Abstract
Background
Exosomes arise from viable cancer cells and may reflect a different biology than circulating cell-free DNA (cfDNA) shed from dying tissues. We compare exosome-derived DNA (exoDNA) to cfDNA in liquid biopsies of patients with pancreatic ductal adenocarcinoma (PDAC).
Patients and methods
Patient samples were obtained between 2003 and 2010, with clinically annotated follow up to 2015. Droplet digital PCR was performed on exoDNA and cfDNA for sensitive detection of KRAS mutants at codons 12/13. A cumulative series of 263 individuals were studied, including a discovery cohort of 142 individuals: 68 PDAC patients of all stages; 20 PDAC patients initially staged with localized disease, with blood drawn after resection for curative intent; and 54 age-matched healthy controls. A validation cohort of 121 individuals (39 cancer patients and 82 healthy controls) was studied to validate KRAS detection rates in early-stage PDAC patients. Primary outcome was circulating KRAS status as detected by droplet digital PCR. Secondary outcomes were disease-free and overall survival.
Results
KRAS mutations in exoDNA, were identified in 7.4%, 66.7%, 80%, and 85% of age-matched controls, localized, locally advanced, and metastatic PDAC patients, respectively. Comparatively, mutant KRAS cfDNA was detected in 14.8%, 45.5%, 30.8%, and 57.9% of these individuals. Higher exoKRAS MAFs were associated with decreased disease-free survival in patients with localized disease. In the validation cohort, mutant KRAS exoDNA was detected in 43.6% of early-stage PDAC patients and 20% of healthy controls.
Conclusions
Exosomes are a distinct source of tumor DNA that may be complementary to other liquid biopsy DNA sources. A higher percentage of patients with localized PDAC exhibited detectable KRAS mutations in exoDNA than previously reported for cfDNA. A substantial minority of healthy samples demonstrated mutant KRAS in circulation, dictating careful consideration and application of liquid biopsy findings, which may limit its utility as a broad cancer-screening method.
Keywords: exosome, liquid biopsy, KRAS, pancreatic cancer, circulating tumor DNA
Introduction
Pancreatic ductal adenocarcinoma (PDAC) composes 85% of all pancreatic malignancies and is associated with a dismal 5-year survival of 6% [1, 2]. While cancer prevention initiatives and advances in targeted therapies have produced tangible survival improvements in breast, colon, and lung cancers, long-term PDAC survival remains poor and the nature of the disease does not readily present opportunities for screening and early detection [3–8]. Under the best of circumstances, resection of early-stage disease at experienced and high-volume centers improves 5-year survival to only 24%–29% [2, 9–11].
Given the aggressive and recalcitrant clinical course of pancreatic cancer, many efforts have focused on identifying novel protein, DNA or RNA biomarkers to serve as a means for early detection or prognostic stratification [12]. Blood-based liquid biopsy is particularly attractive in the context of PDAC, as the primary tumor itself is not routinely accessible in its retroperitoneal location, and sampling of the tissue is not without morbidity. Circulating tumor DNA and KRAS genetic mutations as a surrogate for PDAC-specific genetic material has been previously studied [13–21], and a study by Bettegowda et al. [22], using a bead-based ultrasensitive PCR assay, demonstrated 48% and 77% detection rates for patients with early- and late-stage tumors, respectively.
Other reservoirs of proteins, DNA, and RNA have recently been identified in the form of microvesicles termed exosomes [23–25]. Exosomes are 40–150 nm lipid bilayer membrane-bound particles derived from specific biogenesis pathways within cells and accessible within the plasma of the circulating peripheral blood [26]. Biologically, exosomes have been shown to be capable of intercellular communication and modulation of the tumor microenvironment [27, 28]. Perhaps more importantly, it is believed that the contents contained within these particles remains distinct from the remainder of the peripheral blood and thus, might represent an enrichment of tumor-specific genomic material [23, 25]. While many have commented on the utility of ‘circulating tumor’ or ‘cell-free’ DNA (cfDNA) in the context of liquid biopsy for cancer, here we tested the potential for exosome-derived DNA (exoDNA) to represent an additional blood-based compartment which may be complementary to cfDNA in the diagnosis and therapeutic stratification of patients with pancreatic cancer.
