Abstract
Mitochondrial activities are linked directly or indirectly to all cellular functions in aerobic eukaryotes. Omics methods enable new approaches to study functional organization of mitochondria and their adaptive and maladaptive network responses to bioenergetic fuels, physiologic demands, environmental challenges and aging. In this review, we consider mitochondria collectively within a multicellular organism as a macroscale “mitochondriome”, functioning to organize bioenergetics and metabolism as an organism utilizes environmental resources and protects against environmental threats. We address complexities of knowledgebase-driven functional mapping of mitochondrial systems and then consider data-driven network mapping using omics methods. Transcriptome-metabolome-wide association study (TMWAS) shows connectivity and organization of nuclear transcription with mitochondrial transport systems in cellular responses to mitochondria-mediated toxicity. Integration of redox and respiratory measures with TMWAS shows central redox hubs separating systems linked to oxygen consumption rate and H2O2 production. Combined redox proteomics, metabolomics and transcriptomics further shows that physiologic network structures can be visualized separately from toxicologic networks. These data-driven integrated omics methods create new opportunities for mitochondrial systems biology.
Keywords: Environmental stressor, Integrative omics, Mitochondrial systems biology, Network structure
1. Introduction
Omics technologies, systems biology and big data create new opportunities to understand mitochondrial biology. Mitochondria have been extensively studied in many biomedical disciplines, with nearly 300,000 mitochondrial publications in PubMed. Substantial detail is available on morphology and macromolecular composition, biophysics and bioenergetics, biochemistry and metabolism, genetics and evolution, and mechanisms of aging, health and disease [1–4]. With this depth of knowledge, research will be served by an interactive knowledgebase with which one can perform in silico experiments to examine connections and predict responses. Doing this in an objective manner is difficult, however, because data is derived mostly from hypothesis-driven research performed with predefined targets and outcomes. Thus, there is an inherent bias in data availability, with knowledgebase hubs determined by perceived importance of a finding and/or research tools and pursuant productivity rather than by centrality of functions.
In the present conceptual review, we use an omics approach to consider mitochondrial networks connecting physicochemical functions with macromolecular structures in oxidative physiology and disease. To provide context, we start with consideration of the molecular logic of mitochondria based upon the principles of the redox code. This provides a scaffold upon which a mitochondrial knowledgebase can be constructed. We then address mitochondria in the omics era. With new integrative omics tools available, terms like “cross-talk” and “complex mixtures” can be replaced by quantitative descriptors of connectivity and hierarchy [5]. We follow with a vision of mitochondria at the gene-environment interface. In this, we note that mitochondrial function is affected both positively and negatively by nutrients and environmental threats. We then examine experimental results demonstrating a hierarchical nature in network interactions of the nuclear transcriptome and metabolome in mitochondrial toxicity, distinct network substructures associated with oxygen consumption rate and mitochondrial H2O2 production, and an integrated redox proteome, metabolome and nuclear transcriptome in cells in response to increasing manganese exposure. We conclude with an optimistic vision that elucidation of network structures will lead to development of improved tools to monitor mitochondrial homeostasis and dysfunction. Creation of an interactive mitochondrial systems knowledgebase with data-driven omics models could facilitate understanding of the integrated functions of mitochondria within the complexities of genetic variation, diet, physical activity and environmental exposures.
2. Mitochondrial networks and the redox code
The redox code complements the genetic code in providing simple principles by which aerobic organisms maintain energetic, metabolic and structural organization to enable reproduction [6]. NAD and NADP are central electron carriers in the bioenergetic and metabolic organization. The steady-state redox potentials of the NAD couple (NADH/NAD+) and NADP couple (NADPH/NADP+) are maintained by near-equilibrium, enzyme catalyzed oxidation-reduction reactions with metabolic fuels. These redox couples serve different functions, with NAD supporting catabolism and high-flux bioenergetic demands, and NADP supporting biosynthetic and detoxification activities. Conventions for expressing the reducing force of these couples in terms of steady-state redox potential (Eh) are considered elsewhere [7–9]; most critically, the NADP couple is maintained at a more negative potential (greater reducing force) to support biosynthetic and detoxification functions while the NAD couple is maintained at a more positive steady state to facilitate oxidation of metabolic fuels for ATP production by oxidative phosphorylation [10–12].
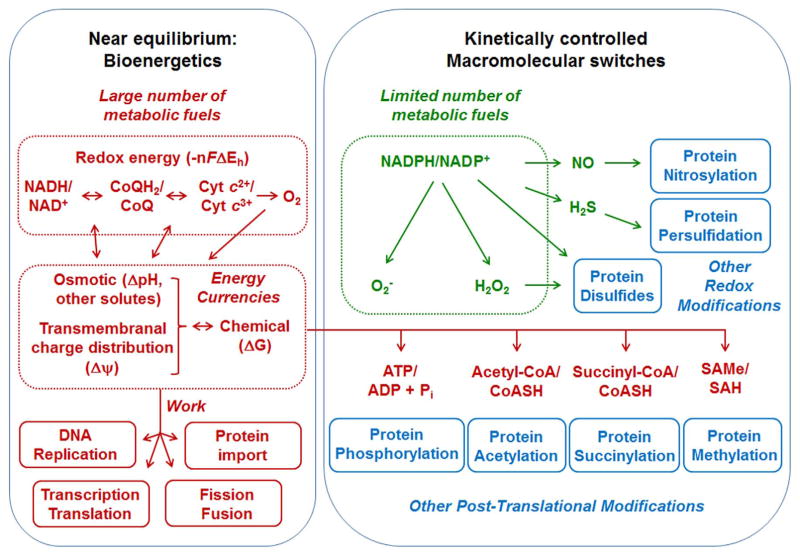
In Fig. 1, we separate the NAD and NADP systems to provide a starting point for elaboration of mitochondrial networks. Mitochondria serve as the powerhouse of aerobic cells, and this central role, with interconvertible energy currencies, is shown in red. These energy currencies are integrated with other cellular systems to support DNA replication, transcription, translation, mitochondrial protein import, mitochondrial repair and replacement and other demands for mitochondrial and cellular homeostasis. In the second principle of the redox code [6], macromolecular structure and function is integrated with bioenergetic systems through reversible switches in the proteome. In Fig. 1, NADP-linked processes supporting redox switches are shown in green while representative examples of the broader spectrum of post-translational modifications linking macromolecular structure and function to bioenergetics and metabolism is shown in blue. In this simplified conceptualization, we have omitted details about production of oxidants by mitochondria, which are included within the redox code [6] and extensively studied in terms of pathology and aging [13–15]. In addition to pathologic consequences from excess production, evidence is accumulating that mitochondria produce oxidants as signaling molecules that integrate mitochondria-cell functions [16–18].
Fig. 1.
Mitochondrial Function and the Redox Code. The first two principles of the redox code address the logic of bioenergetics and metabolism and their integration with macro-molecular structure and function. In red, metabolic fuels are oxidized through near equilibrium reactions linked to the NAD couple provide energy to maintain asymmetric chemical and ion distributions across the mitochondrial inner membrane and potential energy in many types of “high energy” molecules [78]. These reactions support chemical and physical work. In green, redox systems linked to the NADP couple support diverse, kinetically controlled signaling and control functions. In blue, both NAD- and NADP-linked electrochemical systems control macromolecular structure, trafficking and biological functions through reversible post-translational modifications of proteins. Based upon [6].
