Bacteria within the skin microbiome of some individuals produce an antimetabolite that inhibits tumor growth.
Abstract
We report the discovery that strains of Staphylococcus epidermidis produce 6-N-hydroxyaminopurine (6-HAP), a molecule that inhibits DNA polymerase activity. In culture, 6-HAP selectively inhibited proliferation of tumor lines but did not inhibit primary keratinocytes. Resistance to 6-HAP was associated with the expression of mitochondrial amidoxime reducing components, enzymes that were not observed in cells sensitive to this compound. Intravenous injection of 6-HAP in mice suppressed the growth of B16F10 melanoma without evidence of systemic toxicity. Colonization of mice with an S. epidermidis strain producing 6-HAP reduced the incidence of ultraviolet-induced tumors compared to mice colonized by a control strain that did not produce 6-HAP. S. epidermidis strains producing 6-HAP were found in the metagenome from multiple healthy human subjects, suggesting that the microbiome of some individuals may confer protection against skin cancer. These findings show a new role for skin commensal bacteria in host defense.
INTRODUCTION
Mammalian skin harbors diverse microbial communities whose growth is influenced by ecological factors on the body surface such as humidity, temperature, pH, lipid content, and the presence of antimicrobials produced by the host (1). Coagulase-negative staphylococcal species, such as Staphylococcus epidermidis, Staphylococcus hominis, and others, are the predominant bacterial species that colonize normal human skin (2), whereas Staphylococcus aureus frequently colonizes abnormal skin such as that found in patients with atopic dermatitis (3). Some of these organisms can alter epidermal function (4, 5) and, thus, interact with the host to maintain or disrupt the local ecology of the skin (1, 6). This interplay of microbe and host appears to be critical for establishing homeostasis but is poorly understood.
Although the specific mechanisms through which skin surface microbes influence host function are incompletely understood, specific strains of coagulase-negative staphylococcal species have been shown to produce proteins that work together with endogenous host antimicrobial peptides (AMPs) to provide direct protection against infectious pathogens (7–9). For example, the production of phenol-soluble modulins (PSMγ and PSMδ) by S. epidermidis can selectively kill bacterial pathogens such as S. aureus and group A Streptococcus (GAS) (7, 8). This species has also been shown to benefit skin immune function by diminishing inflammation after injury (10), enhancing development of cutaneous T cells (11–13) and promoting expression of host AMPs such as cathelicidins and β-defensins (11, 12, 14–16). Germ-free mice are more susceptible to skin infection than mice maintained under specific pathogen-free conditions or monoassociated with S. epidermidis (12). Further evidence that commensal Staphylococcus species provide host defense has come from observations that nasal colonization with either a specific strain of S. epidermidis that produces a serine protease or a strain of Staphylococcus lugdunensis that produces a thiazolidine-containing cyclic peptide can inhibit nasal colonization by S. aureus (17, 18). More recently, several strains of S. epidermidis, S. hominis, and other coagulase-negative staphylococcal species that produce a variety of previously unknown AMPs were found to be deficient in atopic dermatitis patients colonized by S. aureus, and a clinical trial evaluating the effect of reintroduction of these strains demonstrated that they directly reduced S. aureus colonization on humans (9). Thus, evidence is increasing that the skin microbiome has an important role in promoting host defense.
Here, we report the results of a continued molecular analysis of the metabolic products of human skin commensal bacteria. Unexpectedly, specific strains of S. epidermidis were observed to produce a nucleobase analog with the capacity to inhibit DNA synthesis. When administered intravenously or topically applied to mice, this molecule or the live S. epidermidis strain itself suppressed tumor growth in vivo. These observations suggest that the skin microbiome may contribute to aspects of host defense that include resistance to tumor growth.
RESULTS
A strain of S. epidermidis produces 6-N-hydroxyaminopurine
In the course of testing coagulase-negative Staphylococcus strains for antimicrobial activity isolated from healthy human skin (9), we identified a strain of S. epidermidis that secreted a bactericidal activity against GAS. However, unlike other organisms identified with this bioactivity, the activity from this strain was heat-stable and protease-insensitive, suggesting that this factor was not a protein (Fig. 1, A and B). Purification was performed through five steps, yielding a single peak associated with antimicrobial activity on high-performance liquid chromatography (HPLC) (fig. S1). The final yield of purified compound was 7 mg from 6.4 liters of culture supernatant. The purified molecule had potent capacity to inhibit growth of GAS in a dose-dependent manner (Fig. 1C). High-resolution electrospray ionization mass spectrometry (HR-ESI-MS) analysis deduced its molecular formula as C5H5N5O (found, 151.0487; calculated, 151.0489) (fig. S2). To determine whether this was a de novo synthetic product of this S. epidermidis strain, we cultured the bacterium in the presence of ammonium-15N chloride. Incorporation of the 15N isotope confirmed that the molecule was produced via de novo synthesis and not by fermentation or breakdown of components in the culture media (Fig. 1D). The proton nuclear magnetic resonance (1H NMR) spectrum of the purified compound displayed two proton signals in the aromatic region (δH = 8.19 and 8.17), whereas six signals in dimethyl sulfoxide (DMSO)–d5 (δH = 12.74, 1H; 10.87 0.7H; 9.50, 1H; 8.08, 1H; 7.75, 1H; and 7.48, 0.4H) (fig. S3A). The gradient-selected heteronuclear multiple-bond correlation (gHMBC) spectrum of purified compound revealed five carbon signals in the aromatic region (δC = 113.60, 144.94, 148.17, 150.28, and 150.45) (fig. S4). The combined data from 1H NMR and gHMBC spectra displayed the presence of a purine moiety with an additional oxygen atom attached to one of the five nitrogen atoms. Given the chemical formula as C5H5N5O, the structure was predicted as 6-HAP (Fig. 1E). Chemical synthesis of 6-HAP and subsequent structural analysis and comparison of bioactivity between purified and synthetic material (figs. S3B and S5) supported the conclusion that 6-HAP was made by this S. epidermidis strain.
Fig. 1. S. epidermidis strains isolated from normal human skin produce 6-HAP.
