Abstract
Background
Reconstruction of clonal evolution is critical for understanding tumor progression and implementing personalized therapies. This is often done by clustering somatic variants based on their cellular prevalence estimated via bulk tumor sequencing of multiple samples. The clusters, consisting of the clonal marker variants, are then ordered based on their estimated cellular prevalence to reconstruct clonal evolution trees, a process referred to as ‘clonal ordering’. However, cellular prevalence estimate is confounded by statistical variability and errors in sequencing/data analysis, and therefore inhibits accurate reconstruction of the clonal evolution. This problem is further complicated by intra- and inter-tumor heterogeneity. Furthermore, the field lacks a comprehensive visualization tool to facilitate the interpretation of complex clonal relationships. To address these challenges we developed ClonEvol, a unified software tool for clonal ordering, visualization, and interpretation.
Materials and methods
ClonEvol uses a bootstrap resampling technique to estimate the cellular fraction of the clones and probabilistically models the clonal ordering constraints to account for statistical variability. The bootstrapping allows identification of the sample founding- and sub-clones, thus enabling interpretation of clonal seeding. ClonEvol automates the generation of multiple widely used visualizations for reconstructing and interpreting clonal evolution.
Results
ClonEvol outperformed three of the state of the art tools (LICHeE, Canopy and PhyloWGS) for clonal evolution inference, showing more robust error tolerance and producing more accurate trees in a simulation. Building upon multiple recent publications that utilized ClonEvol to study metastasis and drug resistance in solid cancers, here we show that ClonEvol rediscovered relapsed subclones in two published acute myeloid leukemia patients. Furthermore, we demonstrated that through noninvasive monitoring ClonEvol recapitulated the emerging subclones throughout metastatic progression observed in the tumors of a published breast cancer patient.
Conclusions
ClonEvol has broad applicability for longitudinal monitoring of clonal populations in tumor biopsies, or noninvasively, to guide precision medicine.
Availability
ClonEvol is written in R and is available at https://github.com/ChrisMaherLab/ClonEvol.
Keywords: clonal evolution, cancer evolution, clonality analysis, clonal evolution visualization, next-generation sequencing
Introduction
Massively parallel sequencing has enabled the efficient detection of somatic variants that serve as clonal markers for the identification of cellular subpopulations in tumors. Given the infinite site assumption [1, 2], by estimating the variant allele frequency (VAF) and adjusting for copy number perturbations, we can approximate the fraction of the sample carrying the variant—its cancer cell fraction (CCF). Subsequent clustering of variants with similar cellular prevalence (VAFs or CCFs) across samples is commonly used to identify subclones [3, 4]. While several methods can cluster variants [1, 2], they do not infer clonal evolution. In simple cases, this is often carried out manually by ordering the clones based on their clusters’ CCF, a process referred to as ‘clonal ordering’. However, intra- and inter-tumor heterogeneity increases the complexity and requires more advanced and automated methods to accurately reconstruct clonal evolution.
Despite the recent development of tools for the reconstruction of clonal evolution, there are still limitations. First, current tools have difficulty incorporating the error and statistical variability in cellular prevalence estimates inherent in sequencing data. For example, several tools adjust for errors in ad hoc manners by requiring users to specify error thresholds [5, 6], using theoretical distributions that underestimate the variability of cellular prevalence [7–12], or relying only on pairwise ordering of the clusters or variants [6–9]. This fails to incorporate the complete clonal lineage and underestimates the uncertainty. Second, while many tools estimate cellular fraction for variants, they do not estimate the clonal frequency, i.e. cellular fraction of the clones [1, 2, 5, 8, 9]. This inhibits subsequent biological interpretation such as identifying the founding clone of a sample and distinguishing polyclonal from monoclonal seeding. Finally, while several tools generate simple and specific visualizations of clonal evolution [6, 13], the field lacks a user-friendly tool that automatically generates multiple visualizations for easily interpreting complex clonal relationships across samples.
