Abstract
Background
We performed whole-exome sequencing of pretreatment biopsies and examined whether genome-wide metrics of overall mutational load, clonal heterogeneity or alterations at variant, gene, and pathway levels are associated with treatment response and survival.
Patients and methods
Two hundred and three biopsies from the NeoALTTO trial were analyzed. Mutations were called with MuTect, and Strelka, using pooled normal DNA. Associations between DNA alterations and outcome were evaluated by logistic and Cox-proportional hazards regression.
Results
There were no recurrent single gene mutations significantly associated with pathologic complete response (pCR), except PIK3CA [odds ratio (OR) = 0.42, P = 0.0185]. Mutations in 33 of 714 pathways were significantly associated with response, but different genes were affected in different individuals. PIK3CA was present in 23 of these pathways defining a ‘trastuzumab resistance-network’ of 459 genes. Cases with mutations in this network had low pCR rates to trastuzumab (2/50, 4%) compared with cases with no mutations (9/16, 56%), OR = 0.035; P < 0.001. Mutations in the ‘Regulation of RhoA activity’ pathway were associated with higher pCR rate to lapatinib (OR = 14.8, adjusted P = 0.001), lapatinib + trastuzumab (OR = 3.0, adjusted P = 0.09), and all arms combined (OR = 3.77, adjusted P = 0.02). Patients (n = 124) with mutations in the trastuzumab resistance network but intact RhoA pathway had 2% (1/41) pCR rate with trastuzumab alone (OR = 0.026, P = 0.001) but adding lapatinib increased pCR rate to 45% (17/38, OR = 1.68, P = 0.3). Patients (n = 46) who had no mutations in either gene set had 6% pCR rate (1/15) with lapatinib, but had the highest pCR rate, 52% (8/15) with trastuzumab alone.
Conclusions
Mutations in the RhoA pathway are associated with pCR to lapatinib and mutations in a PIK3CA-related network are associated with resistance to trastuzumab. The combined mutation status of these two pathways could define patients with very low response rate to trastuzumab alone that can be augmented by adding lapatinib or substituting trastuzumab with lapatinib.
Keywords: breast cancer NeoALTTO trial, whole-exome sequencing, regulation of RhoA activity, PIK3CA-related network
Introduction
The anti-human epidermal growth factor receptor 2 (HER2) monoclonal antibody trastuzumab and the small molecule tyrosine kinase inhibitor lapatinib have several non-overlapping mechanisms of action. The neoALTTO trial showed that dual HER2 blockade including both drugs increases pathologic complete response (pCR) rates in early stage, HER2-positive breast cancers [1]. In this trial, patients were randomly assigned to lapatinib (n = 154), or trastuzumab (n = 149), or lapatinib plus trastuzumab (n = 152) each concomitant with weekly paclitaxel × 12 treatments as neoadjuvant therapy. The pCR (ypT0/is, ypN0) rates were 47% in the lapatinib plus trastuzumab arm compared with 28% and 20% in the trastuzumab and lapatinib arms [1]. Baseline tumor biopsies were collected before starting therapy for predictive biomarker discovery. In a previous report, activating mutations in the phosphatidylinositol 3-kinase (PI3K) catalytic subunit (PIK3CA, seen in 23% of cancers), was significantly associated with lower pCR rates in each arm [2]. Tumor infiltrating lymphocytes (TIL) were also assessed and TIL counts >5% were associated with higher pCR rates and better survival independent of treatment arms [3]. Similar observations were made in numerous other neoadjuvant clinical trials, establishing PIK3CA mutations [4] and extensive immune infiltration [5] as negative and positive predictors of pCR- to HER2-targeted therapies, respectively. Several other molecular predictors of response have also been proposed based on biological insights into the mechanisms of resistance to HER2-targeted therapies. These include altered expression and structure of the HER2 receptor, constitutive activation of the downstream signaling pathways, switching to alternative growth and survival pathways, and alterations in the tumor immune microenvironment [6–8]. However, because of their modest positive and negative predictive values, none of these markers is clinically useful in ruling out patients from receiving HER2-targeted therapies or in selecting one therapy over another.