2.1. Mitochondrial network structures maintained by physical and functional connections
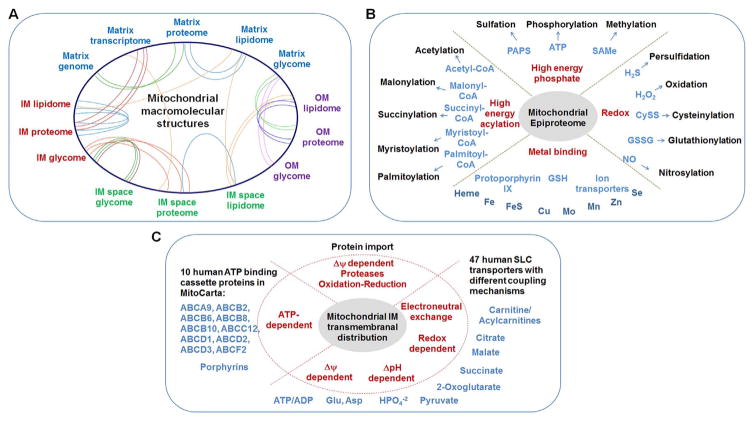
Using the depiction of Fig. 1 as a starting point, one can develop an expansive network structure by linking known processes and structures, as illustrated with circos plots in Fig. 2A. Macromolecular networks can be visualized in terms of physical or functional connections. For instance, multiprotein complexes I to V were isolated from the inner membrane [19–21] and reconstituted in lipid vesicles in elucidation of the mechanisms of oxidative phosphorylation [22,23]. Protein-lipid interactions vary in specificity, with some protein structures requiring specific cardiolipins [24]. The recent study of human platelet lipidomics showed that mitochondrial lipidome is changed acutely in association with phospholipase and COX-1-dependent increased energy metabolism upon platelet activation [25], supporting the critical role for mitochondrial protein-lipid networks in controlling inflammation. Informative relationships of proteins, lipids and other macromolecules can be visualized with such an approach. Similarly, subcompartmental interactions also vary, illustrated by distribution of cytochrome c association/dissociation with the inner membrane [26,27]. Thus, interactive network tools with capability to zoom in to examine detailed interactions can considerably enhance abilities to examine and understand mitochondrial functions.
Fig. 2.
Expanded mitochondrial network structures. Central aspects of bioenergetic, metabolic and macromolecular structures in Fig. 1 can be expanded to include subnetwork structures and elements. A. The central energy currencies support biosynthesis and processing of macromolecules within the physical spaces of mitochondria. B. Systems for post-translational modification of proteins integrate bioenergetic and metabolic functions with the activities, interactions and trafficking of macromolecules. C. Transport systems utilize energy currencies to maintain macromolecular, metabolic and ionic composition of the mitochondrial compartments.
Post-translational modifications of proteins also exist as an epiproteome network, linked by common compartments, enzymes, precursors, binding partners and opposing enzymes (Fig. 2B). Phosphorylation, sulfation, methylation and adenosylation have common links to bioenergetic currencies. Redox networks, including oxidation, glutathionylation, nitrosylation, and a range of other redox elements, have been described [28]; these are commonly linked through thioredoxin, glutathione, H2O2 and other central redox hubs [29–33]. Networks of acetylation, malonylation and other acylation reactions occur; these are linked by acyl-CoA precursors, acyltransferases and de-acylases. Mitochondrial sirtuins represent an important group of deacetylases, de-malonylases, and related enzymes under active investigation due to associations with disease and aging [34–36].
2.2. Mitochondrial transporters play central roles in controlling mitochondrial metabolic networks
Mitochondrial metabolic networks are extensively interconnected through highly conserved families of mitochondrial protein import machinery and transporters for small molecules, especially ABC transporters (ATP-binding cassette proteins) and SLC25 transporters (Fig. 2C). Steps in mitochondrial protein import have been elucidated [37], and associated networks have been well characterized [38–40]. ABC transporters function in porphyrin transport for heme metabolism, thereby creating a functional link within mitochondria as well as between mitochondria and non-mitochondrial hemoproteins. Recent description of an ABC transporter that exports glutathione polysulfide for cytosolic cofactor biosynthesis [41] also illustrates mitochondrial-cytosolic network connectivity. Importantly, many of the ABC transporters have unknown activities so that network connections cannot be projected. A larger number of SLC transporters are known, and central functions of the SLC25 family in mitochondrial metabolism have been described [42,43]. Surprisingly from the standpoint of mitochondrial transport physiology, they have common structural features, with six transmembrane α-helices and a 3-fold repeated signature, but differences in energy coupling through functions as uniporters, symporters and antiporters [44,45]. Thus, some are linked to Δψ, others to ΔpH, and others are electroneutral carriers. In some cases, intervening redox reactions are coupled to NADH/NAD+. Consequently, mitochondrial metabolites exist in network structures with transporters having multiple types of connections, creating a complexity that is difficult to diagram or use for predictive purposes.
With the exception of high-flux systems controlling NAD, NADP and central energy metabolism, these processes (Fig. 2A–C) are kinetically controlled, limited both by catalytic activity and/or precursor availability. The third and fourth principles of the redox code provide logic to these functions [6]. Activation/deactivation cycles of the molecular switches in the proteome control the temporal sequencing and spatial distribution of biologic processes. Thus, network responses can be viewed temporally and spatially. In effect, this is mitochondrial systems biology, examined microscopically within network substructures. Components within this structure are undergoing continuous repair, and molecular machinery detects excessive damage, eliminating and replacing malfunctioning elements [46–50]. Collectively, the entire macromolecular and metabolic system functions as an adaptive network to respond to environmental resources and challenges. Mitochondria in different tissues reflect genetically encoded developmental programs of the tissue. In principle, specific mitochondria in different tissues and under different environmental conditions also differ in composition, structure and function due to current environmental exposures and/or the history of previous exposures. Communally, the mitochondria function as a bioenergetic system to support an individual, and this creates an additional layer of complexity to models for mitochondrial systems biology.
2.3. Mitochondria as a redox interface between the genome and exposome
The central role of the redox proteome and metabolome as an interface between the functional genome and environment has previously been described [51,52]. Mitochondria are a major component of this interface because of their primary role in redox homeostasis and use of redox energy to maintain electrochemical membrane gradients and forms of available chemical energy. Mitochondria contain parallel, non-redundant thiol antioxidant systems dependent upon glutathione (GSH) and thioredoxin-2 (Trx2) [53,54]. The mitochondrial GSH system is maintained at a more negative (reduced) steady state Eh than the cytoplasmic pool [55]. Under steady-state conditions without toxicologic challenge, mitochondrial Trx2 is maintained at a more negative steady-state Eh than cytoplasmic Trx1and is more susceptible to oxidation [56]. With limited energy precursors, mitochondrial Trx2 become oxidized [57], and exposure to toxic environmental metals, arsenic, cadmium and mercury, causes selective oxidation of Trx2 compared to cytoplasmic Trx1 [58]. Thus, mitochondria are maintained at a relatively reducing set point and are selectively vulnerable to oxidation in the presence of toxic metals. This interaction with metals brings in to focus a critical role of mitochondria in responding to variable low levels of environmental exposures to metals, such as present in the diet.
Many environmental toxicants such as insecticides, fungicides, herbicides and metals enter the food chain through plants. Plants have considerable capacity to package and store a range of nutritionally important as well as toxic metals. Mineral content in soil varies considerably, so metals like manganese (Mn) and cadmium (Cd), as well as metalloids like selenium (Se), vary considerably in animal diets and drinking water. In prior studies, we have varied environmental chemical exposures in animal and cell models to study molecular and functional responses of mitochondria [59–62]. In the following sections, we use results from these studies to illustrate data-driven analysis of omics data to advance understanding of mitochondrial networks.