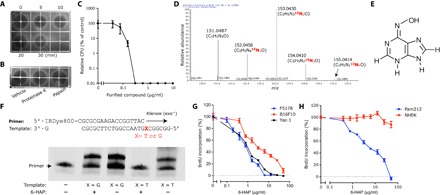
(A and B) Stability of antimicrobial molecules from S. epidermidis against GAS after heat-treatment for the indicated time (A) and incubation with indicated protease (B). The black area represents zone of growth inhibition of GAS. (C) Dose-dependent antimicrobial activity of the purified antimicrobial compound against GAS. Data are means ± SEM of three individual experiments. CFU, colony-forming unit. (D) 15N isotope incorporation into the antibiotic molecule after culturing S. epidermidis MO34 in tryptic soy broth (TSB) containing ammonium-15N chloride (12.5 mM). (E) The determined chemical structure of the active molecule, 6-HAP. (F) Capacity of 6-HAP to block in vitro DNA extension by Klenow fragment polymerase. A template that required adenosine (X = T) or cytidine (X = G) at the initial base for extension was used. (G) 5-Bromo-2′-deoxyuridine (BrdU) incorporation into tumor cell line, L5178, YAC-1 lymphoma, B16F10 melanoma, and Pam212 SCC after a 24-hour incubation in suitable media containing indicated concentrations of 6-HAP. Data are means ± SEM of four individual experiments. (H) BrdU incorporation into nontransformed human keratinocytes (NHEKs) and Pam212 cutaneous squamous cell carcinoma after 24-hour incubation in suitable media containing indicated concentrations of 6-HAP. Data are means ± SEM of four individual experiments.
6-HAP exerts selective antiproliferative activity against tumor cell lines
Given that the structure of 6-HAP was similar to adenine, we next investigated the influence of 6-HAP on DNA synthesis. To directly examine the action of 6-HAP on DNA polymerase, we designed a 25-bp (base pair) template and matching 18-bp fluorescence primer to measure DNA extension in vitro by Klenow (exo−) DNA polymerase. In the presence of 6-HAP, DNA extension occurred normally when the template required cytosine for extension (X = G) but was blocked when adenosine was required (X = T) (Fig. 1F). These data show that 6-HAP directly inhibits DNA synthesis by interfering with adenosine-thymidine base pairing. Consistent with this observation, 6-HAP inhibited BrdU incorporation in several tumor cell lines, including B16F10 melanoma and L5178 and YAC-1 lymphoma (Fig. 1G). By contrast, NHEKs were not affected by 6-HAP (Fig. 1H). These data suggest that 6-HAP exerts selective antiproliferative activity against several tumor cells.
Selective antiproliferative activity of 6-HAP is mediated by mARCs
To understand why 6-HAP showed selective inhibition of transformed cells, we first examined the potential for selective cell penetration. 6-HAP did not directly affect plasma membrane permeability of the primary or transformed cell lines tested (fig. S6). Next, we evaluated the potential of target cells to metabolize 6-HAP. The expression of mitochondrial amidoxime reducing components (mARC1 and mARC2) was much higher in primary cultured skin cells than in cancer cell lines that were sensitive to 6-HAP (Fig. 2A). mARCs 1 and 2 reduce N-hydroxylated nucleobase analogs to nucleobases (19). To validate whether this expression of mARCs in the primary cells (NHEK) conferred resistance to 6-HAP, we performed gene silencing with small interfering RNA (siRNA) to mARC1 and mARC2 (Fig. 2B). This partially eliminated resistance to 6-HAP (Fig. 2C) and suggested that the expression of mARC1 or mARC2 can partially explain selective resistance in nontransformed keratinocytes.
Fig. 2. Selective antiproliferative activity of 6-HAP is mediated by mARCs.
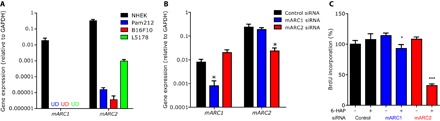
(A) Expression of mARC1 and mARC2 in NHEKs, squamous cell carcinoma (Pam212), melanoma (B16F10), and lymphoma cell lines (L5178). Data are means ± SEM of five individual experiments. UD, undetectable. (B) Expression of mARC1 and mARC2 in NHEKs treated with control siRNA, mARC1 siRNA, and mARC2 siRNA. Data are shown as relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression and represent means ± SEM of six independent assays. (C) Effect of gene silencing with mARC1 and mARC2 siRNA on sensitivity to 6-HAP in NHEKs. Data are means ± SEM of eight individual experiments (*P < 0.05, ***P < 0.0001 by two-tailed independent t test).
Some nucleobase analogs have the potential to exert mutagenic activity. Previous studies of 6-HAP derived from synthetic chemical libraries of N-hydroxylated derivatives of purines concluded that it could induce gene mutations (20, 21). However, these studies did not test for tautomeric isoforms. Therefore, to more carefully evaluate mutagenic potential with defined 6-HAP, we measured mutagenic events by two independent approaches: a well-established assay of the thymidine kinase (tk) locus of L5178Y tk+/− mouse lymphoma cells (22) and the Ames test in Salmonella typhimurium (TA100). Neither sensitive assay of mutagenesis detected mutagenic activity for 6-HAP (table S1, A and B).
6-HAP selectively suppresses melanoma in mice
We next evaluated the systemic activity of 6-HAP in vivo. Toxicity was first assessed in mice by administration of repeated intravenous injections of 6-HAP at the maximal soluble dose of 20 mg/kg every 48 hours for 2 weeks. This resulted in no apparent systemic toxic effects as assessed by visual appearance and weight (Fig. 3A), and no change was observed in complete blood count or liver function as assessed by aspartate aminotransferase activity (table S2). Given the apparent lack of systemic toxicity, 6-HAP was administered intravenously to mice inoculated with the rapidly growing isogenic melanoma cell line B16F10. Tumor size of this aggressively growing tumor was suppressed by >60% in mice receiving 6-HAP compared to those that received injections with vehicle alone (Fig. 3, B and C).
Fig. 3. Systemic administration of 6-HAP suppresses melanoma growth in mice.
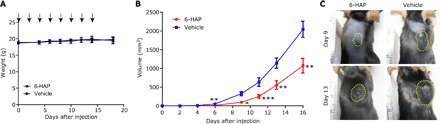
(A) Systemic toxicity of repeated intravascular administration with 6-HAP (20 mg/kg) or with an equal volume of vehicle (2.5% DMSO in 0.9% NaCl) every 48 hours for 2 weeks (arrows) in mice. To observe toxicity of 6-HAP, we determined mouse weight at the indicated time points. Data are means ± SEM of 10 mice. (B and C) Effect of repeated intravascular administrations with 6-HAP on growth of melanoma in mice. Data are means ± SEM from 10 individual mice (*P < 0.05, **P < 0.01, and ***P < 0.001 by two-tailed independent t test versus vehicle control) (B). Representative images of tumor (yellow dashed line) in mouse treated with 6-HAP or vehicle at day 9 and day 13 are shown (C).