To address these limitations, we present a unified software tool, ClonEvol, to reconstruct and visualize clonal evolution to understand inter- and intra-tumor heterogeneity and evolution. ClonEvol performs clonal ordering and tree construction by leveraging the output of existing tools that identify subpopulations of cancer cells via variant clustering [1, 2]. ClonEvol employs a bootstrapping technique that enables estimation of the confidence interval (CI) of the CCF of each clone and a probabilistic evaluation of clonal ordering constraints that tolerates the statistical variability in cellular prevalence. Furthermore, this allows for the identification of founding clones of a sample to interpret monoclonal and polyclonal seeding. To facilitate analysis and interpretation of clonal evolution, ClonEvol outputs multiple visualizations popularized in previously published studies [3, 13–15].
ClonEvol outperformed LICHeE, Canopy and PhyloWGS [6, 10, 12] in our simulation, showing more robust error tolerance and producing more precise trees. ClonEvol also rediscovered relapsed subclones in two published acute myeloid leukemia patients and reconstructed the metastatic progression using 16 tumor and plasma samples of a breast cancer patient. Overall, ClonEvol has applicability for longitudinal monitoring of clonal populations in tumor biopsies, or noninvasively, to guide precision medicine.
Materials and methods
Overview of ClonEvol
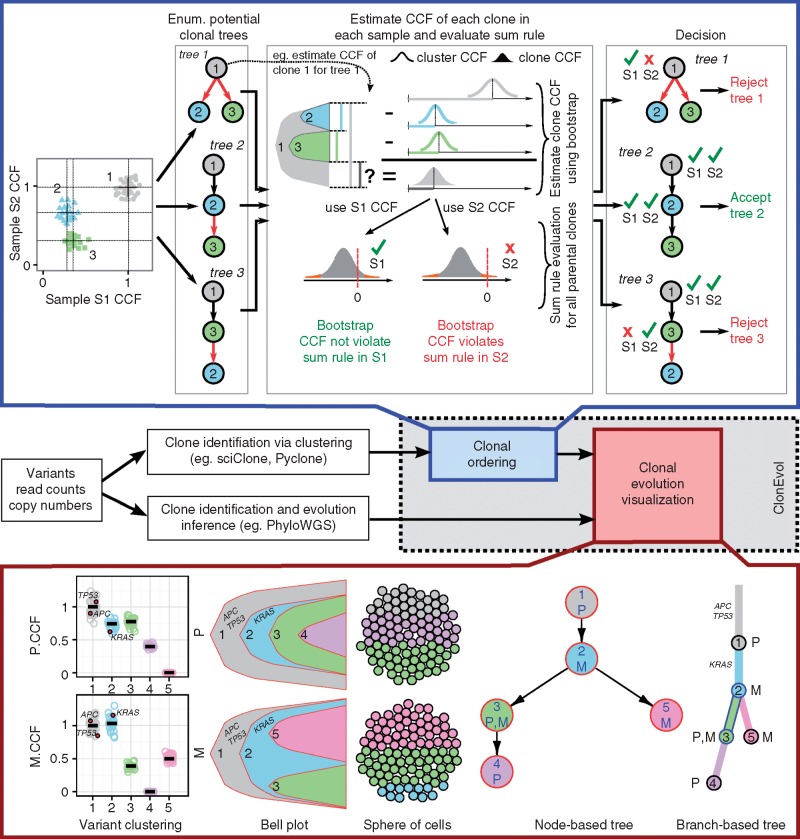
ClonEvol uses clustering of heterozygous variants as input to perform clonal ordering and outputs multiple visualizations (Figure 1; supplementary Methods, available at Annals of Oncology online), including bell plots to present clonal dynamics over time (building upon our recently published Fishplot [3, 13]), sphere of cells to present clonal subpopulations of a sample [14], and annotated node-based and branch-based trees to present clonal relationships and seeding patterns between samples [15].
Figure 1.
Overview of ClonEvol. Top panel, ClonEvol sum rule evaluation of parental clones in three potential trees. Strict sum rule violation is observed in all trees (red branches). ClonEvol corrects for these errors by using bootstrapping to generate the CI of CCF of all clones in all samples given the clonal orderings in the tree. ClonEvol rejects a tree only when a clonal CCF has high probability of being negative (bootstrap CCF violates sum rule). Middle panel, ClonEvol clonal evolution inference pipeline. Bottom panel, several ClonEvol automated graphical presentations of the clonal evolution in a pair of primary (P) and metastatic (M) samples. Driver mutations (e.g. in APC, TP53, KRAS) are automatically mapped into the visualizations to assist the interpretation.