3. Data-driven mitochondrial network analysis: integrative measure of molecular and functional responses of cells to stressor (s)
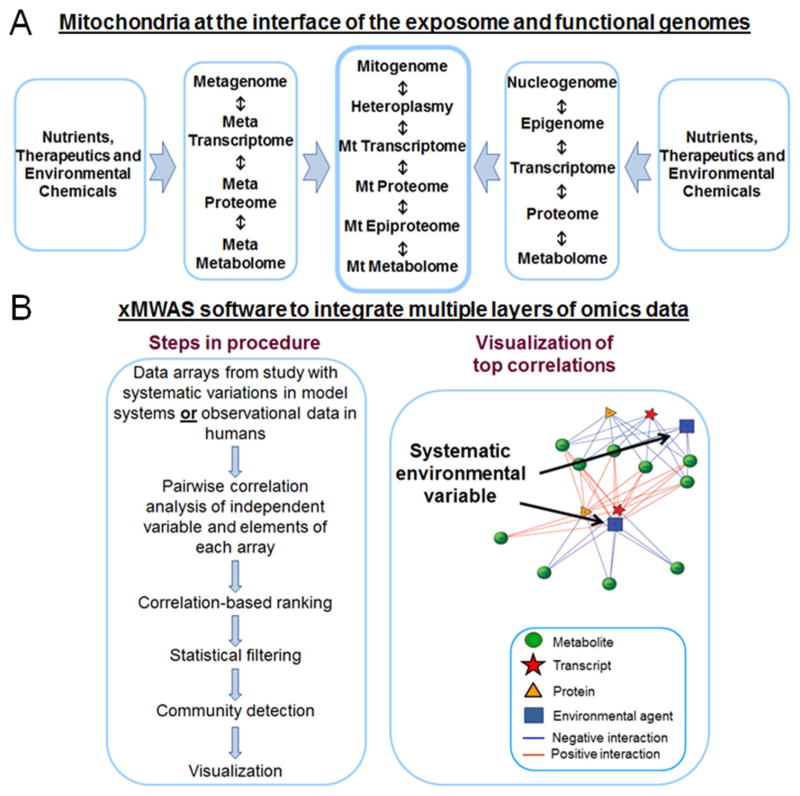
Omics study offers ways to overcome some of the barriers to mitochondrial systems biology by allowing simultaneous views of mitochondrial macromolecular and metabolic functions through multiple omics layers (Fig. 3). By extending this to resolve mitochondrial systems by time and location, quantitative mitochondrial systems biology models can be developed. In Fig. 3A, we outline the mitochondrial omics hierarchy, with the mitochondrial genome (mitogenome) top and center. In this, we delineate nuclear omics and intestinal microbial omics as separate entities, but these are extensively integrated with mitochondrial systems at multiple levels as nutrients and metabolic fuels from food are assimilated and delivered as energy precursors for use by mitochondria. Extensive evidence is available, for instance, on functional interactions of the mitogenome with the nuclear genome (nucleogenome), [63,64] and metagenome of the intestines with mitochondrial function in metabolic diseases of fatty liver and type 2 diabetes [65,66].
Fig. 3.
Data-driven mitochondrial network analysis using multiple omics layers and xMWAS software. A. Systems biology of mitochondria is complex because of the interactions of multiple genetic systems and position at the interface of the interaction of an aerobic organism with its nutrient precursors and environmental exposures. B. xMWAS is a tool for integration of multiple layers of omics data to enable systems level analysis resolved by time and space.
3.1. xMWAS software to integrate multiple omics data and cellular functions
Karan Uppal developed xMWAS software to integrate multiple omics layers with independent variables and phenotypic measures, visualize the association networks, identify communities/clusters of highly connected nodes, and compare integrative networks under different biological conditions [60]. This enables a top-down, data-driven approach to identify and visualize the hierarchy of network structures of mitochondrial systems; by using very high stringency for correlation and signicance criteria, the most central network hubs can be identified (Fig. 3B). xMWAS allows input of up to four omics layers, along with phenotypic measures or independent variables, from a series of samples from observational or experimental studies. The approach was developed to use high-resolution metabolomics and other omics data to provide the detailed molecular phenotyping needed for precision medicine [67,68]. A more detailed logic and workflow for use of xMWAS to study integrated mitochondria-cell signaling is available [68].
3.2. Network analysis by integration of transcriptomic and metabolomic responses to mitochondrial toxicant
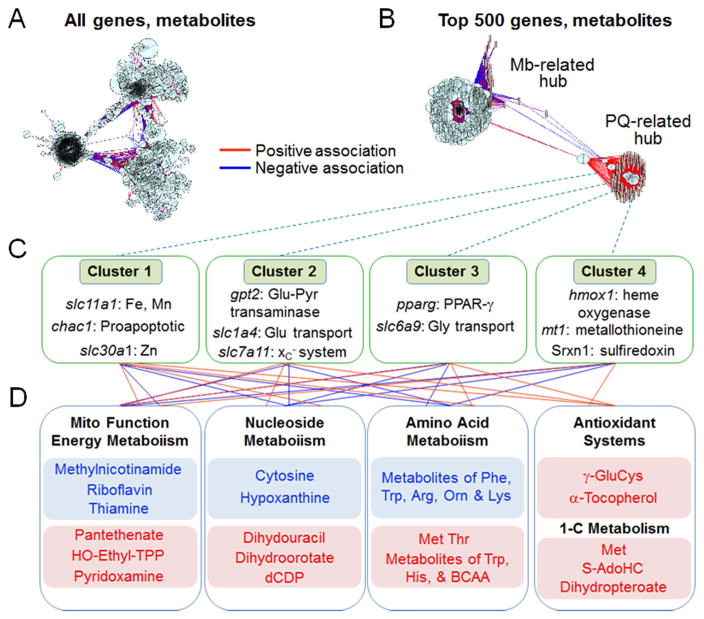
In initial development of this strategy, we used a transcriptome-metabolome wide association study (TMWAS) of the fungicide maneb (MB) in combination with the mitochondrial toxicant, herbicide paraquat (PQ) in a human locus ceruleus cell line, where each alone was minimally toxic but the combination caused 50% cell death after 24 h. Results at 4 h (prior to cell death) showed complex interaction networks (Fig. 4A) that were greatly simplified by using stringency criteria such as statistical filtering and ranking by strength of association (Fig. 4B). With this simplified structure, two hubs were present, a major hub associated with MB exposure and a smaller hub associated with PQ exposure. Earlier studies had shown that MB and PQ act by different mechanisms [69]. MB caused nuclear translocation of Nrf-2 and increased cellular GSH without oxidation of cytoplasmic Trx1 or mitochondrial Trx2 while PQ at minimally toxic dose oxidized Trx2 and mitochondrial peroxiredoxin-3 without oxidation of cytoplasmic Trx1, cytoplasmic peroxiredoxin-1 or cellular GSH [69]. Importantly, in the TMWAS, PQ alone resulted in weak changes in many components without strong network associations. Thus, the results showed that transcript-metabolite networks associated with toxicity could be visualized from integrative network analysis with filtering by statistical criteria and ranking according to strength of association.
Fig. 4.
Transcriptome-Metabolome-Wide Association Study (TMWAS) shows complex network responses of human locus ceruleus (CAD) cell line to combined exposure to the fungicide maneb (MB) and herbicide paraquat (PQ). A. Pair-wise correlation of all transcripts and metabolites using sparse Partial Least Squares regression and visualization with mixOmics. B. Same as A including only top 500 hub genes with correlation threshold > 0.5. The PQ-related clusters were only present in cells treated with MB+PQ. C. Transcripts in 4 central transcript clusters associated with PQ + MB. D. Functional categories of metabolites varying in association with the transcripts in C. Connections between transcripts and metabolites are included to represent complexity of interactions; these are incomplete and original publication should be consulted for more detail.
Modified from Roede et al. [59].
Four transcriptome-metabolome clusters, labeled 1–4 were presented with 13 transcripts in the larger PQ-associated cluster (Fig. 4C). These 13 included diverse hub genes, most of which are understandable in terms of existing knowledge but not likely to be deduced as central hubs using network structures outlined in Fig. 2. Cluster 1 was a cation-response hub which included a gene encoding a proapoptic protein, Chac1, and two cation transporters. This hub could contribute to toxicity due to manganese (Mn) or iron (Fe) transport, PQ transport, activation of apoptosis, ferroptosis [70] or other mechanism. Some of the transcripts in other hubs would be more predictable because of known protective functions. Cluster 2 contained slc7a11, an amino acid/anti-oxidant response hub important in cellular uptake of cystine, a precursor for GSH synthesis. Cluster 3 included ppar-γ, a central component of an adaptive response network to adjust fatty acid and glucose in energy metabolism. Cluster 4 included hmox-1, an early stress-response gene, as well as the metal-detoxifying protein metallothionein-1 and redox recovery protein, sulfiredoxin-1.