The goal of this analysis was to perform whole-exome sequencing (WES) of DNA from pretreatment biopsies and examine whether genome-wide metrics of overall mutational load and clonal heterogeneity or DNA sequence alterations at variant, gene, and pathway levels are associated with pCR and survival.
Methods
Patients and samples
Of the 455 randomized patients, 423 had baseline frozen core needle tumor biopsy collected. Sixty-six of these biopsies had <10% tumor cellularity or yielded poor-quality or low-quantity DNA for further molecular analysis. Of the remaining 357 specimens, 227 had >1 μg genomic DNA left after completion of the prior correlative science projects [2, 3, 9] and was available for this study. Twenty cases failed quality control metrics of library preparation or sequencing resulting in 207 specimens with compete WES data (n = 67 lapatinib, n = 67 trastuzumab, n = 73 lapatinib + trastuzumab). The distribution of tumor size, grade, hormone receptor status, and the pCR rates across treatment arms were similar between patients included in the WES study and the whole-NeoALTTO population (supplementary Table S1, available at Annals of Oncology online). The median follow-up was 3.63 years for event-free survival (EFS) and 3.72 for overall survival (OS). Pathologic complete response was defined as no residual invasive cancer in the breast and lymph nodes.
Library preparation, exome capture and sequencing
Mutations were called with MuTect, and Strelka, using pooled normal DNA, and genes were identified as significantly mutated using MutSigCV at a false discovery rate <0.1 (supplementary Methods, available at Annals of Oncology online).
Statistical analysis
Calculation of mutational load, clonal heterogeneity, and pathway level mutations are described in the supplementary Methods, available at Annals of Oncology online. Associations between pCR and gene or pathway level mutations were assessed using estrogen receptor (ER)-adjusted logistic regression or the Fisher’s exact test when ER-positive and -negative cases were examined separately. Differences in mutation sequence context, mutation type, and mutation spectrum were analyzed using a two-tailed Fisher’s exact test. We used the Kaplan–Meier survival estimator and Cox proportional hazards regression to assess the associations of mutation status as well as other factors including patients’ age, histologic grade, and clinical stage with EFS and OS. We adjusted for multiple hypotheses testing for pathway level mutation analysis using a permutation approach [9] as described in the supplementary Methods, available at Annals of Oncology online.
Results
Association between single gene mutations and pCR or survival
The mean coverage was 150× with > 90% of target bases showing ≥30× coverage in >99% of samples (supplementary Figure S1, available at Annals of Oncology online). The median number of somatic variants was 65 per sample, and the median number of predicted high functional impact variants was 34. Overall, 12 genes had significantly higher than background mutation rates. Among these, only PIK3CA was associated with response in the full cohort including all three treatment arms combined [pCR odds ratio (OR) = 0.42, ER-adjusted logistic regression test P = 0.0185) and also in each arm separately (supplementary Figure S2A and B, available at Annals of Oncology online). When we restricted analysis to known PIK3CA hotspots (E542A/K, E545A/K/V/G, and H1047R/L/Y), the same association was observed (N = 54 mutant cases; OR = 0.47; P = 0.07). We also tested the association between any somatic mutations that occurred in ≥10 cancers (23 genes, supplementary Table S2A, available at Annals of Oncology online) and pCR, EFS, and OS; no gene demonstrated significant association with outcome.