Sum rule and cross rule as biological constraints in clonal ordering
(Sub)clones are frequently identified in cancer samples by clustering heterozygous somatic variants with similar cellular prevalence. Each cluster then represents the clonal marker variants of a clone. Clonal ordering reconstructs the temporal order of the clones based on cellular prevalence of the clusters.
Because each cluster represents a clone it is important to distinguish between the CCF of a clone (also called clonal frequency [12]) and the CCF of a cluster. In particular, the marker variants of a clone are inherited by its descendant clones, thus we equate the CCF of a parental cluster Y as the total CCFs of its clone and all of its (direct and indirect) subclones Xi:
| (1) |
Applying (1) recursively upward the clonal evolution tree gives rise to the sum rule that must be obeyed by all valid clonal orderings of clone Y and its direct subclones Xi:
| (2) |
Because the CCF of any clone must not be negative, the sum rule in (2) requires that the CCF of a parental cluster must not be less than the sum of the CCFs of its direct subclusters. Additionally, the cross rule requires clonal orderings to be consistent across all samples.
The sum rule is often strictly violated when applied to the point estimates (i.e. total mean CCFs of clusters corresponding to the subclones is greater than the mean CCF of their parental clone) due to errors in sequencing data/analysis. As such, ClonEvol applies the sum rule probabilistically (as defined below) when performing clonal ordering.
Bootstrapping model of the sum and cross rules
We define the CCF of a parental clone Y in terms of the CCFs of its cluster and direct subclusters, by rearranging the sum rule in (2), as follows (a graphical example is shown in supplementary Figure S1, available at Annals of Oncology online).
| (3) |
To account for statistical variability, we use a bootstrap approach [16] to resample variants from the clusters to estimate the sampling distribution of the CCF of clone Y, following (3). This allows for calculation of the CI of the CCF of clone Y and the probability that it violates the sum rule, i.e. CCF of clone Y is negative (Pneg). Inversely, the probability that a clone satisfies the sum rule is 1− Pneg (see supplementary Methods, available at Annals of Oncology online).
ClonEvol simultaneously enumerates and evaluates all potential clonal orderings while constructing trees for individual samples (Figure 1, supplementary Methods, available at Annals of Oncology online). It discards trees whose clonal ordering of a parental clone has a high probability of violating the sum rule, defined as having a high Pneg (in contrast to strict violation). The remaining trees are then evaluated across all samples to determine whether they obey the cross rule and subsequently merged into consensus trees that preserve the clonal orderings in individual sample trees.
Additionally, the bootstrapping approach allows for the determination of the founding clones of a sample to interpret clonal seeding and pruning distinct trees sharing seeding models to simplify interpretation (see supplementary Methods, available at Annals of Oncology online).
Results
ClonEvol corrects for errors in cellular prevalence estimates
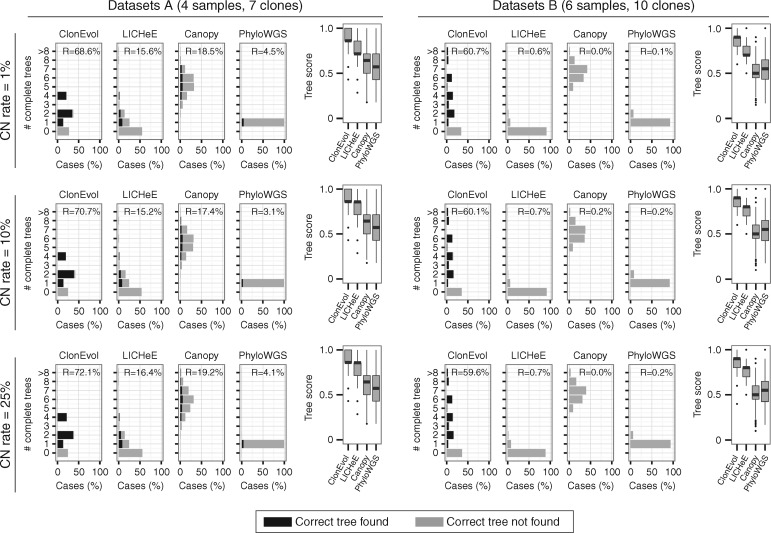
We evaluated the ability of ClonEvol to handle errors in cellular prevalence estimates using 24 simulated datasets (supplementary Table S1, available at Annals of Oncology online). Each dataset had 1000 cases sequenced to mean depth of 100× with 1% error rate, representing 100 random trees from one of the two preset clonal mixtures: 7 clones across 1 primary and 3 metastasis samples (datasets A; supplementary Table S2, available at Annals of Oncology online); 10 clones across 2 primary and 4 metastasis samples (datasets B; supplementary Table S3, available at Annals of Oncology online). Our datasets included both small and non-small