The importance of data-driven network discovery methods is evident from the metabolites associated with the transcripts (Fig. 4D). These included diverse groups of metabolites, including energy, nucleoside, amino acid, 1-carbon and antioxidant metabolism. These varied considerably in the strengths of association with the different genes, but together showed a community of metabolic systems functioning together in response to the toxicologic challenge [59]. The data-driven results are interpretable but not readily predicted from existing knowledge.
Metabolites associated with oxidative stress and mitochondrial energy metabolism supported prior findings of mitochondrial oxidative stress and mitochondrial toxicity of PQ. Effects on amino acid metabolism and nucleoside metabolism are expected under conditions of mitochondrial damage and cell stress. Amino acid and energy metabolite associations with ppar-γ can be anticipated because of the function of PPAR-γ in fatty acid and glucose metabolism. The overall results of the TMWAS show the benefit of a complex systems approach in studies of human pathobiology [71].
As outlined by Loscalzo et al. [71], diseases with mechanisms involving one cause for one disease are probably non-existent. Simple additive and synergistic cause-effect mechanisms do not occur in complex systems because biologic systems are designed to utilize a range of energy precursors [52], have multiple defense mechanisms and be adaptable to environmental conditions [6]. With availability of powerful omic tools and newly developed data-driven integration tools like xMWAS software [60], investigation of network level interactions enables a new level of understanding of disease mechanisms.
3.3. Mitochondrial networks linked to mitochondrial respiration and H2O2 production
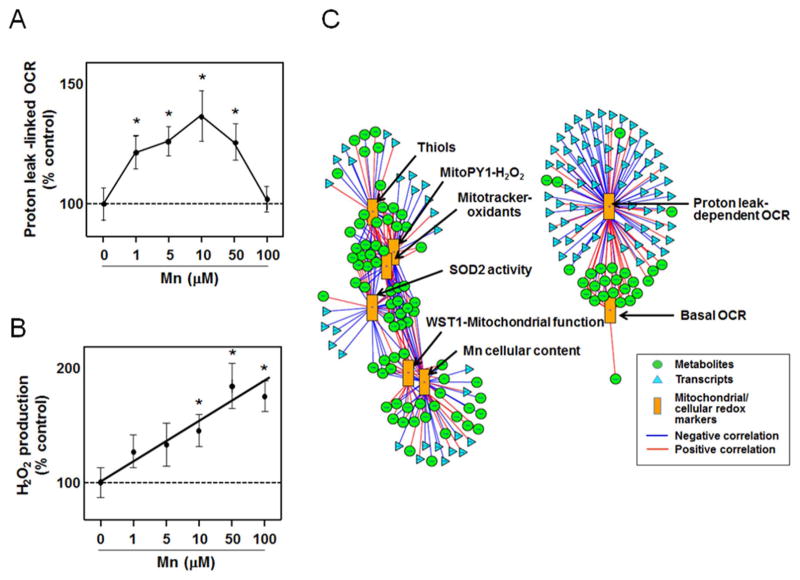
Variation in Mn exposure serves as a useful model to experimentally examine mitochondrial networks. Mn is an essential nutrient, required as a cofactor for many enzymes, including mitochondrial superoxide dismutase-2 (SOD2) [72]. At higher exposures, Mn causes manganism, a Parkinson disease-like movement disorder with mitochondrial dysfunction and neuronal cell death [73,74]. Exposure of human neuroblastoma (SH-SY5Y) cells to increasing Mn concentrations showed that concentrations up to 10 μM resulted in increased cellular Mn similar to values in normal human brain without causing cellular toxicity [75]. Basal mitochondrial respiration as well as proton leak-linked respiration increased over this range. At 50 and 100 μM, Mn accumulated to cellular levels found in human brain of individuals with Mn toxicity, and mitochondrial respiration progressively decreased [75]. Over the entire range, mitochondrial H2O2 production increased as a function of cellular Mn content. Thus, the results showed that mitochondrial respiration varied with a different Mn dose response characteristic compared to mitochondrial H2O2 production (Figs. 5A, 5B).
Fig. 5.
Transcriptome-metabolome network structures linked to mitochondrial respiratory and oxidative stress measures in SH-SY5Y cells. A. Proton leak-dependent oxygen consumption rate (OCR) as a function of Mn concentration in the culture medium measured by Seahorse Bioenergetics Analyzer. B. Mitochondrial H2O2 production rate as a function of Mn concentration measured by MitoPy1. C. Top hubs of transcriptome-metabolome interaction networks associated with OCR and other mitochondrial oxidative stress measures. Original data on mitochondrial oxidative stress and OCR measures from Fernandes et al. [75].
Data-driven analysis of transcriptome and metabolome networks with xMWAS showed that the most central network hubs associated with respiration measures were separated from those for H2O2 production and thiol measures (Fig. 5C). Proton leak-dependent oxygen consumption rate (OCR) and basal OCR clustered together and correlated with many of the same metabolites and transcripts. Proton leak-dependent OCR and basal OCR were positively associated with free fatty acids, palmitate and stearate, and negatively associated with acyl carnitines. These changes are consistent with mobilization of fatty acids for β-oxidation and stimulated use of acyl-carnitines in transfer to mitochondria for oxidation. Proton leak-dependent OCR and basal OCR also highly associated with abundance of more than 100 transcripts, including genes for cell signaling, proliferation and energy regulation.
At the level of stringency shown, transcripts and metabolites associated with MitoPy1 signal (H2O2 production) were completely separate from those associated with OCR measures (Fig. 5C). Associations with H2O2 production included metabolites and transcripts associated with MitoTracker, an indicator of mitochondrial oxidants; somewhat weaker associations occurred with thiols and SOD2 activity (Fig. 5C). Importantly, the thiol and WST-1 signals were negatively associated with the H2O2 production. A relatively small number of annotated metabolites were strongly positively associated with thiols; these included cysteine, acetyl-glutamate and acetyl-lysine. In contrast, more than 100 transcripts were positively associated with thiols, including signaling molecules, transporters and oxidoreductases. Examples include the protooncogene fyn, nicotinamide nucleotide adenylyltransferase 2 (nmnat2), an 11-beta-hydroxysteroid dehydrogenase 1-like variant, and very long chain acyl-CoA dehydrogenase (acadvl). A relatively small number of annotated metabolites positively associated with H2O2 (MitoPy1 signal) also associated with cellular Mn content and included deoxyinosine diphosphate (dIDP) and a C21 sterol. More than 100 transcripts strongly associated with H2O2, including those for signaling molecules, acyltransferases and regulatory systems, such as microRNAs. The relatively poor annotation of the metabolites and the extensive range of transcript associations impedes detailed interpretation. On the other hand, the visualization of distinct metabolite-transcript clusters associated with OCR and H2O2 production emphasizes that mitochondrial network structures are present to integrate diverse activities into common functions. The data-driven mitochondrial network structures separate network substructures for energy production from those related to redox signaling by H2O2 and involving thiol oxidation and thereby provide a useful separation for more specific testing of hypotheses.
3.4. Molecular networks visualized by integrated redox proteome, metabolome and transcriptome can separate physiological and toxicological mitochondrial responses
Separation of network structures for physiologic 0–10 μM Mn) concentrations from toxicologic concentrations that led to subsequent cell death (50, 100 μM Mn) (Fig. 6) provides a very different perspective on mitochondrial network structures. Although one must recognize that association analyses based on correlation alone do not establish cause-effect relationships, prior demonstration of H2O2 production as a function of Mn concentration and inverse correlation of mitochondrial thiols with H2O2 production supports the interpretation that Mn causes oxidation of protein thiols. Thus, in an interaction study of the redox proteome, transcriptome and metabolome, one can infer that protein oxidation resulted in metabolic effects and changes in transcription.