Methods
Study populations
Discovery cohort
Whole blood samples were collected at MD Anderson Cancer Center (MDACC) through informed consent following institutional review board (IRB) approval (PA14-0552). Patients with all stages of pancreatic cancer were included in the study. Healthy control samples were obtained from volunteers in the clinic waiting rooms, and for the most part, are relatives of the patients. Demographic information and personal medical history was collected from these volunteers, but samples were de-identified after collection, so follow-up of these volunteers was not possible. Individuals with diabetes, a history of pancreatitis, or a family history of pancreatic cancer were excluded from the discovery cohort. Whole blood was collected in green top (Sodium Heparin, BD Vacutainer) tubes. Blood samples were centrifuged at 2500×g for 10 min for plasma isolation and then stored at −80°C until the time of exosome isolation. Samples were collected between 2003 and 2010, and between 0.9 and 1.5 ml of plasma were available per patient for both cfDNA and exoDNA analysis. Medical records were queried for the American Joint Committee on Cancer staging, treatment status, and clinical outcomes. Staging considerations were supplemented with National Comprehensive Cancer Network guidelines with regard to borderline-resectable tumors. A total of 68 patients with PDAC of all clinical stages, an additional 20 PDAC patients initially staged with localized disease, with blood drawn after resection for curative intent, and 54 age-matched healthy controls were included in this cohort (Table 1).
Table 1.
Characteristics of patients from the discovery and validation cohorts
| Characteristic | Subclass | PDAC—all stages | CA19-9 mean (range) | Healthy controls |
|---|---|---|---|---|
| Discovery cohort | ||||
| Total | 88 | 54 | ||
| Gender | Male | 51 | 34 | |
| Female | 37 | 20 | ||
| Age in years (range) | 61 (41–79) | 65 (55–83) | ||
| Stage of disease | Localized | 33 | 105 (1–1604) | – |
| Localized postsurgical | 20 | 159 (1–1193) | – | |
| Locally advanced | 15 | 118 (1–7800) | – | |
| Metastatic | 20 | 193 (2–125 000) | – | |
| Validation cohort | ||||
| Total | 39 | 82 | ||
| Gender | Male | 21 | 43 | |
| Female | 18 | 39 | ||
| Age in years (range) | (37–79) | (38–84) | ||
| Stage of disease | Localized | 39 | – | |
| Localized postsurgical | – | – | ||
| Locally advanced | – | – | ||
| Metastatic | – | – |
Validation cohort
A total of 39 early-stage PDAC patients and 82 age-matched healthy controls were recruited through an International Agency for Research on Cancer (IARC) case–control study coordinated in the Czech Republic and Slovakia following informed consent. Researchers were blinded to the cancer-status of the clinical samples at the time of processing and data analysis. Peripheral blood was collected in EDTA tubes at the time of consent and processed as rapidly as possible. Blood samples were centrifuged at 2000×g for 10 min for plasma isolation and then stored at −80°C until the time of exosome isolation, where 200 µl of plasma were available for exoDNA analysis.
Exosomes isolation and characterization
Exosomes were isolated using serial ultracentrifugation and characterized with electron microscopy, flow cytometry and particle analysis as previously described [25].
DNA isolation and mutation detection
CfDNA was isolated using the QIAmp Circulating Nucleic Acid Kit (Qiagen) as described in supplementary Methods (available at Annals of Oncology online). DNA was extracted using the MagAttract High Molecular Weight DNA kit (Qiagen) as described in supplementary Methods (available at Annals of Oncology online). Droplet digital polymerase chain reaction (ddPCR) was applied to detect KRAS mutations with a multiplex assay from BioRad as described in supplementary Methods (available at Annals of Oncology online). KRAS mutant DNA titration experiments were performed and a lower limit for mutant allele frequencies at 0.01% was confirmed for calling positivity (see supplementary Figure S2, available at Annals of Oncology online).
Statistical analyses
Statistical analyses were performed using the R and SAS programming languages. Descriptive comparisons of study variables used the Fisher’s exact test for categorical data and the Wilcoxon rank sum test for continuous data. Univariate analyses using Cox proportional hazard models were performed to examine potential clinical and molecular factors contributing to survival. Survival curves were generated using the Kaplan–Meier method, and log-rank tests were used to compare survival curves. Clinical outcomes were established as defined by the National Cancer Institute [29]. KRAS sensitivity and specificity were determined as related to the patient’s cancer status.