Association between pathway mutations and pCR or survival
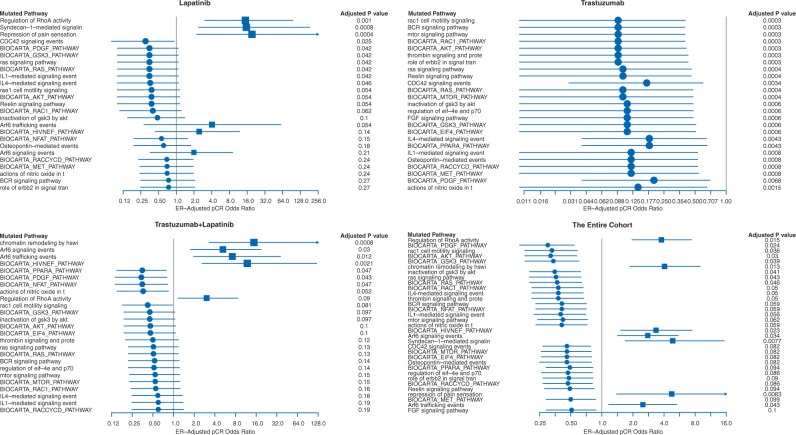
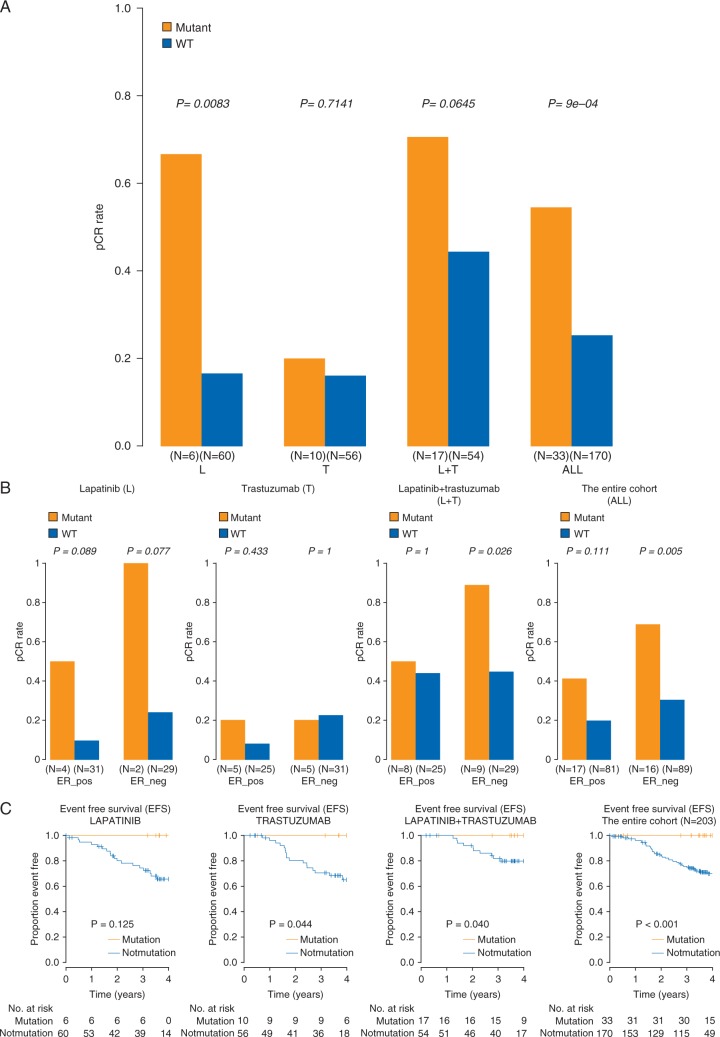
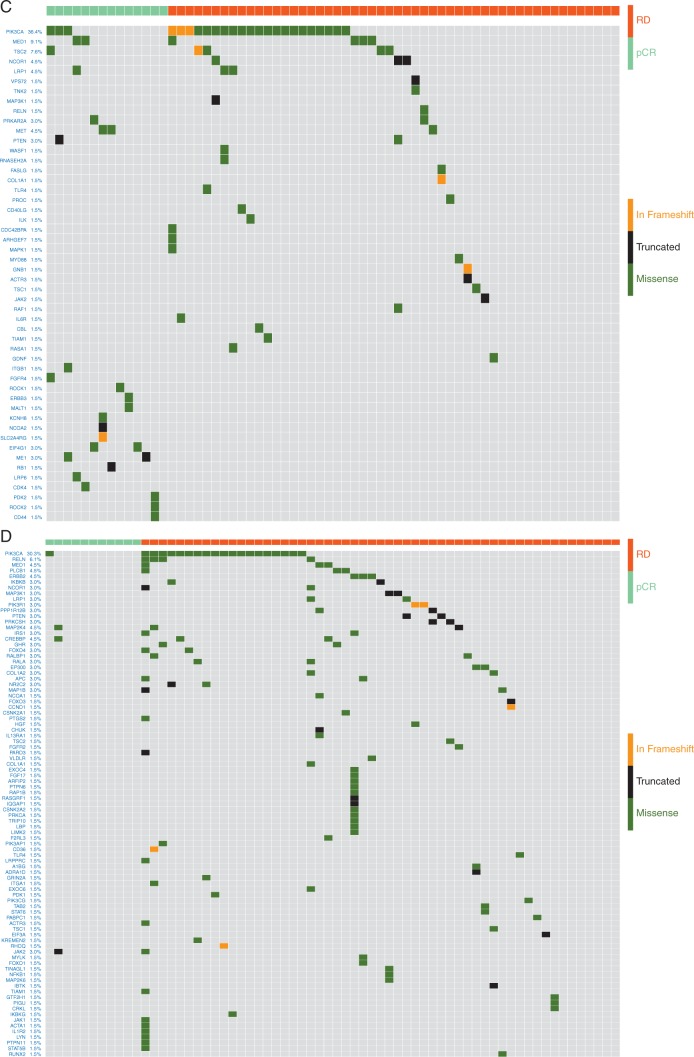
Mutations in 33 of 714 pathways corresponding to major biological processes showed significant association with response in the full cohort (adjusted P ≤ 0.1 by ER status-adjusted logistic regression, supplementary Tables S2B and S3A, available at Annals of Oncology online). Mutations in seven pathways were associated with higher pCR (OR > 1), the remaining with residual disease (RD) (Figure 1). In the trastuzumab arm, no pathways were associated with pCR, but 23 pathways showed significant association with RD (adjusted P < 0.001, Figure 1, supplementary Table S3C, available at Annals of Oncology online). Each of these 23 pathways included PIK3CA, but PIK3CA itself was not mutated in most cases. In the lapatinib arm, three pathways were significantly associated with higher pCR including ‘Regulation of RhoA activity’, ‘Syndecan-1 mediated signaling’, and ‘Repression of pain sensation by transcriptional regulator dream’ (adjusted P ≤0.001, Figure 1, supplementary Table S3B, available at Annals of Oncology online) and none was significantly associated with RD. The ‘Regulation of RhoA Activity’ pathway (n = 48 genes) mutations had the strongest association with pCR (OR = 14.8, P = 0.0083, adjusted P =0.001) in the lapatinib arm and in the full cohort (OR = 3.77, P = 0.0009, adjusted P =0.02) and also showed a similar trend in the lapatinib + trastuzumab arm (supplementary Figure S3D, available at Annals of Oncology online) (OR = 3.0, P = 0.06, adjusted P = 0.09) but not in the trastuzumab alone arm. In the lapatinib arm, cases with RhoA pathway mutations had 67% pCR rate compared with 17% in wild type (P = 0.0083, adjusted P =0.001) (Figure 2A/B). Patients with