Fig. 6.
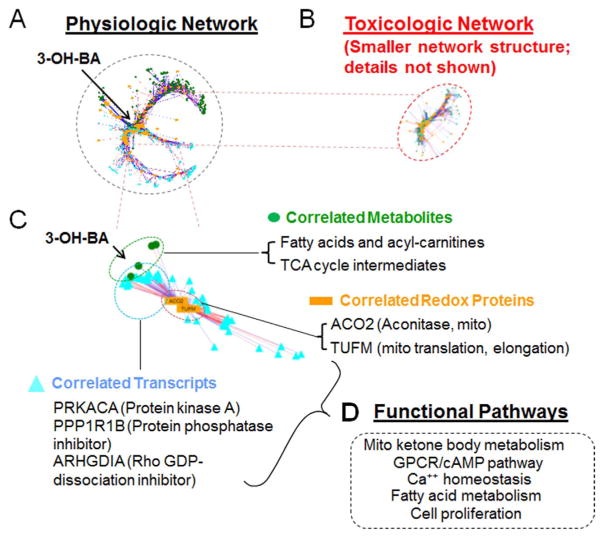
Integrated Redox Proteome-Metabolome-Transcriptome analysis of physiologic variation in Mn concentration in SH-SY5Y cells. A. xMWAS analysis (sparse partial least squares; sPLS, r ≥ 0.6) of redox proteome, metabolome and transcriptome correlations as a function of added Mn 0–10 μM, 5 h). B. xMWAS as in A with toxicologic Mn 50–100 μM, 5 h). Note that the network size is much smaller and includes far fewer elements. C. Expanded view of metabolites, transcripts and redox-sensitive proteins changing in association with central mitochondrial metabolite, 3-hydroxybutyric acid (3-OH-BA). Correlated mitochondrial metabolites included tricarboxylic acid cycle intermediates, fatty acids and acyl carnitines. Correlated redox proteins included mitochondrial aconitase and mitochondrial translation elongation factor, TUFM. Correlated transcripts included regulatory kinase and phosphatase. D. Pathway mapping using Kyoto Encyclopedia of Genes and Genomes (KEGG) showed associations of central clusters with functional pathways.
Transcriptome, metabolome and redox proteome measurements for SH-SY5Y cells were performed at 5 h, when no cell death was apparent. No cell death was apparent at 24 or 48 h for cells incubated with ≤10 μM Mn for 5 h and returned to normal culture medium. In contrast, cells incubated 5 h with 50 and 100 μM Mn and then placed back in culture with normal culture medium showed extensive cell death after 24 h [75]. The network structures at 5 h for physiologic Mn concentrations were extensive (Fig. 6A), including more than 280 metabolites and large numbers of transcripts and redox proteins. In contrast, the network structures at 5 h for toxicologic Mn concentrations were much smaller, with only 80 metabolites and relatively small numbers of transcripts and redox proteins (Fig. 6B). The results show that the healthy, physiologic system has extensive connectivity, as expected for maximal flexibility and adaptability, while the toxicologic system is stressed, with limited connectivity as expected for systems with limited flexibility and adaptability [76].
In an expanded view of the physiologic network structure (Fig. 6C), a central mitochondrial metabolite, 3-hydroxybutyric acid (3-OH-BA; β-hydroxybutyrate), becomes apparent as one of the hub metabolites. 3-OH-BA is directly linked to the mitochondrial NADH/NAD+ couple, one of the central redox couples for coupling of redox energy to bioenergetics and metabolism in the redox code [6]. 3-OH-BA is associated with other mitochondrial metabolites including tricarboxylic acid cycle intermediates, fatty acids and acyl carnitines. Correlated redox proteins with 3-OH-BA include the tricarboxylic acid pathway enzyme, mitochondrial aconitase. Correlation with oxidation of mitochondrial translation elongation factor, TUFM, provides an unanticipated connection between energetics and mitochondrial protein synthesis. The data suggest that mitochondrial translation is linked to protein thiol oxidation that varies with Mn. This concept is further extended by the examination of transcripts varying with 3-OH-BA, which included regulatory kinase, phosphatase and G-protein regulatory protein (Fig. 6C). These network structures can be further analyzed with knowledgebase tools to gain insight into associated mechanistic and metabolic pathways (Fig. 6D).
Key observations from integrated omics analysis of redox proteomic, metabolomic and transcriptomic responses to physiologic variation in the essential nutrient, Mn, show that mitochondria respond with a network structure in which elements of different omics layers are extensively associated across omics layers. In combination with the mitochondrial phenotypic measures, such as OCR and H2O2 production rate described above, this implies that functional pathways as currently exist in bioinformatics tools may need to be reconstructed to reflect functional network structures. Such visualization would more correctly describe biology and also could yield improved capabilities to evaluate system health and response to interventions.
4. Perspective on mitochondrial systems biology
Mitochondrial biologists have accumulated an extensive knowledge of molecular composition, structure, activities and homeostatic mechanisms of mitochondria. Much of this has involved studies of mitochondrial fragments in artificial reconstituted systems in which integrated mitochondrial systems biology is largely ignored. Mitochondria are not homogenous in time or space, so understanding of mitochondrial contributions to chronic and age-related disease requires consideration of the functional mitochondriome as a collection of related particles functioning as a community of elements distributed heterogeneously within a complex organism.
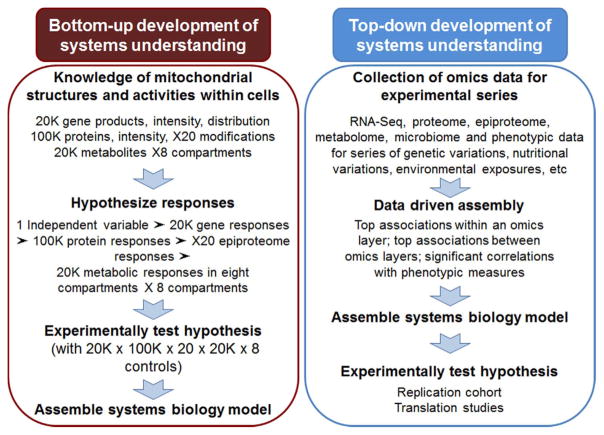
Assembly of the extensive knowledge of mitochondria into a systems view, i.e., systems biology of mitochondria, represents a new frontier. There are two engineering approaches to describe a system, termed simply “bottom-up” and “top-down” approaches (Fig. 7). In a bottom-up approach, one starts with all of the components and assembles the functional system from the components. For mitochondria, the nuclear genome encodes most of the mitochondrial proteins, so knowledge of the abundances and distributions of each of the products of each element is needed to assemble the system. This must include mRNA, microRNA, non-coding RNA, as well as subsequent translation products and modified proteins of the epiproteome. Using targeted, hypothesis driven research, this becomes a very large series of hypotheses and controls, most of which must be ignored for practical reasons (Fig. 7, left).
Fig. 7.
Integration of top-down and bottom-up approaches will facilitate development of mitochondrial systems biology. Extensive knowledge about mitochondrial structure and function is available, with tens of thousands to hundreds of thousands of experimental results from hypothesis-driven research. A bottom-up approach to assemble the extensive array of combinations and interactions of components creates a challenging task for mitochondrial systems biology. Top-down analysis of omics data provides a powerful complement to this knowledge-driven approach, and together with the mitochondrial knowledgebase, provides a foundation for rapid progress. Data-driven assembly, integration and visualization of multiple omics layers allows identification of top hubs of interaction associated with mitochondrial phenotypic measures. These hubs can be used with available experimental data to advance functional models and develop systems models for experimental testing.
