the Medical Research Council, UK for fellowship funding. R.J.S. gratefully acknowledges funding from the Rothermere Foundation, the National Science and Engineering Research Council Canada and the Canadian Centennial Scholarship Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Arrington ED, Smith WJ, Chambers HG, Bucknell AL, Davino NA. Complications of iliac crest bone graft harvesting. Clin Orthop Relat Res. 1996:300–309. doi: 10.1097/00003086-199608000-00037. [DOI] [PubMed] [Google Scholar]
- Aubin JE, Liu F, Malaval L, Gupta AK. Osteoblast and chondroblast differentiation. Bone. 1995;17:77S–83S. doi: 10.1016/8756-3282(95)00183-e. [DOI] [PubMed] [Google Scholar]
- Bielby RC, Boccaccini AR, Polak JM, Buttery LD. In vitro differentiation and in vivo mineralization of osteogenic cells derived from human embryonic stem cells. Tissue Eng. 2004;10:1518–1525. doi: 10.1089/ten.2004.10.1518. [DOI] [PubMed] [Google Scholar]
- Boraiah S, Paul O, Hawkes D, Wickham M, Lorich DG. Complications of recombinant human BMP-2 for treating complex tibial plateau fractures: a preliminary report. Clin Orthop Relat Res. 2009;467:3257–3262. doi: 10.1007/s11999-009-1039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruder SP, Kurth AA, Shea M, Hayes WC, Jaiswal N, Kadiyala S. Bone regeneration by implantation of purified, culture-expanded human mesenchymal stem cells. J Orthop Res. 1998;16:155–162. doi: 10.1002/jor.1100160202. [DOI] [PubMed] [Google Scholar]
- Buttery LD, Bourne S, Xynos JD, Wood H, Hughes FJ, Hughes SP, Episkopou V, Polak JM. Differentiation of osteoblasts and in vitro bone formation from murine embryonic stem cells. Tissue Eng. 2001;7:89–99. doi: 10.1089/107632700300003323. [DOI] [PubMed] [Google Scholar]
- Cho YM, Kwon S, Pak YK, Seol HW, Choi YM, Park do J, Park KS, Lee HK. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem Biophys Res Commun. 2006;348:1472–1478. doi: 10.1016/j.bbrc.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Dressel R, Guan K, Nolte J, Elsner L, Monecke S, Nayernia K, Hasenfuss G, Engel W. Multipotent adult germ-line stem cells, like other pluripotent stem cells, can be killed by cytotoxic T lymphocytes despite low expression of major histocompatibility complex class I molecules. Biol Direct. 2009;4:31. doi: 10.1186/1745-6150-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Facucho-Oliveira JM, Alderson J, Spikings EC, Egginton S, St John JC. Mitochondrial DNA replication during differentiation of murine embryonic stem cells. J Cell Sci. 2007;120:4025–4034. doi: 10.1242/jcs.016972. [DOI] [PubMed] [Google Scholar]
- Facucho-Oliveira JM, St John JC. The relationship between pluripotency and mitochondrial DNA proliferation during early embryo development and embryonic stem cell differentiation. Stem Cell Rev. 2009;5:140–158. doi: 10.1007/s12015-009-9058-0. [DOI] [PubMed] [Google Scholar]
- Fink T, Lund P, Pilgaard L, Rasmussen JG, Duroux M, Zachar V. Instability of standard PCR reference genes in adipose-derived stem cells during propagation, differentiation and hypoxic exposure. BMC Mol Biol. 2008;9:98. doi: 10.1186/1471-2199-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedenstein AJ, Chailakhjan RK, Lalykina KS. The development of fibroblast colonies in monolayer cultures of guinea-pig bone marrow and spleen cells. Cell Tissue Kinet. 1970;3:393–403. doi: 10.1111/j.1365-2184.1970.tb00347.x. [DOI] [PubMed] [Google Scholar]
- Gadeau AP, Chaulet H, Daret D, Kockx M, Daniel-Lamaziere JM, Desgranges C. Time course of osteopontin, osteocalcin, and osteonectin accumulation and calcification after acute vessel wall injury. J Histochem Cytochem. 2001;49:79–86. doi: 10.1177/002215540104900108. [DOI] [PubMed] [Google Scholar]