S. epidermidis strain producing 6-HAP suppresses UV-induced skin tumor in mice
Our observation that 6-HAP was constitutively produced by a skin commensal bacterium led us to hypothesize that the presence of these strains on the skin surface could be protective against skin tumor formation. To address this hypothesis, we used a two-stage ultraviolet (UV) carcinogenesis model (23, 24). SKH-1 hairless mice were treated with 7,12-dimethylbenz[a]anthracene (DMBA) for 1 week, followed by UV-B irradiation twice a week. During UV exposure, these mice were colonized by topical application of either a strain of S. epidermidis that produced 6-HAP or equal amounts of S. epidermidis 1457 that was used as a control. The density of S. epidermidis applied was similar to that found on normal human skin (25). Mice colonized with the control strain of S. epidermidis had the expected high incidence of tumor formation (Fig. 4, A to C). By contrast, mice colonized with S. epidermidis that produced 6-HAP had a significantly decreased incidence and number of tumors (Fig. 4, A, B, and D). Histopathogenic examination of samples of 81 of these tumors in 19 control mice identified all tumors as squamous papillomas (Fig. 4E). Microscopic evidence of papilloma formation was not seen in normal-appearing skin of mice that received epicutaneous application of the 6-HAP–producing strain (Fig. 4F). S. epidermidis was observed to enter the skin as previously described (25), and both the control and the 6-HAP–producing strains were seen within the dermis at similar frequencies (Fig. 4, G and H). The 6-HAP–producing strain of S. epidermidis and the control 1457 strain equally induced a small increase in expression of Il-6, Cxcl2, and Camp in the skin (fig. S7). This suggested that the antitumor activity of the 6-HAP–producing MO34 strain was not due to a difference in immunomodulatory activity from this strain compared to control.
Fig. 4. Skin colonization by S. epidermidis strain producing 6-HAP protects from UV-induced neoplasia in mice.
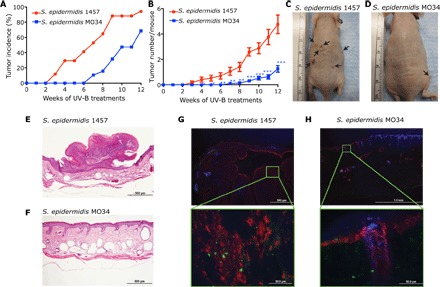
(A to D) Effect of colonization by S. epidermidis MO34 strain producing 6-HAP on tumor incidence (A) and number (B) in SKH-1 hairless mice treated with DMBA, followed by repeated UV-B irradiation. S. epidermidis 1457 was used as a control strain that does not produce 6-HAP. Tumor incidence and tumor number in each mouse were recorded every week. Data are means ± SEM of 19 mice (*P < 0.05, **P < 0.01, and ***P < 0.001 by two-tailed independent t test). Representative images of UV-induced tumor formation in mouse treated with S. epidermidis 1457 (C) or MO34 (D) at week 12 are shown. (E and F) A representative hematoxylin and eosin staining of UV-induced skin tumor or skin obtained from SKH-1 mice colonized by S. epidermidis 1457 (E) or MO34 (F), respectively, treated with UV-B for 12 weeks. (G and H) Immunostaining for S. epidermidis (green) and keratin-14 (red) in the UV-induced tumor or skin of SKH-1 mice treated with S. epidermidis 1457 (G) or MO34 (H), respectively. The sections were counter stained with 4′,6-diamidino-2-phenylindole (blue).
S. epidermidis strains producing 6-HAP are common in human skin
6-HAP was detected by HPLC of culture supernatant from two distinct clinical isolates of S. epidermidis but not from the reference laboratory S. epidermidis strains ATCC12228 and 1457 strains (Fig. 5, A to D). This suggested that 6-HAP production was not rare but universal among all S. epidermidis strains. To further explore the frequency of 6-HAP production in human commensal S. epidermidis stains, we performed and used whole-genome sequencing of the MO34 strain for analysis of existing metagenomic data sets of the human skin microbiome (26). Sequence analysis frequently identified S. epidermidis strains similar to the 6-HAP–producing isolate within the human skin microbiome but detected similar strains at a different frequency at distinct body sites (Fig. 5E).
Fig. 5. S. epidermidis strain producing 6-HAP is commonly distributed on the human skin.
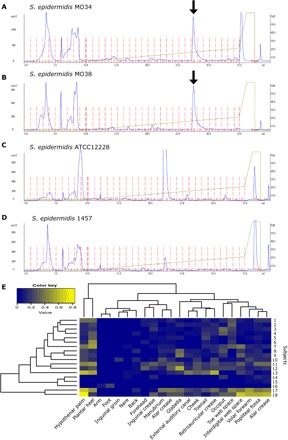
(A to D) Productions of 6-HAP by skin isolate strains, MO34 (A) and MO38 (B), and laboratory strains of S. epidermidis, ATCC12228 (C) and 1457 (D). Production of 6-HAP was evaluated by HPLC. Arrow indicates elution time of 6-HAP. The data are representative of three independent experiments. mAU, milli absorbance units. (E) Heat map showing relative abundance of putative S. epidermidis strains producing 6-HAP in the metagenome of skin microbiome samples from 22 distinct body sites of 18 healthy subjects.
DISCUSSION
Here, we describe identification of unique strains of S. epidermidis that produce a chemical compound that impairs tumor growth. Our data suggest that this compound, 6-HAP, inhibits DNA synthesis and has selective antiproliferative function against transformed tumor cell lines, as well as the ability to suppress growth induced de novo by UV exposure. This observation suggests that commensal skin bacteria may assist in defending the host against neoplasia of the skin.