In a bottom-up analysis, one quickly recognizes that the mitochondrial genome cannot be considered in isolation because the mitogenome is complemented by the functions of the nucleogenome and intestinal metagenome (Fig. 3). The connectivity of the mitochondrial proteome to the nuclear genome is directly evident from the import of proteins translated within the cytoplasm. However, the mitochondria also depend upon essential dietary components (niacin, riboflavin) as well as products of the intestinal microbiome (pantothenate). Even though assembly of molecular information provides a tractable place to begin, this rapidly becomes overly complex as one connects elements of the different mitochondrial compartments, integrates post-translational modifications of the epiproteome, or maps transmembranal metabolite distributions responding to Δψ, ΔpH, ATP or other driving force (See Fig. 2). Thus, the systems biology of mitochondria requires a holistic approach that includes diverse influences on structure and function.
An alternative top-down approach is enabled by powerful omics technologies (Fig. 7, right). Omics methods provide enhanced data capture, and powerful new data-driven bioinformatics tools allow mitochondria to be studied as integrated systems. This can be performed in both experimental and observational studies and also at cell, organ or whole organism levels. In an experimental approach, an independent variable, such as Mn, is systematically changed and associated responses are measured. This approach can be used to evaluate mitochondrial responses to nutritional, environmental or other factors. In an observational approach, a parameter such as Mn content of tissues, can be used to test for associated mitochondrial responses. The latter cannot establish cause-effect relationships, however, and care must be taken to avoid inappropriate conclusions.
xMWAS is an integrative network analysis and visualization tool that allows pairwise association analysis among elements in data arrays, including both phenotypic, functional and omics data. This data-driven approach can visualize network associations of omics data with any phenotypic or functional measure, dose-response, or time dependent process at different association thresholds [60]. Use for TMWAS in a toxicologic model provided evidence for simultaneous adverse outcome responses as well as adaptive or protective system responses (Fig. 4). Application to mitochondrial function showed separation of the most central network hubs associated with respiration from those associated with H2O2 production (Fig. 5). Separate application to physiologic and toxicologic ranges of Mn exposure showed that toxic networks are smaller and have more limited connectivity, consistent with decreased flexibility and adaptability (Fig. 6). Together, the studies show that data-driven approaches with multiomics data provide a powerful approach to understand mitochondrial systems biology. Importantly, this cannot be expected to replace targeted molecular approaches, but rather to complement these approaches to advance systems level understanding of mitochondrial biology.
Data-driven approaches to mitochondria systems biology places increased demand on the omics platforms. For instance, less than half of the high-resolution mass spectral features associated with mitochondria have accurate mass matches to known metabolites. Consequently, there is a need for chemistry and biochemistry to deliver improved curation of the mitochondrial metabolome. For the epiproteome, routine analysis provides only shallow and incomplete analysis of the broad range of post-translational modifications of the mitochondrial proteome. At the same time, subjects such as the mitochondrial glycome, proteoglycome, glycolipidome and lipidoproteome, are substantially underdeveloped. Additionally, as experimental studies of integrative omics of mitochondria accumulate, development of smart bioinformatics tools to learn common network structures will be needed to allow these to be used for predictive and therapeutic purposes.
5. Conclusion
Science is poised at the brink of mitochondrial systems biology, a point at which we begin to understand the mitochondriome as a continuous, dynamic entity within complex multicellular organisms. This is greatly facilitated by detailed knowledgebases because molecular structures and functions of elements are essential to make sense of hubs and edges in networks. Thus, extension of knowledgebases, such as MitoCarta [77], is essential to achieve a systems level understanding. At the same time, systematic studies of data-driven mitochondrial network structures are needed to assemble this knowledge according functional relationships. Each step is advancing science with strategies and tools to interrogate and visualize the complex, interactive molecular structures and functions of mitochondrial systems biology.
Acknowledgments
This study was supported by NIEHS Grants R01 ES023485 (DPJ and YMG), R21 ES025632, (DPJ and YMG), P30 ES019776, and NIH S10 OD018006.
Abbreviations
- 3-OH-BA
3-hydroxybutyric acid
- ABC
ATP-binding cassette proteins
- Cd
cadmium
- GSH
glutathione
- Mb
maneb
- Mn
manganese
- MWAS
metabolome-wide association study
- NAD
nicotinamide adenine dinucleotide
- OCR
oxygen consumption rate
- PQ
paraquat
- SOD
superoxide dismutase
- TMWAS
transcriptome-metabolome-wide association study
- Trx
thioredoxin
- Se
selenium
Footnotes
Based upon a lecture by DP Jones given at the Oxygen Club of California meeting in Berlin, Germany, June 2017.
References
- 1.Dunham-Snary KJ, Ballinger SW. Mitochondrial genetics and obesity: evolutionary adaptation and contemporary disease susceptibility. Free Radic Biol Med. 2013;65:1229–1237. doi: 10.1016/j.freeradbiomed.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill S, Van Remmen H. Mitochondrial stress signaling in longevity: a new role for mitochondrial function in aging. Redox Biol. 2014;2:936–944. doi: 10.1016/j.redox.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chacko BK, Kramer PA, Ravi S, Benavides GA, Mitchell T, Dranka BP, Ferrick D, Singal AK, Ballinger SW, Bailey SM, Hardy RW, Zhang J, Zhi D, Darley-Usmar VM. The Bioenergetic health index: a new concept in mitochondrial translational research. Clin Sci (Lond) 2014;127:367–373. doi: 10.1042/CS20140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin F, Boveris A, Cadenas E. Mitochondrial energy metabolism and redox signaling in brain aging and neurodegeneration. Antioxid Redox Signal. 2014;20:353–371. doi: 10.1089/ars.2012.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patil KR, Nielsen J. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc Natl Acad Sci USA. 2005;102:2685–2689. doi: 10.1073/pnas.0406811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones DP, Sies H. The redox code. Antioxid Redox Signal. 2015;23:734–746. doi: 10.1089/ars.2015.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 8.Krebs HA. The redox state of nicotinamide adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Adv Enzym Regul. 1967;5:409–434. doi: 10.1016/0065-2571(67)90029-5. [DOI] [PubMed] [Google Scholar]
- 9.Williamson DH, Lund P, Krebs HA. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem J. 1967;103:514–527. doi: 10.1042/bj1030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bucher T, Brauser B, Conze A, Klein F, Langguth O, Sies H. State of oxidation-reduction and state of binding in the cytosolic NADH-system as disclosed by equilibration with extracellular lactate-pyruvate in hemoglobin-free perfused rat liver. Eur J Biochem. 1972;27:301–317. doi: 10.1111/j.1432-1033.1972.tb01840.x. [DOI] [PubMed] [Google Scholar]
- 11.Sies H. Nicotinamide nucleotide compartmentation. In: Sies H, editor. Metabolic Compartmentation. Academic Press; London: 1982. pp. 205–231. [Google Scholar]
- 12.Sies H, Berndt C, Jones DP. Oxidative stress. Annu Rev Biochem. 2017;86:715–748. doi: 10.1146/annurev-biochem-061516-045037. [DOI] [PubMed] [Google Scholar]