RhoA pathway mutations (n = 33) had similar high pCR rates with lapatinib alone or with lapatinib + trastuzumab (67% versus 70%) but had lower pCR with trastuzumab alone (20%). In multivariate analysis of the full cohort, ER-negative status (OR = 2.1, P = 0.003), combined HER2 blockade (OR = 5.23, P < 0.001), and RhoA pathway mutation status (OR = 3.37, P = 0.005) were independently associated with higher pCR rate. The interaction term between treatment and RhoA pathway mutation status was not significant likely due to the small sample size. Twenty-seven of the 48 RhoA pathway genes had at least one mutation in 33 patients but different genes were affected in different individuals (Figure 3A). Consequently, none of the genes alone showed significant association with pCR. Patients with mutations in the RhoA pathway also had significantly better EFS (Figure 2C) and OS compared to wild-type cancers (supplementary Figure S3, available at Annals of Oncology online).
Figure 1.
Mutations in 33 of 713 biological pathways were associated with response in all arms combined and in the three individual treatment arms. P-values were derived from ER status-adjusted logistic regression.
Figure 2.
Pathologic complete response (pCR) rates by mutation status in the RhoA activity pathway in the full cohort and in each treatment arm (A) and by estrogen receptor (ER) status (B). Event-free survival according to mutation status of the RhoA activity pathway (C). P-values were calculated using ER-adjusted logistic regression (A) and the Fisher’s exact test (B). L = lapatinib, T = trastuzumab, WT = wild type; no high functional impact somatic mutation in any of the member genes. Mutant = somatic mutation in at least one of the genes in the pathway. (C) Kaplan–Meier survival curves are shown for each treatment arm and for the full cohort (all arms combined).