clusters and incorporated three levels (1%, 10%, and 25%) of small and uncorrected copy number alterations (supplementary Methods, available at Annals of Oncology online). Given the diversity of the clonal architecture in the simulated experiments, all simulations yielded cellular prevalence estimates that strictly violated the sum rule imposed by the ground truth models in at least one clone (supplementary Figure S2, available at Annals of Oncology online). Moreover, the majority of datasets were overdispersed showing larger variance than expected by a binomial model, when copy number rates increased and/or the number of variants within clusters decreased (supplementary Figure S2, available at Annals of Oncology online). Assuming the correct clusters were identified, ClonEvol effectively corrected for the sum rule violation producing at least one tree in ∼80% and ∼70%, and the correct tree in ∼70% and ∼60% of the cases for datasets A and B, respectively (Figure 2 and supplementary Figure S3, available at Annals of Oncology online). ClonEvol tree prediction was not impaired by increased level of dispersion (also see supplementary Discussion, available at Annals of Oncology online).
Figure 2.
ClonEvol outperforms LICHeE, Canopy, and PhyloWGS in simulated data. Six datasets with non-small cluster sizes are shown. Bar plots show percentage of cases having the indicated number of complete trees predicted by each method (R indicates percent cases with correct trees found). Box plots show the precision of the complete trees predicted by each method. Tree scores are calculated as the ratio of number of correct branches to total number of branches in predicted trees.
ClonEvol distinguishes polyclonal from monoclonal seeding
Next, we evaluated if ClonEvol can distinguish between polyclonal and monoclonal seeding by constraining parental-child relationship to the clones when random trees were generated such that the seeding clones from primary tumors to metastasis were predefined (supplementary Methods, available at Annals of Oncology online). ClonEvol correctly predicted the ground truth tree in ∼71%–87% of the cases for 12 datasets (supplementary Figure S4, available at Annals of Oncology online). Among the predicted trees, ClonEvol correctly inferred monoclonal seeding for ∼86%–99% of the cases and polyclonal seeding in ∼62%–99% of the cases (supplementary Figure S4, available at Annals of Oncology online). ClonEvol clonal seeding prediction was robust against random copy number perturbation but slightly decreased when cluster size was small (supplementary Figure S4, available at Annals of Oncology online). This demonstrated that our bootstrap resampling is effective in determining the existence of clones in individual samples, and distinguishing polyclonal from monoclonal seeding models.
Evaluating the performance of ClonEvol with other methods
We compared ClonEvol with LICHeE, Canopy and PhyloWGS by evaluating predicted trees having all clones mapped (complete trees) in 12 simulated datasets without parental constraints (supplementary Methods, available at Annals of Oncology online). We found that ClonEvol nominated the correct tree as one of the predicted trees at least ∼3.6 times more frequently compared with other methods (Figure 2, supplementary Figure S3, available at Annals of Oncology online). The percentage of cases with correct trees predicted decreased in more complex datasets (B). However, ClonEvol still identified correct trees in ∼60% cases while other methods failed to produce correct trees in most cases. ClonEvol and Canopy tended to report more trees than LICHeE and PhyloWGS. However, ClonEvol obtained higher precision among its trees compared with all other methods (P < 3.8 ×10−7, Figure 2; supplementary Figure S3, available at Annals of Oncology online). Two example cases where ClonEvol outperformed other methods are shown in supplementary Figure S5, available at Annals of Oncology online.