Biosynthesis of 6-HAP likely requires successive oxidation of the amine group at the carbon C-6 of adenine to N-hydroxyl group, as well as several additional catalytic steps that involve multiple enzymes such as flavin-dependent N-oxygenases, cytochrome P450s, and iron- and copper-containing N-oxygenases (27). Further studies are required to understand the biosynthetic pathway of 6-HAP and the mechanisms by which 6-HAP production is regulated. 6-HAP appears to act by competing with adenine to inhibit DNA synthesis, a well-known antiproliferative mechanism for nucleobase analogs. For example, 6-mercaptopurine is converted in vivo to 6-thioguanine and is then incorporated into DNA in place of guanine (28). 8-Azaguanine also suppresses DNA synthesis by a similar mechanism (29). Nucleobase analogs constitute an important class of antimetabolites used for the treatment of hematological malignancies and solid tumors (30). In 6-HAP, the amino group at the carbon C-6 position of the purine ring is replaced with an N-hydroxyl group. This is a critical position for DNA synthesis because the hydrogen of the amino group at the carbon C-6 position of adenine is required to bind with oxygen at the carbon C-4 position of thymine (31).
The mechanism for selective activity of 6-HAP has not been definitively proven for all cell types, but our data suggest that reduction of the N-hydroxyl group by mARC enzymes can rescue primary keratinocytes from the cytotoxic activity of 6-HAP. Previous reports have also reported that activity of synthetic 6-HAP against HeLa cells could be repaired by inosine triphosphate pyrophosphatase (32). Other studies have also reported that synthetic 6-HAP exhibited no toxicity to a wild-type strain of Escherichia coli, whereas it inhibited growth of mutant strains deficient in genes involved in molybdenum cofactors (33, 34). This suggests that molybdoenzymes such as xanthine oxidase may also be capable of detoxifying 6-HAP to protect some cells. Thus, it is likely that multiple mechanisms for inactivation of 6-HAP occur in nature, a finding consistent with our discovery that bacteria producing this molecule are commonly present in the normal skin microbiome. The inability of transformed cells to inactivate the compound is the most likely explanation for selective sensitivity to 6-HAP, although additional work is required to firmly establish this conclusion. Given the selective antiproliferative activity on tested cancer cell lines, 6-HAP may have therapeutic potential as a selective antimetabolite for some tumors.
No mutagenic activity could be detected in 6-HAP produced by S. epidermidis or in our preparations made by chemical synthesis. However, previous studies of 6-HAP derived from synthetic chemical libraries of N-hydroxylated derivatives of purines concluded that it could induce mutations in yeast and bacterial cells (20, 21). Because our results in standard mutagenesis assays did not agree with these previous observations, we speculate that the previous positive results may have been due to the presence of tautomeric isoforms of 6-HAP that were not detected in the former synthetic preparations. Migration of hydrogen accompanied by a switch of the adjacent double bond would not have been detected by mass analysis. Therefore, we believe it is highly unlikely that this common skin commensal produces a potent mutagen. However, if this was the case, then the current observations would remain highly significant because this would identify a previously undetected risk factor for cancer. These observations of the capacity of a commensal skin microbe to produce a 6-HAP are important either way and need further evaluation.
Previous observations have reported that dysbiosis (a state of altered microbiome) may promote cancer. For example, observations associating bacteria in the gut with an increase in carcinogenesis suggested that this effect was dependent on inflammation (35, 36). Intestinal inflammation has also been reported to promote development of tumors through increasing the capacity of microbiota to produce genotoxins that elicit DNA damage (37). The present findings suggest an entirely new concept that some members of our skin microbiome may suppress tumor growth, and dysbiosis is potentially detrimental because of loss of a protective function instead of (or in addition to) a gain of a detrimental microbial community. Other observations that support this perspective include previous observations from the intestinal microbiome that suggested microbes may suppress tumor growth by the production of short-chain free fatty acids (38–40). In addition, it has been demonstrated that skin microflora potentially produces cis-urocanic acid by degrading l-histidine, which affects immune suppression induced by UV exposure and suppresses melanoma growth (41, 42). Overall, in addition to the well-known mutagenic potential of several human viral pathogens, several lines of evidence support the concept that skin bacteria could promote tumor development. This study now shows for the first time, to our knowledge, that a skin bacterium could protect against neoplasia. It should be a high priority to better define the molecular basis for this association to establish causation.
In conclusion, we report here the discovery that a strain of S. epidermidis, a common microbe on healthy human skin, produces 6-HAP. This interesting small molecule inhibits DNA synthesis and has the potential to convey protection against neoplasia. A beneficial role for skin bacteria in host defense is consistent with observations of a role for commensal bacteria to resist S. aureus infection (9) but extends this concept to host defense functions against cancer. Further study is needed to examine whether a loss of S. epidermidis strains producing 6-HAP increases a risk of skin cancer in humans or could be used as a preventative treatment.
MATERIALS AND METHODS
Characterization and purification of 6-HAP
S. epidermidis MO34 strain was cultured in TSB at 37°C for 24 hours and was removed from culture supernatant with 0.22-μm filter. To examine heat stability and resistance to protease, we treated the filtrated conditioned media at 100°C for 0, 5, 10, 20, and 30 min or incubated it with proteinase K or papain (2 mg/ml) at 37°C for 3 hours, followed by a 5-min incubation at 90°C to inactivate enzyme. The antimicrobial activity in the treated conditioned media was measured by radial diffusion assay as described below. To isolate the antimicrobial component, we lyophilized the filtrated culture supernatant, and we suspended the residue in methanol to precipitate proteins. The supernatant was dried under vacuum, and the residual substance was dissolved in water. Because 6-HAP is weakly retained in the C18 reverse phase column, the solution was applied on Sep-Pak cartridge (Waters Co.), washed with H2O, and eluted with 5% acetonitrile in H2O. The elution was lyophilized and suspended in 90% acetonitrile/10% water. The supernatant was separated by HPLC. After each purification step, activity was determined by radial diffusion assay against GAS (NZ131). The purified compound was characterized by mass spectrometry and NMR. Purified 6-HAP was lyophilized, and dry weight was measured to measure specific activity.
Antimicrobial assays
Radial diffusion assay was performed using GAS as previously described (43). Briefly, melted Todd-Hewitt broth agar (10 ml) was mixed with GAS (1 × 106 CFU) and poured in a 10-cm petri dish. Two to 4 μl of test samples was applied in a small well punched on the agar plate. Plates were incubated at 37°C overnight to allow visible growth of bacteria. Antibacterial activity was indicated by the clear zone (no bacterial growth) around the well.
Antimicrobial activity of 6-HAP was determined by incubating 1 × 105 CFU/ml bacteria with twofold serial dilutions of synthetic 6-HAP in Mueller-Hinton broth at 37°C for 24 hours. After incubation, the number of viable bacteria was measured by counting CFU after spreading 10-fold serial dilutions of bacteria on suitable agar plates.