- 13.Cortassa S, Sollott SJ, Aon MA. Mitochondrial respiration and ROS emission during beta-oxidation in the heart: an experimental-computational study. PLoS Comput Biol. 2017;13:e1005588. doi: 10.1371/journal.pcbi.1005588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Judge S, Jang YM, Smith A, Hagen T, Leeuwenburgh C. Age-associated increases in oxidative stress and antioxidant enzyme activities in cardiac interfibrillar mitochondria: implications for the mitochondrial theory of aging. FASEB J. 2005;19:419–421. doi: 10.1096/fj.04-2622fje. [DOI] [PubMed] [Google Scholar]
- 15.He Y, Ge J, Burke JM, Myers RL, Dong ZZ, Tombran-Tink J. Mitochondria impairment correlates with increased sensitivity of aging RPE cells to oxidative stress. J Ocul Biol Dis Info. 2010;3:92–108. doi: 10.1007/s12177-011-9061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaplin NL, Nieves-Cintron M, Fresquez AM, Navedo MF, Amberg GC. Arterial smooth muscle mitochondria amplify hydrogen peroxide microdomains functionally coupled to L-type calcium channels. Circ Res. 2015;117:1013–1023. doi: 10.1161/CIRCRESAHA.115.306996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gill T, Levine AD. Mitochondria-derived hydrogen peroxide selectively enhances T cell receptor-initiated signal transduction. J Biol Chem. 2013;288:26246–26255. doi: 10.1074/jbc.M113.476895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bao L, Avshalumov MV, Patel JC, Lee CR, Miller EW, Chang CJ, Rice ME. Mitochondria are the source of hydrogen peroxide for dynamic brain-cell signaling. J Neurosci. 2009;29:9002–9010. doi: 10.1523/JNEUROSCI.1706-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhuchenko O, Wehnert M, Bailey J, Sun ZS, Lee CC. Isolation, mapping, and genomic structure of an X-linked gene for a subunit of human mitochondrial complex I. Genomics. 1996;37:281–288. doi: 10.1006/geno.1996.0561. [DOI] [PubMed] [Google Scholar]
- 20.Nalecz MJ, Bolli R, Azzi A. Molecular conversion between monomeric and dimeric states of the mitochondrial cytochrome b-c1 complex: isolation of active monomers. Arch Biochem Biophys. 1985;236:619–628. doi: 10.1016/0003-9861(85)90666-6. [DOI] [PubMed] [Google Scholar]
- 21.Sato R, Atsuta Y, Imai Y, Taniguchi S, Okuda K. Hepatic mitochondrial cytochrome P-450: isolation and functional characterization. Proc Natl Acad Sci USA. 1977;74:5477–5481. doi: 10.1073/pnas.74.12.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schagger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pollak JK. The maturation of the inner membrane of foetal rat liver mitochondria. Biochem J. 1975;150:477–488. doi: 10.1042/bj1500477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang M, Mileykovskaya E, Dowhan W. Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J Biol Chem. 2005;280:29403–29408. doi: 10.1074/jbc.M504955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slatter DA, Aldrovandi M, O’Connor A, Allen SM, Brasher CJ, Murphy RC, Mecklemann S, Ravi S, Darley-Usmar V, O’Donnell VB. Mapping the human platelet lipidome reveals cytosolic phospholipase A2 as a regulator of mitochondrial bioenergetics during activation. Cell Metab. 2016;23:930–944. doi: 10.1016/j.cmet.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirkinezos IG, Bacman SR, Hernandez D, Oca-Cossio J, Arias LJ, Perez-Pinzon MA, Bradley WG, Moraes CT. Cytochrome c association with the inner mitochondrial membrane is impaired in the CNS of G93A-SOD1 mice. J Neurosci. 2005;25:164–172. doi: 10.1523/JNEUROSCI.3829-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mikhailov V, Mikhailova M, Degenhardt K, Venkatachalam MA, White E, Saikumar P. Association of Bax and Bak homo-oligomers in mitochondria. Bax requirement for Bak reorganization and cytochrome c release. J Biol Chem. 2003;278:5367–5376. doi: 10.1074/jbc.M203392200. [DOI] [PubMed] [Google Scholar]
- 28.Go YM, Jones DP. Thiol/disulfide redox states in signaling and sensing. Crit Rev Biochem Mol Biol. 2013;48:173–181. doi: 10.3109/10409238.2013.764840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalle-Donne I, Rossi R, Giustarini D, Colombo R, Milzani A. S-glutathionylation in protein redox regulation. Free Radic Biol Med. 2007;43:883–898. doi: 10.1016/j.freeradbiomed.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 30.Hess DT, Stamler JS. Regulation by S-nitrosylation of protein post-translational modification. J Biol Chem. 2012;287:4411–4418. doi: 10.1074/jbc.R111.285742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tebay LE, Robertson H, Durant ST, Vitale SR, Penning TM, Dinkova-Kostova AT, Hayes JD. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic Biol Med. 2015;88:108–146. doi: 10.1016/j.freeradbiomed.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hwang JW, Yao H, Caito S, Sundar IK, Rahman I. Redox regulation of SIRT1 in inflammation and cellular senescence. Free Radic Biol Med. 2013;61:95–110. doi: 10.1016/j.freeradbiomed.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Becker K, Gromer S, Schirmer RH, Muller S. Thioredoxin reductase as a patho-physiological factor and drug target. Eur J Biochem. 2000;267:6118–6125. doi: 10.1046/j.1432-1327.2000.01703.x. [DOI] [PubMed] [Google Scholar]
- 34.Tseng AH, Shieh SS, Wang DL. SIRT3 deacetylates FOXO3 to protect mitochondria against oxidative damage. Free Radic Biol Med. 2013;63:222–234. doi: 10.1016/j.freeradbiomed.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Poulose N, Raju R. Sirtuin regulation in aging and injury. Biochim Biophys Acta. 2015;1852:2442–2455. doi: 10.1016/j.bbadis.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osborne B, Cooney GJ, Turner N. Are sirtuin deacylase enzymes important modulators of mitochondrial energy metabolism? Biochim Biophys Acta. 2014;1840:1295–1302. doi: 10.1016/j.bbagen.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 37.Pfanner N, Geissler A. Versatility of the mitochondrial protein import machinery. Nat Rev Mol Cell Biol. 2001;2:339–349. doi: 10.1038/35073006. [DOI] [PubMed] [Google Scholar]
- 38.Dudek J, Rehling P, van der Laan M. Mitochondrial protein import: common principles and physiological networks. Biochim Biophys Acta. 2013;1833:274–285. doi: 10.1016/j.bbamcr.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 39.Becker T, Bottinger L, Pfanner N. Mitochondrial protein import: from transport pathways to an integrated network. Trends Biochem Sci. 2012;37:85–91. doi: 10.1016/j.tibs.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Harbauer AB, Zahedi RP, Sickmann A, Pfanner N, Meisinger C. The protein import machinery of mitochondria-a regulatory hub in metabolism, stress, and disease. Cell Metab. 2014;19:357–372. doi: 10.1016/j.cmet.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 41.Schaedler TA, Thornton JD, Kruse I, Schwarzlander M, Meyer AJ, van Veen HW, Balk J. A conserved mitochondrial ATP-binding cassette transporter exports glutathione polysulfide for cytosolic metal cofactor assembly. J Biol Chem. 2014;289:23264–23274. doi: 10.1074/jbc.M114.553438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clemencon B, Babot M, Trezeguet V. The mitochondrial ADP/ATP carrier (SLC25 family): pathological implications of its dysfunction. Mol Asp Med. 2013;34:485–493. doi: 10.1016/j.mam.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 43.Taylor EB. Functional properties of the mitochondrial carrier system. Trends Cell Biol. 2017;27:633–644. doi: 10.1016/j.tcb.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson AJ, Overy C, Kunji ER. The mechanism of transport by mitochondrial carriers based on analysis of symmetry. Proc Natl Acad Sci USA. 2008;105:17766–17771. doi: 10.1073/pnas.0809580105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmieri F. Mitochondrial transporters of the SLC25 family and associated diseases: a review. J Inherit Metab Dis. 2014;37:565–575. doi: 10.1007/s10545-014-9708-5. [DOI] [PubMed] [Google Scholar]