- Gentleman E, Swain RJ, Evans ND, Boonrungsiman S, Jell G, Ball MD, Shean TA, Oyen ML, Porter A, Stevens MM. Comparative materials differences revealed in engineered bone as a function of cell-specific differentiation. Nat Mater. 2009;8:763–770. doi: 10.1038/nmat2505. [DOI] [PubMed] [Google Scholar]
- Giannoudis PV, Dinopoulos H, Tsiridis E. Bone substitutes: an update. Injury. 2005;36(Suppl 3):S20–27. doi: 10.1016/j.injury.2005.07.029. [DOI] [PubMed] [Google Scholar]
- Giannoudis PV, Einhorn TA, Marsh D. Fracture healing: The diamond concept. Injury. 2007;38(Suppl 4):S3–6. doi: 10.1016/s0020-1383(08)70003-2. [DOI] [PubMed] [Google Scholar]
- Govindarajan V, Overbeek PA. FGF9 can induce endochondral ossification in cranial mesenchyme. BMC Dev Biol. 2006;6:7. doi: 10.1186/1471-213X-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenloh W, Kambal A, Sondergaard C, McGee J, Nacey C, Kalomoiris S, Pepper K, Olson S, Fierro F, Nolta JA. Characterization and In Vivo Testing of Mesenchymal Stem Cells Derived from Human Embryonic Stem Cells. Tissue Eng Part A. 2011;17:1517–25. doi: 10.1089/ten.tea.2010.0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussmann J, Russell RC, Kucan JO, Khardori R, Steinau HU. Soft-tissue calcifications: differential diagnosis and therapeutic approaches. Ann Plast Surg. 1995;34:138–147. [PubMed] [Google Scholar]
- Kainer MA, Linden JV, Whaley DN, Holmes HT, Jarvis WR, Jernigan DB, Archibald LK. Clostridium infections associated with musculoskeletal-tissue allografts. N Engl J Med. 2004;350:2564–2571. doi: 10.1056/NEJMoa023222. [DOI] [PubMed] [Google Scholar]
- Karp JM, Ferreira LS, Khademhosseini A, Kwon AH, Yeh J, Langer RS. Cultivation of human embryonic stem cells without the embryoid body step enhances osteogenesis in vitro. Stem Cells. 2006;24:835–843. doi: 10.1634/stemcells.2005-0383. [DOI] [PubMed] [Google Scholar]
- Kawaguchi J, Mee PJ, Smith AG. Osteogenic and chondrogenic differentiation of embryonic stem cells in response to specific growth factors. Bone. 2005;36:758–769. doi: 10.1016/j.bone.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Komarova SV, Ataullakhanov FI, Globus RK. Bioenergetics and mitochondrial transmembrane potential during differentiation of cultured osteoblasts. Am J Physiol Cell Physiol. 2000;279:C1220–1229. doi: 10.1152/ajpcell.2000.279.4.C1220. [DOI] [PubMed] [Google Scholar]
- Lai RC, Choo A, Lim SK. Derivation and Characterization of Human ESC Derived Mesenchymal Stem Cells. Methods Mol Biol. 2011;698:141–150. doi: 10.1007/978-1-60761-999-4_11. [DOI] [PubMed] [Google Scholar]
- Leucht P, Kim JB, Amasha R, James AW, Girod S, Helms JA. Embryonic origin and Hox status determine progenitor cell fate during adult bone regeneration. Development. 2008;135:2845–2854. doi: 10.1242/dev.023788. [DOI] [PubMed] [Google Scholar]
- Lieber CA, Mahadevan-Jansen A. Automated method for subtraction of fluorescence from biological Raman spectra. Appl Spectrosc. 2003;57:1363–1367. doi: 10.1366/000370203322554518. [DOI] [PubMed] [Google Scholar]
- McCullen SD, Zhan J, Onorato ML, Bernacki SH, Loboa EG. Effect of varied ionic calcium on human adipose-derived stem cell mineralization. Tissue Eng Part A. 16:1971–1981. doi: 10.1089/ten.TEA.2009.0691. [DOI] [PubMed] [Google Scholar]
- Olivier EN, Rybicki AC, Bouhassira EE. Differentiation of human embryonic stem cells into bipotent mesenchymal stem cells. Stem Cells. 2006;24:1914–1922. doi: 10.1634/stemcells.2005-0648. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips BW, Belmonte N, Vernochet C, Ailhaud G, Dani C. Compactin enhances osteogenesis in murine embryonic stem cells. Biochem Biophys Res Commun. 2001;284:478–484. doi: 10.1006/bbrc.2001.4987. [DOI] [PubMed] [Google Scholar]