High-performance liquid chromatography
The active fraction from the Sep-Pak cartridge was separated by HPLC in a hydrophilic interaction mode with Venusil XBP NH2 (5 μm, 100 Å, 10 × 250 mm) (Agela Technologies) with a linear gradient of H2O from 5 to 35% in acetonitrile at 4 ml/min. The active fraction was fractionated, lyophilized, and dissolved in 90% acetonitrile in H2O. The active fraction was further cleaned with PolyHYDROXYETHYL A (5 μm, 60Å, 9.4 × 250 mm) with a linear gradient of H2O from 5 to 35% in acetonitrile at 3 ml/min. Elution profile was monitored with absorbance at 270 nm. After each purification step, activity was determined by radial diffusion assay against GAS (NZ131). The purified 6-HAP was lyophilized, and dry weight was measured to measure specific activity.
Mass spectrometry
A Thermo Finnigan MAT900XL mass spectrometer (Thermo Fisher Scientific) was used for both low-resolution electron impact–MS (LR-EI-MS)analysis and HR-ESI-MS using direct insertion probe for sample introduction. The electron energy was set at 70 eV with an emission current of 1.0 mA. HR-ESI-MS analysis was performed on a Thermo LTQ Orbitrap XL mass spectrometer. The source voltage was set at 4500 V with a heated capillary temperature of 250°C and a sheath gas flow rate of 60 U.
Proton nuclear magnetic resonance
1H NMR spectra of 6-HAP were recorded on a Mercury Plus 500 MHz Varian instrument. Chemical shifts (δ) are quoted in parts per million referenced to the appropriate residual solvent peak (DMSO-d6 or D2O), with abbreviations s and br s denoting singlet and broad singlet. The 1H NMR spectrum of 6-HAP in AcOD-D2O (1:5 v/v) displayed two proton signals in the aromatic region, whereas that in DMSO-d6 displayed six signals. 1H NMR (500 MHz, AcOD-D2O) δ 8.19 (s, 1H), 8.17 (s, 1H). 1H NMR (500 MHz, DMSO-d6) δ 12.74 (br s, 1H), 10.87 (br s, 0.7H), 9.50 (br s, 1H), 8.08 (br s, 1H), 7.75 (br s, 1H), 7.48 (br s, 0.4H).
Synthesis of 6-HAP
6-HAP was prepared according to the previously reported procedure with slight modifications (Preparation of nucleobases and nucleosides as antiparasitic agents, Loakes, D.; Too, K., PCT Int. Apl., 2007135380, 29 Nov. 2007). Hydroxylamine hydrochloride (1.20 g, 17.3 mmol) was dissolved in 20 ml of boiling absolute ethanol, and a solution of potassium hydroxide (1.12 g, 20.0 mmol) in 4 ml of hot absolute ethanol was added. The precipitated KCl was filtered and washed three times with 2 ml of hot ethanol. Then, 6-chloropurine (300 mg, 1.94 mmol; Sigma), dissolved in 7 ml of absolute ethanol, was added to the hydroxylamine solution. The reaction was refluxed for 2 hours then cooled to room temperature and allowed to stand overnight. The white precipitate formed was filtered and washed thoroughly with water and then ethanol and was dried under high vacuum to provide 6-HAP (230 mg, 78%) as a white solid. 1H NMR (500 MHz, DMSO-d6) δ 12.84 (br s, 1H), 10.92 (br s, 0.5H), 9.48 (br s, 1H), 8.08 (br s, 0.6H), 7.77 (s, 1H), 7.48 (br s, 0.7), in agreement with those reported. The generated 6-HAP was purified by HPLC using Venusil XBP NH2 and PolyHYDROXYETHYL A, as described above.
In vitro DNA polymerization assay
To examine whether 6-HAP disrupts adenosine-thymidine base pair matching in DNA extension, IRDye800-labeled 18-bp primer and 25-bp templates, which required adenosine (X = T) or cytidine (X = G) at the initial base of overhang for extension, were designed (Fig. 3E). The reaction mixture contained 100 nM primer/template, 0.1-U Klenow fragment (exo−) DNA polymerase (Promega), and 1 μM deoxynucleotide triphosphates in DNA polymerase buffer. The mixture was incubated at 37°C for 10 min. The reaction was terminated by adding stop solution (98% formaldehyde and 20 mM EDTA). The extended DNA was separated from primer by electrophoresis on a 20% denaturing polyacrylamide gel containing 7 M urea. Fluorescence was visualized with the Odyssey Imaging System (LI-COR Biosciences).
Cell culture and cell proliferation assay
B16F10, Pam212, L5178, and YAC-1 cell lines were obtained from American Type Culture Collection (ATCC). Pam212, L5178, and YAC-1 cell lines were maintained directly from the authenticated vial from ATCC in RPMI 1640 supplemented with sodium pyruvate (1 mM), nonessential amino acids (0.1 mM), penicillin (100 unit/ml), streptomycin (100 μg/ml), and 10% heat-inactivated fetal bovine serum (FBS) or horse serum at 37°C under atmosphere of 5% (v/v) CO2 in air. B16F10 cell line was maintained directly from the authenticated vial from ATCC in Dulbecco’s modified Eagle’s medium supplemented with penicillin (100 unit/ml), streptomycin (100 μg/ml), and 10% heat-inactivated FBS. NHEKs were obtained from Invitrogen (Life Technologies) and maintained in EpiLife medium (Life Technologies) supplemented with 60 μM calcium, epidermal growth factors, penicillin, and streptomycin. After a 4-hour (tumor cell lines) or 24-hour (NHEK) incubation with 6-HAP, proliferative activity of cells was colorimetrically determined by monitoring BrdU incorporation with Cell Proliferation kit according to the manual (Roche).
Gene silencing of mARCs with siRNA
NHEK was cultured in EpiLife media containing predesigned siRNA for mARC1 or mARC2 (Thermo Fisher Scientific) and RNAiMAX reagent for 24 hours. Cells were maintained in EpiLife for 72 hours and incubated with 6-HAP (10 μg/ml) for 24 hours. Antiproliferative activity of 6-HAP was determined by measuring BrdU incorporation as described above.
Animals
All experiments involving animal work were in accordance with the approval of the Institutional Animal Care and Use Guidelines of the University of California, San Diego (protocol number: S09074).