Figure 3.
Mutations in member genes of the RhoA activity pathway in the lapatinib arm (A, n = 66), the trastuzumab arm (B, n = 66), and mutations in the PIK3CA network genes in the lapatinib arm (C, n = 66) and the trastuzumab arm (D, n = 66). Each column represents a patient, and types of mutations are color coded as indicated. The mutation frequency is given as % of cases affected in the study cohort after the gene name. RD = residual disease, pCR = pathologic complete response. In the RhoA activity pathway, members are also color coded for their roles in wild-type form, red corresponds to activating and blue to inhibiting RhoA function.
Next, we constructed a PIK3CA gene network that included all unique genes (n = 459) from the 23 pathways associated with RD to trastuzumab and excluded genes that overlapped with the RhoA pathway (supplementary Table S2C, available at Annals of Oncology online). In the trastuzumab arm, patients with ≥1 mutation in the PIK3CA network genes (n = 50 patients) had 4% pCR rate compared to 56% in patients with no mutations (n = 16, P = 0.0001, adjusted P < 0.001, ER-adjusted logistic regression, Figure 3D). The association between network-level mutation and pCR in the trastuzumab arm remained significant even if mutations in the PIK3CA gene itself were excluded (P = 0.001, adjusted P < 0.001). Mutation in the PIK3CA network was not associated with response in the lapatinib alone arm (pCR = 20% in both mutant and wild type).
We also examined the combined effect of mutations in these two mutually exclusive gene sets on response and survival (supplementary Figure S4A, available at Annals of Oncology online). Patients who had mutation in the PIK3CA network but not in the RhoA pathway (n = 124) had 2% pCR rate with trastuzumab alone, adding lapatinib to trastuzumab increased the pCR rate to 45% (OR = 0.03, P < 0.0005) (supplementary Figure S4A, available at Annals of Oncology online), EFS and OS were also significantly improved if treated with the combined HER2 blockade compared with trastuzumab alone (supplementary Figure S4B, available at Annals of Oncology online). Patients (n = 46) with no mutations either in the PIK3CA network or in the RhoA pathway had low pCR rate with lapatinib alone (6%) and the highest pCR rate with trastuzumab (52%). In a Cox multivariate model including the mutation status of RhoA pathway, stage and grade as covariates for EFS, we found that the only significant variable was age and pathway mutation status [HR 0.97; 95% confidence interval (CI), 0.94–1.00; P = 0.035]. For OS, no clinical variable remained significant except pathway mutation status. In a similar multivariate analysis, including the mutation status of the ‘trastuzumab resistance network’, age remained a significant factor affecting EFS (HR 0.97; 95% CI, 0.94–1.00; P = 0.031) and also OS (HR 0.95; 95% CI, 0.90–0.99; P = 0.02).
Association of exome-wide metrics with pCR and survival
The overall mutation load was similar between cases with pCR or RD, and there was also no association between mutation load and survival (supplementary Figure S5B, C and E, available at Annals of Oncology online). The mean clonal heterogeneity (MATH) score was significantly higher in cases with RD compared with pCR in the full cohort (65.3 versus 59.6, P = 0.036, un-adjusted for multiple comparisons) and in the lapatinib/trastuzumab combination arm (67.0 versus 55.8, P = 0.0048 un-adjusted for multiple comparisons) suggesting that greater clonal heterogeneity is associated with greater resistance to therapy (supplementary Figure S5A, available at Annals of Oncology online). However, survival in the low versus high MATH groups, dichotomized at the median, was not significantly different (supplementary Figure S5D and F available at Annals of Oncology online).
Discussion
In this study, we examined whether mutations in any of approximately 20 000 protein coding genes was associated with response to HER2-targeted therapies. We found only 12 genes to be mutated above background mutation rate, and among these, only the PIK3CA gene was significantly associated with lower response. This is consistent with prior observations that very few genes have somatic mutation frequencies >15% and the rest represent a long tail of individually rare mutations [10]. This implies that any study that includes only a few hundred patients will have limited statistical power to detect a significant association between clinical outcome and rarely mutated genes that account for the majority of mutations. We hypothesized that pathway level alterations may be more important than recurrent single gene mutations since similar deleterious effect on pathway output could be caused by mutations in different genes.