ClonEvol detects emerging subclones in acute myeloid leukemia patients during relapse
We next evaluated the performance of ClonEvol using well-characterized patients from previous publications. We first reconstructed the clonal evolution model of a relapsed AML patient (AML1) [3]. Using whole genome sequencing and targeted validation, the authors discovered that a rare subclone in the primary tumor that survived chemotherapy continued to evolve and founded the relapsed tumor (supplementary Figure S6A, available at Annals of Oncology online). Even with high depth of coverage (∼4000×), a strict violation of the sum rule between the two subclones corresponding to clusters 3 and 4 in the relapse sample was still observed (supplementary Figure S6B, available at Annals of Oncology online), and had to be adjusted manually in the original publication. However, ClonEvol was able to infer one unique clonal evolution model identical to the published model (supplementary Figure S6, available at Annals of Oncology online). Noticeably, ClonEvol correctly inferred that although cluster 4 had a greater mean CCF in the relapse than cluster 3, the true pattern of descent was captured in the primary tumor (where the CCF of cluster 3 far exceeded cluster 4). This example, although simple, demonstrated the utility of the bootstrap approach in tolerating error in cellular prevalence.
To further demonstrate the utility of ClonEvol, we reanalyzed another case (AML31) from our recent study [4]. Each sample was sequenced to high coverage (∼312× genome, ∼433× exome), followed by targeted validation (∼1000×). Similar to AML1 we observed a strict violation of the sum rule between the founding clone 1 and its subclone 3 in the relapse sample despite deep coverage (supplementary Figure S7A and B, available at Annals of Oncology online) that required manual adjustment in the published model [4]. Therefore, we reanalyzed this patient using our latest version of ClonEvol, which uses bootstrapping, and resolved the error automatically. Five models were inferred matching published results. Single-cell sequencing data ruled out three models, leaving two models, differing only in whether clone 6 is derived from clone 2 or 4 (a private event of the primary tumor). Both are equally possible, given the existing data. One of these models is shown in supplementary Figure S7A–E, available at Annals of Oncology online. Furthermore, by pruning sample specific subclones, ClonEvol produced a unique pruned tree that captured the inter-sample clonal lineage in which subclone 3 originated from the primary tumor and later survived chemotherapy and expanded in the relapse, similar to the original findings (supplementary Figure S7F, available at Annals of Oncology online).
Additionally, the original study monitored driver variants at two additional time points between initial presentation of primary disease and relapse using ultra-deep sequencing (10 000×) and ddPCR (250 000×). Notably, two variants in TP53 and KCNT1 were detectable at significant frequency whereas all other founding driver variants were not seen at these intermediate time points. Therefore, we included these additional time points and allowed polyclonal cancer initiation in a ClonEvol analysis and discovered two pruned trees; one of which agreed with the published model supported by single cell sequencing data. This model highlighted the emergence of an independent clone 7 carrying TP53 and KCNT1 variants that was undetectable in the primary tumor but together with subclone 3 progressed and expanded in the relapse (supplementary Figure S7G and H, available at Annals of Oncology online).
Application to monitoring breast cancer progression using liquid biopsies
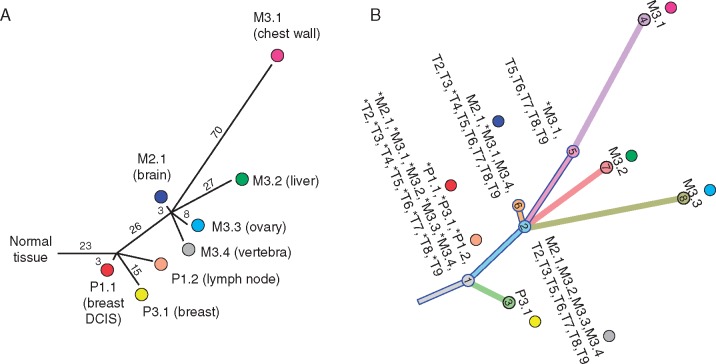
We focused on a recent study of breast cancer metastasis [17] to further demonstrate the applicability of ClonEvol using a large quantity of samples and its utility to solid tumors and liquid biopsies. The authors carried out exome sequencing and targeted validation of 17 specimens collected throughout disease progression from primary tumor (2), lymph node (1), distant metastases (5), and plasma (9). Eight clones were identified, and a tree was constructed using the absence/presence of variants across tumor samples. Notably, the original publication excluded the plasma samples from tree construction due to high level of noise and low quantity of variants detected in the plasma [17]. Here we reanalyzed all eight tumor and eight plasma samples (one has bad quality not analyzed by the authors) and inferred two pruned trees, both match the published phylogenetic tree, one is shown in Figure 3.