Results
Exosome size and concentration
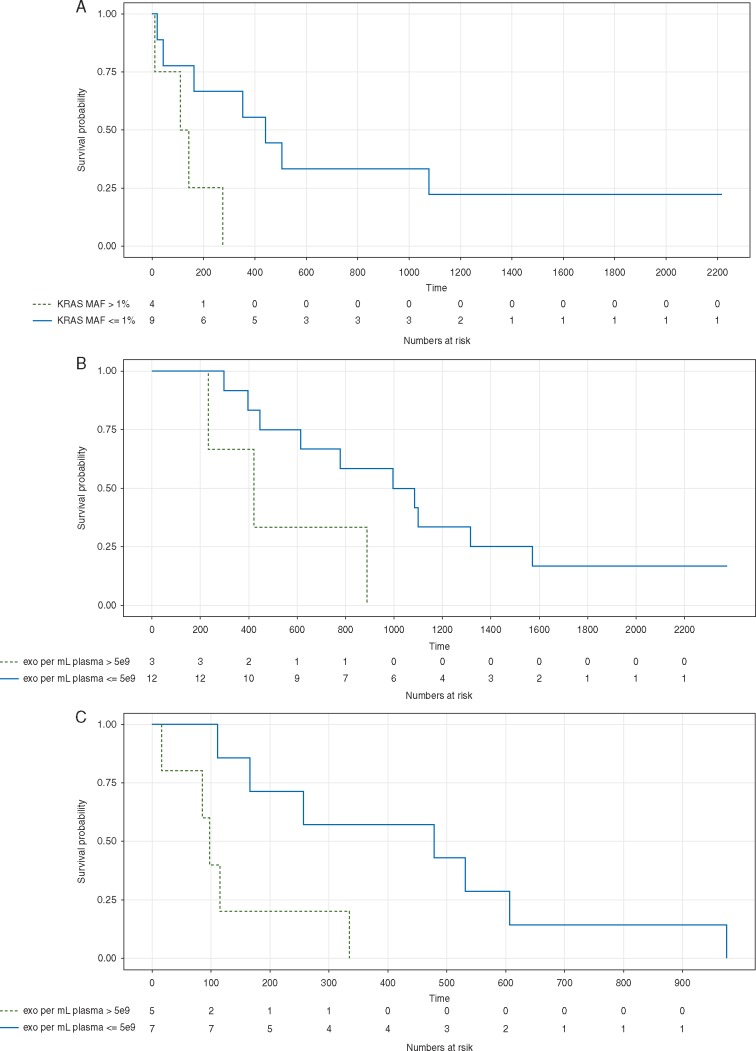
The presence of extracellular vesicles in exosome isolations was confirmed by means of Zetaview nanoparticle tracking analysis, western blot for exosomal markers, and scanning and transmission electron microscopy, with the latter in selected samples (supplementary Figure S1, available at Annals of Oncology online). Average particle size was greater among patients with PDAC compared with healthy controls. Further, average particle size was observed to be greater with more advanced disease (supplementary Figure S1B, available at Annals of Oncology online), particularly, those particles that were between 141 and 220 nm (supplementary Figure S1C, available at Annals of Oncology online). Exosome concentration was defined as number of exosomes per mL plasma. A cutoff value of 5.0 × 109 exosomes was identified through this discovery cohort and found to be associated with overall survival for both localized and metastatic patients, with a higher exosome concentration predicting worse survival (Figure 1B and C). Localized pre-surgical patients with less than 5.0 × 109 exosomes per ml plasma had a median survival of 1040 days compared with 421 days for those with higher exosome concentrations (P = 0.047). Similarly, metastatic patients with less than 5.0 × 109 exosomes per ml plasma had a median survival of 479 days compared with 97 days for those with higher exosome concentrations (P = 0.015).
Figure 1.
Liquid biopsy Kaplan–Meier curves. (A) Stratification of exoKRAS at a mutant allele frequency of 1% was associated with disease free survival in patients with localized disease who were treatment naïve at the time of blood draw (n = 13) with a median survival of 441 days compared to 127 days (P = 0.031). Two treatment naïve patients with no KRAS mutant droplets were excluded from this survival analysis to account for the possibility that they have a KRAS mutation that is not a target of the KRAS multiplex ddPCR kit used. (B and C) Exosome concentration of 5 × 109 per ml of plasma was associated with overall survival in treatment naïve blood draws in patients with (B) localized disease (n = 15, median survival 616 versus 233 days, P = 0.048) and (C) metastatic disease (n = 12, median survival 479 versus 97 days, P = 0.015).