We identified 33 pathways in which mutations were significantly associated with response with adjusted P < 10% in the combined study population. The majority of these (n = 27) were associated with lower pCR rate. Importantly, there was no pathway mutation associated with pCR in the trastuzumab arm, and all pathway mutations that were significantly associated with RD in this arm included the PIK3CA gene. However, different genes were mutated in different individuals and PK3CA itself was often not mutated. When we combined all unique genes from these pathways, but excluded PIK3CA, the association with RD remained statistically significant, indicating that mutations in genes other than PIK3CA could confer lower sensitivity to trastuzumab. These observations support the role that PIK3CA mutations play in reducing sensitivity to trastuzumab and also indicate phenotypic convergence of mutations at pathway level [11, 12].
We also found that mutations in the RhoA pathway were associated with higher pCR rate with lapatinib therapy, whereas these mutations did not confer sensitivity to trastuzumab. This association was mediated by individually rare mutations affecting 27 of the 48 member genes. The biological impact of the mutations in these 27 genes has not yet been studied in the laboratory. RhoA itself had two recurrent previously reported gain-of-function mutations (Gly17, Ref [13]) that both occurred in cases with RD. RhoA mediates microtubule-dependent signal transduction from plasma membrane receptors and plays an important role in HER2-dependent stabilization of microtubules and regulates cell adhesion and cell motility [14, 15]. Inhibition of RhoA activity decreases cell motility, invasiveness, and proliferation of breast cancer cells [16, 17]. Our results suggest that inhibition of the RhoA pathway activity can also selectively increase sensitivity to lapatinib therapy, but the underlying biological mechanism is yet to be elucidated.
There are several inherent limitations of large throughput sequencing studies that use cancer biopsies from randomized clinical trials of combination treatment regimens. The tumor cellularity of the biopsies varies, which limits the ability to capture rare sub-clonal anomalies. Costs and tissue availability often prevent sequencing matching normal tissues, which makes it challenging to distinguish somatic from germline variants. To minimize this problem, we used a normal cohort as reference and also applied a series of filters. The multidrug nature of adjuvant treatment regimens, required for curative treatment, makes it difficult to tease out drug-specific predictive markers. Each of these factors introduces variability into the results that lower the power to identify molecular markers. The large variable space (the whole exome = 44.1 Mb of the genome) also carries the risk for false discoveries and therefore even carefully controlled analysis results should be considered hypothesis generating until independently confirmed. Despite these limitations, robust, high-frequency genomic anomalies that are shared by many samples in a given outcome group can be discovered. Previous studies have shown that mutations in PIK3CA are associated with RD to trastuzumab, and we extend this observation to a network of PIK3CA-related genes and also identified a new biological pathway, the ‘Regulation of RhoA activity’ that confers sensitivity to lapatinib but not to trastuzumab therapy. Patients with mutation in the PIK3CA network but wild-type RhoA pathway had a pCR rate of 2% with trastuzumab alone and adding lapatinib to trastuzumab increased pCR rate to 45%. Patients who had no mutations either in the PIK3CA network or in the RhoA pathway genes had 6% pCR rate with lapatinib alone but had the highest pCR rate with trastuzumab (52%).
This is one of the first whole-exome sequencing efforts to identify new predictive markers from prospectively collected baseline cancer biopsies of a randomized clinical trial. The results indicate that there is no single DNA sequence abnormality that is uniformly shared by highly treatment sensitive versus resistant cancers. Different genes are affected in different individuals and cancers appear to acquire resistance through different mutations that converge at the pathway level. This suggests that biomarkers could be designed around pathway level instead of gene level mutations. Mutations in the RhoA pathway are associated with high sensitivity to lapatinib therapy, and mutations in a PIK3CA gene network are associated with relative resistance to trastuzumab. The mutation status of these two pathways, in combination, could define a patient population with very low response rate to trastuzumab alone that can be augmented by adding lapatinib or substituting trastuzumab with lapatinib.