In vivo tumor growth assay
B16F10 were suspended in sterile phosphate-buffered saline. Shaved mouse dorsal skin was intradermally injected with 3 × 105 cells per 50 μl. The C57BL6 mice were subsequently injected with 6-HAP dissolved in 2.5% DMSO/0.9% NaCl solution (2 mg/ml) at the dose of 20 mg/kg per mouse via intravascular route every 48 hours for 2 weeks. Control mice received an equal volume of vehicle. Tumor size was measured as the two perpendicular diameters with a caliper, and volume was estimated by the following formula: width2 × length/2. The mice were sacrificed when tumor size reached >2 cm according to the animal protocol.
UV-induced tumor formation in SKH-1 mice
Female SKH-1 hairless mice (4 weeks old) were purchased from Charles River Laboratories. The back skin of each mouse was topically treated with a single application of DMBA (200 nmol/100 μl acetone) as a tumor initiator. A week after tumor initiation, mice were irradiated with 180 mJ/cm2 of UV-B twice a week. S. epidermidis MO34 or 1457 strains were cultured in TSB and diluted to 1 × 107 CFU/10 μl in TSB. Ten microliters of either bacterial suspension was spread with a sterile spatula on the entire skin of mouse back six times a week for 12 weeks. Tumor incidence and tumor number in each mouse were recorded every week.
Immunostaining
To detect S. epidermidis in UV-induced tumor, we fixed, blocked, and incubated cryostat sections of each tumor with anti–S. epidermidis monoclonal immunoglobulin G (IgG) (5 μg/ml; Clone 17–5, Santa Cruz Biotechnology) for 1 hour, followed by anti-mouse IgG-Alexa 488 conjugate (2 μg/ml; Invitrogen) for 30 min. To visualize epithelia, we stained the sections with Goat–anti-K14 IgG (1 μg/ml; Santa Cruz Biotechnology), followed by rabbit anti-goat IgG-Alexa 568 (1 μg/ml; Invitrogen).
Metagenomics
Genomic DNA was purified from S. epidermidis MO34 strain using an UltraClean Microbial DNA Isolation kit (MO Bio). Whole-genome DNA sequencing libraries were constructed using the Nextera XT DNA Sample Prep Kit (Illumina) following the vendor’s protocol. The final library was sequenced paired-end (300 × 300 bp) on an Illumina MiSeq. Sequenced reads were de novo assembled using SPAdes 2.5.1 with k-mers of lengths 21, 33, 55, 77, and 127 and the flag for “careful” turned on. On all of the produced scaffolds, a six-frame translation was performed using translate Whole Genome Multi Chromosome.pl (http://proteomics.ucsd.edu/).
To identify exclusive marker genes in S. epidermidis strains producing 6-HAP, we predicted protein-coding genes from the genomes of 2 6-HAP–producing strains and 62 strains that did not produce 6-HAP using prodigal v2.6.3 (https://bmcbioinformatics.biomedcentral.com) with default parameters. The amino acid sequences of the predicted genes were used as input for the BPGA (Bacterial Pan Genome Analysis tool) comparative genomics pipeline v1.3 (44), which identified unique 28 marker genes that are present in 6-HAP–producing strains but not in any of the other strains (table S3). Sequence reads from 694 human skin metagenomic shotgun samples collected from 18 healthy individuals at 22 body sites (45) were individually aligned to the 6-HAP strain-specific gene markers using Bowtie 2 v2.2.9 (46) under very sensitive mode. We report the fraction of bases in the 28 specific gene markers that has at least 1× coverage as an indicator of the presence of putative 6-HAP–producing strains in the corresponding sample.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5 software (GraphPad). Independent t test was used. Independent two-tailed t tests were used to compare experimental and control groups for significance of differences (P < 0.05).
Supplementary Material
Acknowledgments
We thank Q. Gao (Fudan University, Shanghai) for collecting S. epidermidis strains, V. Nizet [University of California, San Diego (UCSD)] for providing pathogenic bacteria, and C. C. Zouboulis (Dessau Medical Center) for providing SZ95 human sebocytes. Funding: This study was supported by NIH R01 AR064781, R01 AI116576, and U19 AI117673, and the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (ZIAAI000904 to M.O.). Author contributions: T.N., T.H.C., A.M.B., and K.T.S. performed most of the experiments and analyzed the results. L.L.T., S.-J.N., and W.F. assisted with the NMR analyses and chemical synthesis experiments and reviewed the manuscript. M.O. provided the clinical isolate strains of S. epidermidis. J.O. and W.Z. identified targeted strain of S. epidermidis in metagenome of skin microbiome. T.N. and R.L.G. designed all experiments and prepared the manuscript. Competing interests: T.N. and R.L.G. are co-inventors on a patent application filed by UCSD on technology related to the bacterial antiproliferative compound discussed herein (provisional no. 62477370, filed 27 March 2017). R.L.G. is cofounder and consultant to MatriSys Bioscience (La Jolla, CA), a company that is developing skin bacteriotherapy, and has also provided consulting services for SENTÉ Inc. All other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. The whole-genome sequence data of S. epidermidis MO34 strain has been submitted in the DNA Data Bank of Japan Mass Submission System (www.ddbj.nig.ac.jp/index-e.html) under the accession code SAMD00099650.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/2/eaao4502/DC1
Supplementary data
table S1. Mutagenic activity of 6-HAP.
table S2. Systemic toxicity of repeated intravascular administration with 6-HAP in mice.
table S3. List of unique marker genes that are present in S. epidermidis strains producing 6-HAP.
fig. S1. Purification of a unique antimicrobial compound from the culture supernatant of S. epidermidis strain MO34 isolated from normal human skin.
fig. S2. Molecular mass of purified antibiotic from the S. epidermidis MO34 strain analyzed by HR-ESI-MS.
fig. S3. Comparison of chemical shifts of purified antibiotic with those of synthetic 6-HAP in 1H-NMR.
fig. S4. The gHMBC spectrum (500 MHz) of 6-HAP in AcOD-D2O.
fig. S5. Comparison of the fragmentation profile of purified antibiotic with that of synthetic 6-HAP on electron impact mass spectrometry.
fig. S6. Capacity of 6-HAP to directly disrupt plasma membrane of human keratinocytes and sebocytes.
fig. S7. Effect of epicutaneous application of S. epidermidis strain producing 6-HAP on the cutaneous immune system in mice.