Liquid biopsy detects exoDNA KRAS mutants by digital PCR
In the discovery cohort, ddPCR analysis of exoDNA detected KRAS mutations in 66.7% (22/33), 80% (12/15), and 85% (17/20) of localized, locally advanced, and metastatic PDAC patients, respectively, and in 7.4% (4/54) of controls (Table 2). For predicting PDAC status, the resultant sensitivity and specificity are 75.4% and 92.6%, respectively. Positive mutant KRAS status from exoDNA was significantly associated with early-stage PDAC when comparing patients with localized disease to healthy individuals (Fisher’s exact test P < 0.001), where an individual with positive KRAS status is 8.17 times (95% CI: 2.46–35.58) more likely to have early-stage pancreatic cancer than to be cancer free. Further, compared with localized pre-resected patients with a mutant KRAS detection rate of 66.7%, in a similar cohort of 20 localized PDAC patients with blood sampled after resection, mutant detection rate was much lower at 5%. Mutant KRAS status was significantly associated with pre-resection blood sampling (Fisher’s exact test, P < 0.001). Of note, healthy controls had a mutant detection rate of 7.4% (4/54). KRAS mutant status in the healthy controls was associated with increased age (mean age of 75 years in mutant KRAS individuals versus 64 years in wild-type KRAS individuals; Wilcoxon rank sum test P = 0.004).
Table 2.
Liquid biopsy mutant KRAS call rates
| Stage of disease | cfKRAS mutant call rate (%) | exoKRAS mutant call rate (%) |
|---|---|---|
| Discovery cohort | ||
| Healthy | 8/54 (14.8) | 4/54 (7.4) |
| Localized | 15/33 (45.5) | 22/33 (66.7) |
| Localized postsurgical | 0/20 (0) | 1/20 (5) |
| Locally advanced | 4/13 (30.8) | 12/15 (80) |
| Metastatic | 11/19 (57.9) | 17/20 (85) |
| Validation cohort | ||
| Healthy | – | 17/82 (20.7) |
| Localized | – | 17/39 (43.6) |
In the validation cohort, 44% (17/39) of early-stage pancreatic cancer patients tested positive for KRAS compared with 20% (17/82) of healthy individuals, confirming that KRAS positivity is associated with pancreatic cancer (Fisher's exact test, P = 0.0163). An individual with KRAS positivity was 2.96 times (95% CI: 1.29–6.76) more likely to have pancreatic cancer than to be healthy. Unlike with the discovery cohort, no association of age with mutant exoKRAS status was found in the healthy controls.
Mean KRAS mutation allele frequencies were higher in metastatic compared with localized samples (mean of 10.09% versus 2.7%, respectively; Wilcoxon rank sum test P = 0.0109).
Stratification of localized patients based on a pre-surgery exoKRAS MAF threshold of 1% was associated with disease-free survival following resection (Figure 1A), with a median disease-free time of 441 versus 127 days for patients with less than 1% MAF compared with those with more than 1% MAF (P = 0.031; Figure 1A). In addition, greater than a 1% MAF was a significant risk factor impacting disease-free survival (RR, 4.68; 95% CI 1.014–21.61). While a slight, yet statistically significant positive correlation existed between KRAS MAF and CA19-9 levels (P = 0.019, r = 0.303), only KRAS MAF was associated with disease-free survival. Cox proportional hazard analyses were also performed on locally advanced and metastatic patients but no clinical factors were found to be significantly associated with overall or progression-free survival.
Performance of cfDNA in liquid biopsy
In the discovery cohort, mutant cfKRAS was detected in 14.8% (8/54), 45.5% (15/33), 30.8% (4/13), and 57.9% (11/19) of healthy controls, localized, locally advanced, and metastatic patients. Of these positive cfDNA calls, respectively, 12.5% (1/8), 73.3% (11/15), 100% (4/4), and 100% (11/11) were also called positive through exoDNA. As opposed to the exoDNA results, KRAS positive status in healthy control cfDNA was not associated with increasing age (data not shown). In the metastatic group, the presence of mutant KRAS cfDNA suggested worse overall survival (median survival of 115 days compared with 506 days for mutant KRAS negative patients), but this was not statistically significant (P = 0.107).