- Place ES, Evans ND, Stevens MM. Complexity in biomaterials for tissue engineering. Nat Mater. 2009;8:457–470. doi: 10.1038/nmat2441. [DOI] [PubMed] [Google Scholar]
- Powers DE, Millman JR, Huang RB, Colton CK. Effects of oxygen on mouse embryonic stem cell growth, phenotype retention, and cellular energetics. Biotechnol Bioeng. 2008;101:241–254. doi: 10.1002/bit.21986. [DOI] [PubMed] [Google Scholar]
- Savitzky A, Golay MJ. Smoothing and Differentiation of data by simplified least squares procedures. Anal Chem. 1964;36:1627–1639. [Google Scholar]
- Shapiro F. Bone development and its relation to fracture repair. The role of mesenchymal osteoblasts and surface osteoblasts. Eur Cell Mater. 2008;15:53–76. doi: 10.22203/ecm.v015a05. [DOI] [PubMed] [Google Scholar]
- Shimko DA, Burks CA, Dee KC, Nauman EA. Comparison of in vitro mineralization by murine embryonic and adult stem cells cultured in an osteogenic medium. Tissue Eng. 2004;10:1386–1398. doi: 10.1089/ten.2004.10.1386. [DOI] [PubMed] [Google Scholar]
- Sottile V, Thomson A, McWhir J. In vitro osteogenic differentiation of human ES cells. Cloning Stem Cells. 2003;5:149–155. doi: 10.1089/153623003322234759. [DOI] [PubMed] [Google Scholar]
- St John JC, Ramalho-Santos J, Gray HL, Petrosko P, Rawe VY, Navara CS, Simerly CR, Schatten GP. The expression of mitochondrial DNA transcription factors during early cardiomyocyte in vitro differentiation from human embryonic stem cells. Cloning Stem Cells. 2005;7:141–153. doi: 10.1089/clo.2005.7.141. [DOI] [PubMed] [Google Scholar]
- Sugden K, Pariante CM, McGuffin P, Aitchison KJ, D'Souza UM. Housekeeping gene expression is affected by antidepressant treatment in a mouse fibroblast cell line. J Psychopharmacol. 24:1253–1259. doi: 10.1177/0269881108099690. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tian L, Catt JW, O'Neill C, King NJ. Expression of immunoglobulin superfamily cell adhesion molecules on murine embryonic stem cells. Biol Reprod. 1997;57:561–568. doi: 10.1095/biolreprod57.3.561. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems E, Mateizel I, Kemp C, Cauffman G, Sermon K, Leyns L. Selection of reference genes in mouse embryos and in differentiating human and mouse ES cells. Int J Dev Biol. 2006;50:627–635. doi: 10.1387/ijdb.052130ew. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Takada T, Narita J, Yamamoto G, Torii R. Osteoblastic differentiation of monkey embryonic stem cells in vitro. Cloning Stem Cells. 2005;7:232–237. doi: 10.1089/clo.2005.7.232. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Urganus AL, Spevak L, Shrestha S, Doty SB, Boskey AL, Pachman LM. Characterization of dystrophic calcification induced in mice by cardiotoxin. Calcif Tissue Int. 2009;85:267–275. doi: 10.1007/s00223-009-9271-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Nieden NI, Kempka G, Ahr HJ. In vitro differentiation of embryonic stem cells into mineralized osteoblasts. Differentiation. 2003;71:18–27. doi: 10.1046/j.1432-0436.2003.700602.x. [DOI] [PubMed] [Google Scholar]
- zur Nieden NI, Kempka G, Rancourt DE, Ahr HJ. Induction of chondro-, osteo- and adipogenesis in embryonic stem cells by bone morphogenetic protein-2: effect of cofactors on differentiating lineages. BMC Dev Biol. 2005;5:1. doi: 10.1186/1471-213X-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Nieden NI, Price FD, Davis LA, Everitt RE, Rancourt DE. Gene profiling on mixed embryonic stem cell populations reveals a biphasic role for beta-catenin in osteogenic differentiation. Mol Endocrinol. 2007;21:674–685. doi: 10.1210/me.2005-0438. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.