REFERENCES AND NOTES
- 1.Grice E. A., Kong H. H., Conlan S., Deming C. B., Davis J., Young A. C.; NISC Comparative Sequencing Program, Bouffard G. G., Blakesley R. W., Murray P. R., Green E. D., Turner M. L., Segre J. A., Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kloos W. E., Musselwhite M. S., Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl. Microbiol. 30, 381–385 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrd A. L., Deming C., Cassidy S. K. B., Harrison O. J., Ng W. I., Conlan S.; NISC Comparative Sequencing Program, Belkaid Y., Segre J. A., Kong H. H., Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci. Transl. Med. 9, eaal4651 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams M. R., Nakatsuji T., Sanford J. A., Vrbanac A. F., Gallo R. L., Staphylococcus aureus induces increased serine protease activity in keratinocytes. J. Invest. Dermatol. 137, 377–384 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chng K. R., Tay A. S., Li C., Ng A. H., Wang J., Suri B. K., Matta S. A., McGovern N., Janela B., Wong X. F., Sio Y. Y., Au B. V., Wilm A., De Sessions P. F., Lim T. C., Tang M. B., Ginhoux F., Connolly J. E., Lane E. B., Chew F. T., Common J. E. A., Nagarajan N., Whole metagenome profiling reveals skin microbiome-dependent susceptibility to atopic dermatitis flare. Nat. Microbiol. 1, 16106 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Sanford J. A., Gallo R. L., Functions of the skin microbiota in health and disease. Semin. Immunol. 25, 370–377 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cogen A. L., Yamasaki K., Sanchez K. M., Dorschner R. A., Lai Y., MacLeod D. T., Torpey J. W., Otto M., Nizet V., Kim J. E., Gallo R. L., Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J. Invest. Dermatol. 130, 192–200 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cogen A. L., Yamasaki K., Muto J., Sanchez K. M., Crotty Alexander L., Tanios J., Lai Y., Kim J. E., Nizet V., Gallo R. L., Staphylococcus epidermidis antimicrobial δ-toxin (phenol-soluble modulin-γ) cooperates with host antimicrobial peptides to kill group A Streptococcus. PLOS ONE 5, e8557 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakatsuji T., Chen T. H., Narala S., Chun K. A., Two A. M., Yun T., Shafiq F., Kotol P. F., Bouslimani A., Melnik A. V., Latif H., Kim J.-N., Lockhart A., Artis K., David G., Taylor P., Streib J., Dorrestein P. C., Grier A., Gill S. R., Zengler K., Hata T. R., Leung D. Y., Gallo R. L., Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 9, eaah4680 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai Y., Di Nardo A., Nakatsuji T., Leichtle A., Yang Y., Cogen A. L., Wu Z.-R., Hooper L. V., Schmidt R. R., von Aulock S., Radek K. A., Huang C.-M., Ryan A. F., Gallo R. L., Commensal bacteria regulate Toll-like receptor 3–dependent inflammation after skin injury. Nat. Med. 15, 1377–1382 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naik S., Bouladoux N., Linehan J. L., Han S.-J., Harrison O. J., Wilhelm C., Conlan S., Himmelfarb S., Byrd A. L., Deming C., Quinones M., Brenchley J. M., Kong H. H., Tussiwand R., Murphy K. M., Merad M., Segre J. A., Belkaid Y., Commensal–dendritic-cell interaction specifies a unique protective skin immune signature. Nature 520, 104–108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naik S., Bouladoux N., Wilhelm C., Molloy M. J., Salcedo R., Kastenmuller W., Deming C., Quinones M., Koo L., Conlan S., Spencer S., Hall J. A., Dzutsev A., Kong H., Campbell D. J., Trinchieri G., Segre J. A., Belkaid Y., Compartmentalized control of skin immunity by resident commensals. Science 337, 1115–1119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laborel-Preneron E., Bianchi P., Boralevi F., Lehours P., Fraysse F., Morice-Picard F., Sugai M., Sato’o Y., Badiou C., Lina G., Schmitt A. M., Redoules D., Casas C., Davrinche C., Effects of the Staphylococcus aureus and Staphylococcus epidermidis secretomes isolated from the skin microbiota of atopic children on CD4+ T cell activation. PLOS ONE 10, e0144323 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai Y., Cogen A. L., Radek K. A., Park H. J., Macleod D. T., Leichtle A., Ryan A. F., Di Nardo A., Gallo R. L., Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J. Invest. Dermatol. 130, 2211–2221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wanke I., Steffen H., Christ C., Krismer B., Gotz F., Peschel A., Schaller M., Schittek B., Skin commensals amplify the innate immune response to pathogens by activation of distinct signaling pathways. J. Invest. Dermatol. 131, 382–390 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Li D., Lei H., Li Z., Li H., Wang Y., Lai Y., A novel lipopeptide from skin commensal activates TLR2/CD36-p38 MAPK signaling to increase antibacterial defense against bacterial infection. PLOS ONE 8, e58288 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwase T., Uehara Y., Shinji H., Tajima A., Seo H., Takada K., Agata T., Mizunoe Y., Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465, 346–349 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Zipperer A., Konnerth M. C., Laux C., Berscheid A., Janek D., Weidenmaier C., Burian M., Schilling N. A., Slavetinsky C., Marschal M., Willmann M., Kalbacher H., Schittek B., Brötz-Oesterhelt H., Grond S., Peschel A., Krismer B., Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535, 511–516 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Plitzko B., Havemeyer A., Kunze T., Clement B., The pivotal role of the mitochondrial amidoxime reducing component 2 in protecting human cells against apoptotic effects of the base analog N6-hydroxylaminopurine. J. Biol. Chem. 290, 10126–10135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozmin S. G., Schaaper R. M., Shcherbakova P. V., Kulikov V. N., Noskov V. N., Guetsova M. L., Alenin V. V., Rogozin I. B., Makarova K. S., Pavlov Y. I., Multiple antimutagenesis mechanisms affect mutagenic activity and specificity of the base analog 6-N-hydroxylaminopurine in bacteria and yeast. Mutat. Res. 402, 41–50 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Pavlov Y. I., Suslov V. V., Shcherbakova P. V., Kunkel T. A., Ono A., Matsuda A., Schaaper R. M., Base analog N6-hydroxylaminopurine mutagenesis in Escherichia coli: Genetic control and molecular specificity. Mutat. Res. 357, 1–15 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Lloyd M., Kidd D., The mouse lymphoma assay. Methods Mol. Biol. 817, 35–54 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Dwivedi C., Valluri H. B., Guan X., Agarwal R., Chemopreventive effects of α-santalol on ultraviolet B radiation-induced skin tumor development in SKH-1 hairless mice. Carcinogenesis 27, 1917–1922 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Dinkova-Kostova A. T., Jenkins S. N., Fahey J. W., Ye L., Wehage S. L., Liby K. T., Stephenson K. K., Wade K. L., Talalay P., Protection against UV-light-induced skin carcinogenesis in SKH-1 high-risk mice by sulforaphane-containing broccoli sprout extracts. Cancer Lett. 240, 243–252 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Nakatsuji T., Chiang H.