Discussion
Exosomes, which have been shown to harbor DNA [25, 30], are the product of specific biogenesis pathways and are shed from viable cells by the tens of millions into circulation. Conversely, traditional cfDNA is derived from apoptosis and necrosis of tumor cells, which are characteristic of later stage disease [31]. It may thus be possible that exoDNA is a significant contributor of the DNA in circulation in patients of earlier clinical stage, before cell death and tumor necrosis begin to occur. In this context, the origin of the circulating DNAs may explain why the detection rate for early-stage patients in this study was slightly higher with exoDNA than that previously described for cfDNA, but also why the identification of late-stage patients was concordant [22].
Most encouraging is the observation of a precipitously lower detection rate in the localized pre and post-resection cohorts, from 66% to 5%. With mutant KRAS being a surrogate for tumor-specific DNA, and resection for curative intent aimed at removing the entirety of the localized disease, pre- and post-procedure liquid biopsies may have utility in determining the early success of resection. It is important to mention though, that the lower KRAS detection may be an overall marker of response to any therapy, and not just surgery alone. We are unable to draw such conclusions from this data set as time points before and after other treatment modalities are not available for our cohorts.
In this study, exoKRAS mutant allele frequency, but not CA19-9, was associated with disease free survival in localized disease. Whereas the presence or the absence of cfDNA and overall amount of DNA has previously been used for stratification, we did not identify such a clinical correlation. In a tumor where oncogenic KRAS gene mutations are believed to be near ubiquitous, to the best of our knowledge, this is the first time KRAS mutant allele frequency in exoDNA has been used for prognostic stratification. While a 1% mutant KRAS fractional abundance was identified in our discovery cohort as being informative towards disease-free survival, further validation is warranted for any such proposed cancer biomarker [32].
CfDNA was detected between 30.8% and 57.9% across stages, which is concordant with the findings of earlier studies [22]. No studies to date have described the detection rate of KRAS mutant alleles within exosomes across a series of PDAC patients across all stages, nor compared these directly with cfDNA. For this reason, we performed a parallel analysis of liquid biopsy for cfDNA KRAS mutations from plasma samples from the same patients to serve as a comparison, in addition to historical cfDNA detection rates in the literature. In our study, rates of detection of KRAS mutants in exosomes were superior to cfDNA across all stages. Of particular interest is our finding that exoDNA revealed a greater detection of patients with localized disease than previously observed using a highly sensitive method of detection [22]. Validation is warranted, but this finding has potential ramifications for liquid biopsy-based diagnostics, especially in tumors where specific mutant detection yields the opportunity for treatment with targeted therapy.
Identification of KRAS mutations in 7.4% of exoDNA and 14.8% of cfDNA healthy controls in the discovery cohort and in 20.7% in the exoDNA of the validation cohort was an unexpected finding with potential implications for using liquid biopsies as a screening tool. Indeed, a survey of the literature shows that KRAS mutations in apparently healthy samples have been previously described (see supplementary Table S1, available at Annals of Oncology online) both in a liquid biopsy setting, and in autopsy series (in non-cancerous pancreata). It is important to mention that in an era where highly sensitive detection techniques are now available, detection rates for ‘background’ oncogenic mutations are likely to increase. It is possible that the finding of such mutations describe a pre-malignant process within the pancreas or a KRAS-mediated malignancy outside the pancreas. Perhaps, these mutations accumulate in organs with increasing age but the rate at which these mutations progress to invasive cancer is unknown. Mutant KRAS findings in the normal controls of the discovery cohort suggests that accumulation of driver mutations may be an age-related phenomena as recently described by Krimmel et al. [33] for TP53 mutations in control patients. However, no association of age and mutant KRAS status in healthy controls was found in our validation cohort. For purposes as an early cancer-screening diagnostic, the specificity of our approach would need to be improved possibly by requiring a minimum KRAS mutation allele frequency to make a positive mutant status call, which is the focus of continued work. Additional biomarkers, such as other cancer DNA mutations or protein biomarkers could also be added into the screening model to increase the sensitivity to make it clinically useful.
In the setting where the patient’s cancer status is known a priori, then the utility of a liquid biopsy lies in the ability to observe serially the response of genetic mutations as a form of personalized biomarkers to therapy. It is necessary to consider that KRAS mutations as a PDAC biomarker may be of particular value in terms of assessing response to therapy in those 5-20% of patients who do not express the Sialyl Lewis-A, or CA 19-9 antigen [34], and furthermore in those patients in whom CA-19-9 becomes unreliable in the frequent setting of obstructive jaundice. Additionally, the radiologic appearance and response of PDAC to therapy on cross-sectional imaging is negligible to the point that non-progression on therapy has become a qualifier to proceed to surgery in borderline-resectable patients [35].