Figure 3.
ClonEvol analysis of metastatic breast cancer progression. (A) Phylogenetic tree reported in the original publication [17] that does not include plasma samples; (B) A matching clonal evolution tree inferred by ClonEvol using all tumor and plasma samples. Blue bordered branches and nodes represent seeding clones between samples (the pruned tree).
There were two significant improvements from our reanalysis using ClonEvol compared with the original analysis. First, unlike the original publication, ClonEvol integrated eight additional plasma samples enabling us to reconstruct the clonal relationship between plasma samples and the primary, lymph node, and metastatic tumors (Figure 3B). Second, the detailed models produced by ClonEvol describe the relationship between the clones and highlight the potential seeding patterns between samples. This was not easily seen in the published phylogenetic tree. For example, clones 2 and 6 arose earlier during progression and therefore were present in multiple metastases. Our incorporation of the plasma samples also enabled us to confirm the presence of these clones across all analyzed time points (T2–T9). In contrast, clone 5 arose later during metastatic progression and only presented in the chest wall metastasis (M3.1). Noninvasive monitoring confirmed this since our ClonEvol results show clone 5 was only seen in the plasma from later time points (T5–T9; Figure 3B). Taken together, these results demonstrate the utility of ClonEvol for analyzing larger quantities of samples as well as its ability to incorporate liquid biopsies to monitor cancer progression.
Discussion
Uncertainty in cellular prevalence estimates is a significant barrier to accurately reconstruct clonal evolution from cancer sequencing. We have shown efficacy of ClonEvol in error tolerance and its superior performance against current methods. However, one source of error is undetectable copy number alterations. Such errors could be vastly reduced if cellular prevalence estimates provided to ClonEvol are already corrected for copy number variation (e.g. using copy number aware clustering tools such as Pyclone). We have also shown ClonEvol performance was robust against random copy number alterations; however, ClonEvol performance may be impaired when there exists an imbalance between gain and loss events.
Most current tools do not calculate, or only deterministically calculate, the CCF of the clones in the samples. This inhibits further statistical interpretation of the presence of the clones in individual samples to distinguish polyclonal from monoclonal seeding. Although polyclonal seeding is rare, several recent studies have highlighted complex polyclonal seeding in metastatic cancers [15, 18]. A polyclonal seeding model is interpreted when more than one clone is present in two related samples (e.g. a primary tumor and its seeded metastasis). However, it is difficult to distinguish a low cellular prevalence estimate from the errors, thus a polyclonal model can be mistakenly inferred while the underlying true model is monoclonal. ClonEvol addresses this issue by bootstrapping estimates of the CCF to identify the presence of a clone in a sample with high confidence.
The visualizations produced by ClonEvol are intended to facilitate the analysis and interpretation of the clonal evolution models. As the first tool automating the generation of many easily interpretable, publication quality visualizations, ClonEvol minimizes the manual curation (often requiring multiple iterations) needed to accurately understand the clonal evolution models.
Our reanalysis of two relapsed acute myeloid leukemia patients and a metastatic breast cancer patients demonstrated that ClonEvol was able to reconstruct the clonal evolution underlying tumor progression in blood and solid cancers, using both tumor and liquid biopsies. Additionally, several recent studies have also utilized ClonEvol to understand metastatic progression and drug resistance in multiple cancer types including myelodysplastic syndrome [19], breast cancer [20, 21], and glioblastoma [22]. Taken together, this highlights the broad applicability of ClonEvol for inferring, visualizing, and interpreting clonal evolution and ultimately improving precision medicine.
Supplementary Material
Acknowledgement
We thank Charles Geyer for insightful discussion on the bootstrap technique.