- 46.Gredilla R. DNA damage and base excision repair in mitochondria and their role in aging. J Aging Res. 2010;2011:257093. doi: 10.4061/2011/257093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boesch P, Weber-Lotfi F, Ibrahim N, Tarasenko V, Cosset A, Paulus F, Lightowlers RN, Dietrich A. DNA repair in organelles: pathways, organization, regulation, relevance in disease and aging. Biochim Biophys Acta. 2011;1813:186–200. doi: 10.1016/j.bbamcr.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 48.Zimmermann M, Reichert AS. How to get rid of mitochondria: crosstalk and regulation of multiple mitophagy pathways. Biol Chem. 2017;399:29–45. doi: 10.1515/hsz-2017-0206. [DOI] [PubMed] [Google Scholar]
- 49.Kubli DA, Gustafsson AB. Mitochondria and mitophagy: the yin and yang of cell death control. Circ Res. 2012;111:1208–1221. doi: 10.1161/CIRCRESAHA.112.265819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bota DA, Davies KJ. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat Cell Biol. 2002;4:674–680. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]
- 51.Go YM, Jones DP. The redox proteome. J Biol Chem. 2013;288:26512–26520. doi: 10.1074/jbc.R113.464131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Go YM, Jones DP. Redox biology: interface of the exposome with the proteome, epigenome and genome. Redox Biol. 2014;2:358–360. doi: 10.1016/j.redox.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 54.Yin F, Sancheti H, Cadenas E. Mitochondrial thiols in the regulation of cell death pathways. Antioxid Redox Signal. 2012;17:1714–1727. doi: 10.1089/ars.2012.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swain L, Kesemeyer A, Meyer-Roxlau S, Vettel C, Zieseniss A, Guntsch A, Jatho A, Becker A, Nanadikar MS, Morgan B, Dennerlein S, Shah AM, El-Armouche A, Nikolaev VO, Katschinski DM. Redox imaging using cardiac myocyte-specific transgenic biosensor mice. Circ Res. 2016;119:1004–1016. doi: 10.1161/CIRCRESAHA.116.309551. [DOI] [PubMed] [Google Scholar]
- 56.Go YM, Jones DP. Redox compartmentalization in eukaryotic cells. Biochim Biophys Acta. 2008;1780:1273–1290. doi: 10.1016/j.bbagen.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Go YM, Ziegler TR, Johnson JM, Gu L, Hansen JM, Jones DP. Selective protection of nuclear thioredoxin-1 and glutathione redox systems against oxidation during glucose and glutamine deficiency in human colonic epithelial cells. Free Radic Biol Med. 2007;42:363–370. doi: 10.1016/j.freeradbiomed.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hansen JM, Zhang H, Jones DP. Differential oxidation of thioredoxin-1, thioredoxin-2, and glutathione by metal ions. Free Radic Biol Med. 2006;40:138–145. doi: 10.1016/j.freeradbiomed.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 59.Roede JR, Uppal K, Park Y, Tran V, Jones DP. Transcriptome-metabolome wide association study (TMWAS) of maneb and paraquat neurotoxicity reveals network level interactions in toxicologic mechanism. Toxicol Rep. 2014;1:435–444. doi: 10.1016/j.toxrep.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uppal K, Ma C, Go YM, Jones DP. xMWAS: a data-driven integration and differential network analysis tool. Bioinformatics. 2017 doi: 10.1093/bioinformatics/btx656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Go YM, Sutliff RL, Chandler JD, Khalidur R, Kang BY, Anania FA, Orr M, Hao L, Fowler BA, Jones DP. Low-dose cadmium causes metabolic and genetic dysregulation associated with fatty liver disease in mice. Toxicol Sci. 2015;147:524–534. doi: 10.1093/toxsci/kfv149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Go YM, Roede JR, Orr M, Liang Y, Jones DP. Integrated redox proteomics and metabolomics of mitochondria to identify mechanisms of cd toxicity. Toxicol Sci. 2014;139:59–73. doi: 10.1093/toxsci/kfu018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Niki Y, Chigusa SI, Matsuura ET. Complete replacement of mitochondrial DNA in Drosophila. Nature. 1989;341:551–552. doi: 10.1038/341551a0. [DOI] [PubMed] [Google Scholar]
- 64.Latorre-Pellicer A, Moreno-Loshuertos R, Lechuga-Vieco AV, Sanchez-Cabo F, Torroja C, Acin-Perez R, Calvo E, Aix E, Gonzalez-Guerra A, Logan A, Bernad-Miana ML, Romanos E, Cruz R, Cogliati S, Sobrino B, Carracedo A, Perez-Martos A, Fernandez-Silva P, Ruiz-Cabello J, Murphy MP, Flores I, Vazquez J, Enriquez JA. Mitochondrial and nuclear DNA matching shapes metabolism and healthy ageing. Nature. 2016;535:561–565. doi: 10.1038/nature18618. [DOI] [PubMed] [Google Scholar]
- 65.Ishikawa H, Takaki A, Tsuzaki R, Yasunaka T, Koike K, Shimomura Y, Seki H, Matsushita H, Miyake Y, Ikeda F, Shiraha H, Nouso K, Yamamoto K. L-carnitine prevents progression of non-alcoholic steatohepatitis in a mouse model with up-regulation of mitochondrial pathway. PLoS One. 2014;9:e100627. doi: 10.1371/journal.pone.0100627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mollica MP, Mattace Raso G, Cavaliere G, Trinchese G, De Filippo C, Aceto S, Prisco M, Pirozzi C, Di Guida F, Lama A, Crispino M, Tronino D, Di Vaio P, Berni Canani R, Calignano A, Meli R. Butyrate regulates liver mitochondrial function, efficiency, and dynamics in insulin-resistant obese mice. Diabetes. 2017;66:1405–1418. doi: 10.2337/db16-0924. [DOI] [PubMed] [Google Scholar]
- 67.Johnson JM, Yu T, Strobel FH, Jones DP. A practical approach to detect unique metabolic patterns for personalized medicine. Analyst. 2010;135:2864–2870. doi: 10.1039/c0an00333f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Go YM, Uppal K, Jones DP. Central mitochondrial signaling mechanisms in response to environmental agents: Integrated omics for visualizatiion. In: Will Y, Dykens JA, editors. Drug-induced mitochondrial toxicity. 2017. [Google Scholar]
- 69.Roede JR, Hansen JM, Go YM, Jones DP. Maneb and paraquat-mediated neurotoxicity: involvement of peroxiredoxin/thioredoxin system. Toxicol Sci. 2011;121:368–375. doi: 10.1093/toxsci/kfr058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wenzel SE, Tyurina YY, Zhao J, St Croix CM, Dar HH, Mao G, Tyurin VA, Anthonymuthu TS, Kapralov AA, Amoscato AA, Mikulska-Ruminska K, Shrivastava IH, Kenny EM, Yang Q, Rosenbaum JC, Sparvero LJ, Emlet DR, Wen X, Minami Y, Qu F, Watkins SC, Holman TR, VanDemark AP, Kellum JA, Bahar I, Bayir H, Kagan VE. PEBP1 wardens ferroptosis by enabling lipoxygenase generation of lipid death signals (e626) Cell. 2017;171:628–641. e626. doi: 10.1016/j.cell.2017.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Loscalzo J, Kohane I, Barabasi AL. Human disease classification in the post-genomic era: a complex systems approach to human pathobiology. Mol Syst Biol. 2007;3:124. doi: 10.1038/msb4100163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith MR, Fernandes J, Go YM, Jones DP. Redox dynamics of manganese as a mitochondrial life-death switch. Biochem Biophys Res Commun. 2017;482:388–398. doi: 10.1016/j.bbrc.2016.10.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aschner M, Erikson KM, Herrero Hernandez E, Tjalkens R. Manganese and its role in Parkinson’s disease: from transport to neuropathology. Neuromolecular Med. 2009;11:252–266. doi: 10.1007/s12017-009-8083-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cersosimo MG, Koller WC. The diagnosis of manganese-induced parkinsonism. Neurotoxicology. 2006;27:340–346. doi: 10.1016/j.neuro.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 75.Fernandes J, Hao L, Bijli KM, Chandler JD, Orr M, Hu X, Jones DP, Go YM. From the cover: manganese stimulates mitochondrial H2O2 production in SH-SY5Y human neuroblastoma cells Over Physiologic as well as Toxicologic range. Toxicol Sci. 2017;155:213–223. doi: 10.1093/toxsci/kfw196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones DP. Redox theory of aging. Redox Biol. 2015;5:71–79. doi: 10.1016/j.redox.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lipmann F. Metabolic Generation and Utilization of Phosphate Bond Energy. Interscience Publishers; New York: 1941. [Google Scholar]