In this study, exoDNA outperformed cfDNA for the detection of mutant KRAS in PDAC patients. Further, the exoDNA detection rate of patients with early-stage tumors is greater than that previously reported. However, a substantial portion of healthy control patients also exhibited KRAS mutations. This suggests that follow-up studies more generally focused on uncovering the prevalence of known cancer mutations (in addition to KRAS mutations) in healthy individuals are needed to try to put these mutations into biological context and to ultimately understand their clinical repercussions. In the context of liquid biopsy, the application for ultrasensitive identification of a single genetic mutation as a predictor for PDAC may be limited.
Funding
This work was supported by the MD Anderson Moonshot Program in Pancreatic Cancer and the Sheikh Ahmed Pancreatic Cancer Research Center; the National Institute of Health [5T32CA009599-27 to KA, U01CA196403 to AM, U01CA200468 to AM, P30CA016672 for the MD Anderson High Resolution Electron Microscopy Facility]; the Translational Molecular Pathology Fellowship at MD Anderson Cancer Center to FS; the Cancer Prevention Research Institute of Texas [RP140106 to VB]; and the MD Anderson Institutional Cancer Center Support Grant CA16672 for the High Resolution Electron Microscopy Facility. The case control study which provided the validation series was funded by the US National Cancer Institute at the National Institutes of Health (R03 CA123546-02), the Ministry of Health of the Czech Republic – Development of Research Organization (MMCI, 00209805; IGA MZ N. 9422-3 & 8090-3), and the Ministry of Health of the Slovak Republic for the Epidemiological study on Pancreatic Cancer, ESNAP (Regional Authority of Public Health in Banská Bystrica, MZSR2007/17-RUVZBB-02).
Disclosure
The authors have declared no conflicts of interest.
Supplementary Material
Footnotes
The first three contributed equally as senior first authors.
Both authors contributed equally as senior last authors.
References
- 1. Ryan DP, Hong TS, Bardeesy N.. Pancreatic adenocarcinoma. N Engl J Med 2014; 371: 1039–1049. [DOI] [PubMed] [Google Scholar]
- 2. Yeo TP. Demographics, epidemiology, and inheritance of pancreatic ductal adenocarcinoma. Semin Oncol 2015; 42: 8–18. [DOI] [PubMed] [Google Scholar]
- 3. Warner E. Clinical practice. Breast-cancer screening. N Engl J Med 2011; 365: 1025–1032. [DOI] [PubMed] [Google Scholar]
- 4. Arteaga CL. Progress in breast cancer: overview. Clin Cancer Res 2013; 19: 6353–6359. [DOI] [PubMed] [Google Scholar]
- 5. Lieberman DA. Clinical practice. Screening for colorectal cancer. N Engl J Med 2009; 361: 1179–1187. [DOI] [PubMed] [Google Scholar]
- 6. Lee MS, Kopetz S.. Current Future Approaches to target the epidermal growth factor receptor and its downstream signaling in metastatic colorectal cancer. Clin Colorectal Cancer 2015; 14: 203–218. [DOI] [PubMed] [Google Scholar]
- 7. Zhou C, Wu YL, Chen G. et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011; 12: 735–742. [DOI] [PubMed] [Google Scholar]
- 8. Smith RA, Manassaram-Baptiste D, Brooks D. et al. Cancer screening in the United States, 2015: a review of current American cancer society guidelines and current issues in cancer screening. CA Cancer J Clin 2015; 65: 30–54. [DOI] [PubMed] [Google Scholar]
- 9. Katz MH, Wang H, Fleming JB. et al. Long-term survival after multidisciplinary management of resected pancreatic adenocarcinoma. Ann Surg Oncol 2009; 16: 836–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Howlader N NA, Krapcho M, Miller D. et al. (eds). SEER Cancer Statistics Review, 1975–2013. Bethesda, MD: National Cancer Institute; 2016. [Google Scholar]
- 11. Siegel RL, Miller KD, Jemal A.. Cancer statistics, 2016. CA Cancer J Clin 2016; 66: 7–30. [DOI] [PubMed] [Google Scholar]
- 12. Chakraborty S, Baine MJ, Sasson AR, Batra SK.. Current status of molecular markers for early detection of sporadic pancreatic cancer. Biochim Biophys Acta 2011; 1815: 44–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vietsch EE, van Eijck CH, Wellstein A.. Circulating DNA and micro-RNA in patients with pancreatic cancer. Pancreat Disord Ther 2015; 5(2): 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mulcahy HE, Lyautey J, Lederrey C. et al. A prospective study of K-ras mutations in the plasma of pancreatic cancer patients. Clin Cancer Res 1998; 4: 271–275. [PubMed] [Google Scholar]