-I., Jiang S. B., Nagarajan H., Zengler K., Gallo R. L., The microbiome extends to subepidermal compartments of normal skin. Nat. Commun. 4, 1431 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh J., Byrd A. L., Deming C., Conlan S.; NISC Comparative Sequencing Program, Kong H. H., Segre J. A., Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waldman A. J., Ng T. L., Wang P., Balskus E. P., Heteroatom–heteroatom bond formation in natural product biosynthesis. Chem. Rev. 117, 5784–5863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lennard L., The clinical pharmacology of 6-mercaptopurine. Eur. J. Clin. Pharmacol. 43, 329–339 (1992). [DOI] [PubMed] [Google Scholar]
- 29.Nelson J. A., Carpenter J. W., Rose L. M., Adamson D. J., Mechanisms of action of 6-thioguanine, 6-mercaptopurine, and 8-azaguanine. Cancer Res. 35, 2872–2878 (1975). [PubMed] [Google Scholar]
- 30.Galmarini C. M., Mackey J. R., Dumontet C., Nucleoside analogues and nucleobases in cancer treatment. Lancet Oncol. 3, 415–424 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Corey R. B., Pauling L., Specific hydrogen-bond formation between pyrimidines and purines in deoxyribonucleic acids. Arch. Biochem. Biophys. 65, 164–181 (1956). [DOI] [PubMed] [Google Scholar]
- 32.Menezes M. R., Waisertreiger I. S. R., Lopez-Bertoni H., Luo X., Pavlov Y. I., Pivotal role of inosine triphosphate pyrophosphatase in maintaining genome stability and the prevention of apoptosis in human cells. PLOS ONE 7, e32313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozmin S. G., Wang J., Schaaper R. M., Role for CysJ flavin reductase in molybdenum cofactor-dependent resistance of Escherichia coli to 6-N-hydroxylaminopurine. J. Bacteriol. 192, 2026–2033 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozmin S. G., Schaaper R. M., Molybdenum cofactor-dependent resistance to N-hydroxylated base analogs in Escherichia coli is independent of MobA function. Mutat. Res. 619, 9–15 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu S., Rhee K.-J., Albesiano E., Rabizadeh S., Wu X., Yen H.-R., Huso D. L., Brancati F. L., Wick E., McAllister F., Housseau F., Pardoll D. M., Sears C. L., A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15, 1016–1022 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshimoto S., Loo T. M., Atarashi K., Kanda H., Sato S., Oyadomari S., Iwakura Y., Oshima K., Morita H., Hattori M., Honda K., Ishikawa Y., Hara E., Ohtani N., Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Arthur J. C., Perez-Chanona E., Mühlbauer M., Tomkovich S., Uronis J. M., Fan T.-J., Campbell B. J., Abujamel T., Dogan B., Rogers A. B., Rhodes J. M., Stintzi A., Simpson K. W., Hansen J. J., Keku T. O., Fodor A. A., Jobin C., Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bindels L. B., Porporato P., Dewulf E. M., Verrax J., Neyrinck A. M., Martin J. C., Scott K. P., Buc Calderon P., Feron O., Muccioli G. G., Sonveaux P., Cani P. D., Delzenne N. M., Gut microbiota-derived propionate reduces cancer cell proliferation in the liver. Br. J. Cancer 107, 1337–1344 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang Y., Chen Y., Jiang H., Robbins G. T., Nie D., G-protein-coupled receptor for short-chain fatty acids suppresses colon cancer. Int. J. Cancer 128, 847–856 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Archer S. Y., Meng S., Shei A., Hodin R. A., p21WAF1 is required for butyrate-mediated growth inhibition of human colon cancer cells. Proc. Natl. Acad. Sci. U.S.A. 95, 6791–6796 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hug D. H., Dunkerson D. D., Hunter J. K., The degradation of l-histidine and trans and cis-urocanic acid by bacteria from skin and the role of bacterial cis-urocanic acid isomerase. J. Photochem. Photobiol. B 50, 66–73 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Laihia J. K., Kallio J. P., Taimen P., Kujari H., Kähäri V.-M., Leino L., Protodynamic intracellular acidification by cis-urocanic acid promotes apoptosis of melanoma cells in vitro and in vivo. J. Invest. Dermatol. 130, 2431–2439 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Zaiou M., Nizet V., Gallo R. L., Antimicrobial and protease inhibitory functions of the human cathelicidin (hCAP18/LL-37) prosequence. J. Invest. Dermatol. 120, 810–816 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Chaudhari N. M., Gupta V. K., Dutta C., BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6, 24373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oh J., Byrd A. L., Park M.; NISC Comparative Sequencing Program, Kong H. H., Segre J. A., Temporal stability of the human skin microbiome. Cell 165, 854–866 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/2/eaao4502/DC1
Supplementary data
table S1. Mutagenic activity of 6-HAP.
table S2. Systemic toxicity of repeated intravascular administration with 6-HAP in mice.
table S3. List of unique marker genes that are present in S. epidermidis strains producing 6-HAP.
fig. S1. Purification of a unique antimicrobial compound from the culture supernatant of S. epidermidis strain MO34 isolated from normal human skin.
fig. S2. Molecular mass of purified antibiotic from the S. epidermidis MO34 strain analyzed by HR-ESI-MS.
fig. S3. Comparison of chemical shifts of purified antibiotic with those of synthetic 6-HAP in 1H-NMR.
fig. S4. The gHMBC spectrum (500 MHz) of 6-HAP in AcOD-D2O.
fig. S5. Comparison of the fragmentation profile of purified antibiotic with that of synthetic 6-HAP on electron impact mass spectrometry.
fig. S6. Capacity of 6-HAP to directly disrupt plasma membrane of human keratinocytes and sebocytes.
fig. S7. Effect of epicutaneous application of S. epidermidis strain producing 6-HAP on the cutaneous immune system in mice.