Supplementary Material
Acknowledgements
Human investigations were performed after approval by Yale review board and in accord with an assurance filed with and approved by the U.S. Department of Health and Human Services
Funding
This work was supported by Breast Cancer Research Foundation (DLR and LP) (no grant number) and National Institutes of Health [P30 CA008748].
Disclosure
The authors have declared no conflicts of interest.
References
- 1. Baselga J, Bradbury I, Eidtmann H. et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): A randomised, open-label, multicentre, phase 3 trial. Lancet 2012; 379: 633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Majewski IJ, Nuciforo P, Mittempergher L. et al. PIK3CA mutations are associated with decreased benefit to neoadjuvant human epidermal growth factor receptor 2-targeted therapies in breast cancer. J Clin Oncol 2015; 33(12): 1334–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Salgado R, Denkert C, Campbell C. et al. Tumor-infiltrating lymphocytes and associations with pathological complete response and event-free survival in her2-positive early-stage breast cancer treated with lapatinib and trastuzumab A secondary analysis of the NeoALTTO Trial. JAMA Oncol 2015; 1(4): 448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Loibl S, Von Minckwitz G, Schneeweiss A. et al. PIK3CA mutations are associated with lower rates of pathologic complete response to anti–human epidermal growth factor receptor 2 (HER2) therapy in primary HER2-overexpressing breast cancer. J Clin Oncol 2014; 32(29): 3212–3220. [DOI] [PubMed] [Google Scholar]
- 5. Perez EA, Thompson EA, Ballman KV. et al. Genomic analysis reveals that immune function genes are strongly linked to clinical outcome in the North Central Cancer Treatment Group N9831 adjuvant trastuzumab trial. J Clin Oncol 2015; 33: 701–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carvajal-Hausdorf D., Schalper K., Pusztai L. et al. Measurement of domain-specific HER2 (ERBB2) expression may classify benefit from trastuzumab in breast cancer. J Natl Cancer Inst 2015; 107(8): djv136. doi: 10.1093/jnci/djv136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Scaltriti M, Nuciforo P, Bradbury I. et al. High HER2 expression correlates with response to the combination of lapatinib and trastuzumab. Clin Cancer Res 2015; 21(3): 569–576. [DOI] [PubMed] [Google Scholar]
- 8. Rimawi MF, Schiff R, Osborne KC. Targeting HER2 for the Treatment of Breast Cancer. Annual Rev Med 2015; 66: 111–128. [DOI] [PubMed] [Google Scholar]
- 9. Subramanian A, Tamayo P, Mootha VK. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005; 102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cancer Genome Atlas Research, Network. “The Cancer Genome Atlas Pan-Cancer analysis project.” Nature Genet 2013; 45(10): 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Berns K, Horlings HM, Hennessy BT. et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell 2007; 12: 395–402. [DOI] [PubMed] [Google Scholar]
- 12. Clark AS, West K, Streicher S. et al. Constitutive and inducible Akt activity promotes resistance to chemotherapy, trastuzumab, or tamoxifen in breast cancer cells. Mol Cancer Ther 2002; 1:707–717. [PubMed] [Google Scholar]
- 13. Kakiuchi M, Nishizawa T, Ueda H. et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nature Genet 2014; 46(6): 583–587. [DOI] [PubMed] [Google Scholar]
- 14. Simpson KJ, Dugan AS, Mercurio AM. Functional analysis of the contribution of RhoA and RhoC GTPases to invasive breast carcinoma. Cancer Res 2004; 64(23): 8694–8701. [DOI] [PubMed] [Google Scholar]
- 15. Zaoui K, Benseddik K, Daou P. et al. ErbB2 receptor controls microtubule capture by recruiting ACF7 to the plasma membrane of migrating cells. Proc Natl Acad Sci USA 2010; 107(43): 18517–18522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pillé JY, Denoyelle C, Varet J. et al. Anti-RhoA and anti-RhoC siRNAs inhibit the proliferation and invasiveness of MDA-MB-231 breast cancer cells in vitro and in vivo. Mol Ther 2005; 11(2): 267–274. [DOI] [PubMed] [Google Scholar]
- 17. Cao XX, Xu JD, Xu JW. et al. RACK1 promotes breast carcinoma migration/metastasis via activation of the RhoA/Rho kinase pathway. Breast Cancer Res Treat 2011; 126(3): 555–563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.