- 15. Dianxu F, Shengdao Z, Tianquan H. et al. A prospective study of detection of pancreatic carcinoma by combined plasma K-ras mutations and serum CA19-9 analysis. Pancreas 2002; 25: 336–341. [DOI] [PubMed] [Google Scholar]
- 16. Maire F, Micard S, Hammel P. et al. Differential diagnosis between chronic pancreatitis and pancreatic cancer: value of the detection of KRAS2 mutations in circulating DNA. Br J Cancer 2002; 87: 551–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uemura T, Hibi K, Kaneko T. et al. Detection of K-ras mutations in the plasma DNA of pancreatic cancer patients. J Gastroenterol 2004; 39: 56–60. [DOI] [PubMed] [Google Scholar]
- 18. Dabritz J, Preston R, Hanfler J, Oettle H.. Follow-up study of K-ras mutations in the plasma of patients with pancreatic cancer: correlation with clinical features and carbohydrate antigen 19-9. Pancreas 2009; 38: 534–541. [DOI] [PubMed] [Google Scholar]
- 19. Chen H, Tu H, Meng ZQ. et al. K-ras mutational status predicts poor prognosis in unresectable pancreatic cancer. Eur J Surg Oncol 2010; 36: 657–662. [DOI] [PubMed] [Google Scholar]
- 20. Castells A, Puig P, Mora J. et al. K-ras mutations in DNA extracted from the plasma of patients with pancreatic carcinoma: diagnostic utility and prognostic significance. J Clin Oncol 1999; 17: 578–584. [DOI] [PubMed] [Google Scholar]
- 21. Diaz LA Jr, Bardelli A.. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014; 32: 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bettegowda C, Sausen M, Leary RJ. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014; 6: 224ra224.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu L, Risch HA.. Exosomes: potential for early detection in pancreatic cancer. Future Oncol 2016; 12: 1081–1090. [DOI] [PubMed] [Google Scholar]
- 24. Melo SA, Luecke LB, Kahlert C. et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015; 523: 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. San Lucas FA, Allenson K, Bernard V. et al. Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. Ann Oncol 2016; 27: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sluijter JP, Verhage V, Deddens JC. et al. Microvesicles and exosomes for intracardiac communication. Cardiovasc Res 2014; 102: 302–311. [DOI] [PubMed] [Google Scholar]
- 27. Skog J, Wurdinger T, van Rijn S. et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008; 10: 1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Demory Beckler M, Higginbotham JN, Franklin JL. et al. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol Cell Proteomics 2013; 12: 343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dictionaries NCI. U.S. Department of Health and Human Services. National Institutes of Health. National Cancer Institute 2015.
- 30. Kahlert C, Melo SA, Protopopov A. et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem 2014; 289: 3869–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jahr S, Hentze H, Englisch S. et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001; 61: 1659–1665. [PubMed] [Google Scholar]
- 32. Diamandis EP. A word of caution on new and revolutionary diagnostic tests. Cancer Cell 2016; 29: 141–142. [DOI] [PubMed] [Google Scholar]
- 33. Krimmel JD, Schmitt MW, Harrell MI. et al. Ultra-deep sequencing detects ovarian cancer cells in peritoneal fluid and reveals somatic TP53 mutations in noncancerous tissues. Proc Natl Acad Sci USA 2016; 113: 6005–6010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Poruk KE, Gay DZ, Brown K. et al. The clinical utility of CA 19-9 in pancreatic adenocarcinoma: diagnostic and prognostic updates. Curr Mol Med 2013; 13: 340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Katz MH, Pisters PW, Evans DB. et al. Borderline resectable pancreatic cancer: the importance of this emerging stage of disease. J Am Coll Surg 2008; 206: 833–846. Discussion 846–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.