Funding
NIH NCI (R21CA185983-01) (to CAM); a Siteman Cancer Center (no grant number applies); Barnes-Jewish Hospital Foundation Cancer Frontier Fund Research Grant (no grant number applies).
Disclosure
The authors have declared no conflicts of interest.
References
- 1. Roth A, Khattra J, Yap D. et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods 2014; 11(4): 396–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Miller CA, White BS, Dees ND. et al. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput Biol 2014; 10(8): e1003665.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ding L, Ley TJ, Larson DE. et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012; 481(7382): 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Griffith M, Miller CA, Griffith OL. et al. Optimizing cancer genome sequencing and analysis. Cell Syst 2015; 1(3): 210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Strino F, Parisi F, Micsinai M, Kluger Y.. TrAp: a tree approach for fingerprinting subclonal tumor composition. Nucleic Acids Res 2013; 41(17): e165.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Popic V, Salari R, Hajirasouliha I. et al. Fast and scalable inference of multi-sample cancer lineages. Genome Biol 2015; 16: 91.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Prandi D, Baca SC, Romanel A. et al. Unraveling the clonal hierarchy of somatic genomic aberrations. Genome Biol 2014; 15(8): 439.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Niknafs N, Beleva-Guthrie V, Naiman DQ, Karchin R.. SubClonal hierarchy inference from somatic mutations: automatic reconstruction of cancer evolutionary trees from multi-region next generation sequencing. PLoS Comput Biol 2015; 11(10): e1004416.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. El-Kebir M, Oesper L, Acheson-Field H, Raphael BJ.. Reconstruction of clonal trees and tumor composition from multi-sample sequencing data. Bioinformatics 2015; 31(12): i62–i70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deshwar AG, Vembu S, Yung CK. et al. PhyloWGS: reconstructing subclonal composition and evolution from whole-genome sequencing of tumors. Genome Biol 2015; 16: 35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Malikic S, McPherson AW, Donmez N, Sahinalp CS.. Clonality inference in multiple tumor samples using phylogeny. Bioinformatics 2015; 31(9): 1349–1356. [DOI] [PubMed] [Google Scholar]
- 12. Miller CA, McMichael J, Dang HX. et al. Visualizing tumor evolution with the Fishplot package for R. BMC Genomics 2016; 17(1): 880.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McPherson A, Roth A, Laks E. et al. Divergent modes of clonal spread and intraperitoneal mixing in high-grade serous ovarian cancer. Nat Genet 2016; 48(7): 758–767. [DOI] [PubMed] [Google Scholar]
- 14. Gundem G, Van Loo P, Kremeyer B. et al. The evolutionary history of lethal metastatic prostate cancer. Nature 2015; 520(7547): 353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang Y, Qiu Y, Minn AJ, Zhang NR.. Assessing intratumor heterogeneity and tracking longitudinal and spatial clonal evolutionary history by next-generation sequencing. Proc Natl Acad Sci USA 2016; 113(37): E5528–E5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Efron B, Tibshirani RJ.. An Introduction to the Bootstrap. Chapman & Hall, New York: CRC Press, 1994. [Google Scholar]
- 17. Murtaza M, Dawson S-J, Pogrebniak K. et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun 2015; 6: 8760.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aceto N, Bardia A, Miyamoto DT. et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014; 158(5): 1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Uy GL, Duncavage EJ, Chang GS. et al. Dynamic changes in the clonal structure of MDS and AML in response to epigenetic therapy. Leukemia 2017; 31(4): 872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miller CA, Gindin Y, Lu C. et al. Aromatase inhibition remodels the clonal architecture of estrogen-receptor-positive breast cancers. Nat Commun 2016; 7: 12498.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoadley KA, Siegel MB, Kanchi KL. et al. Tumor evolution in two patients with basal-like breast cancer: a retrospective genomics study of multiple metastases. PLoS Med 2016; 13(12): e1002174.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johanns TM, Miller CA, Dorward IG. et al. Immunogenomics of hypermutated glioblastoma: a patient with germline POLE deficiency treated with checkpoint blockade immunotherapy. Cancer Discov 2016; 6(11): 1230–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.